Impaired Molecular Mechanisms Contributing to Chronic Pain in Patients with Hidradenitis Suppurativa: Exploring Potential Biomarkers and Therapeutic Targets
Abstract
:1. Introduction
2. Results
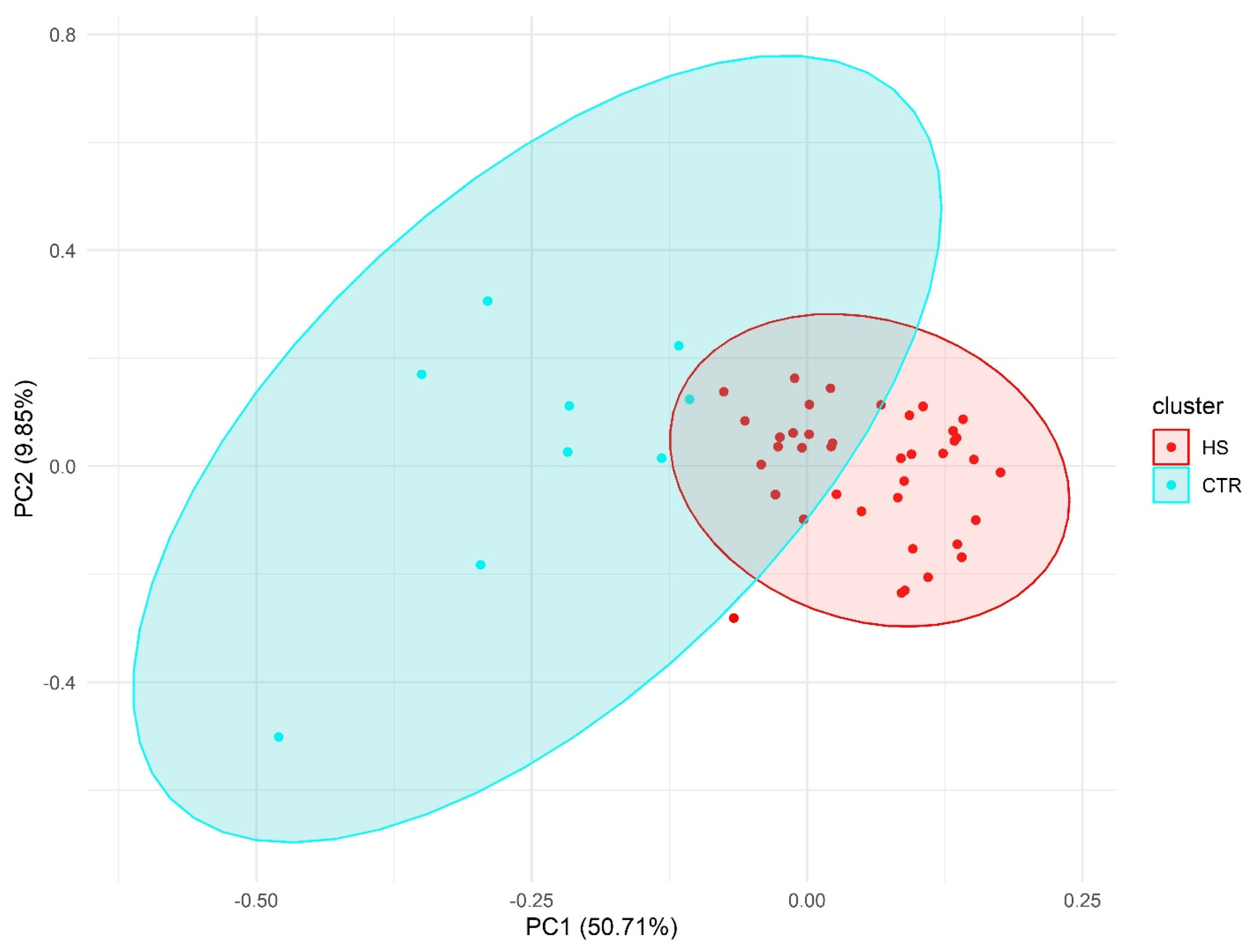
2.1. Heatmaps and Principal Component Analysis
2.2. Principal Component Analysis
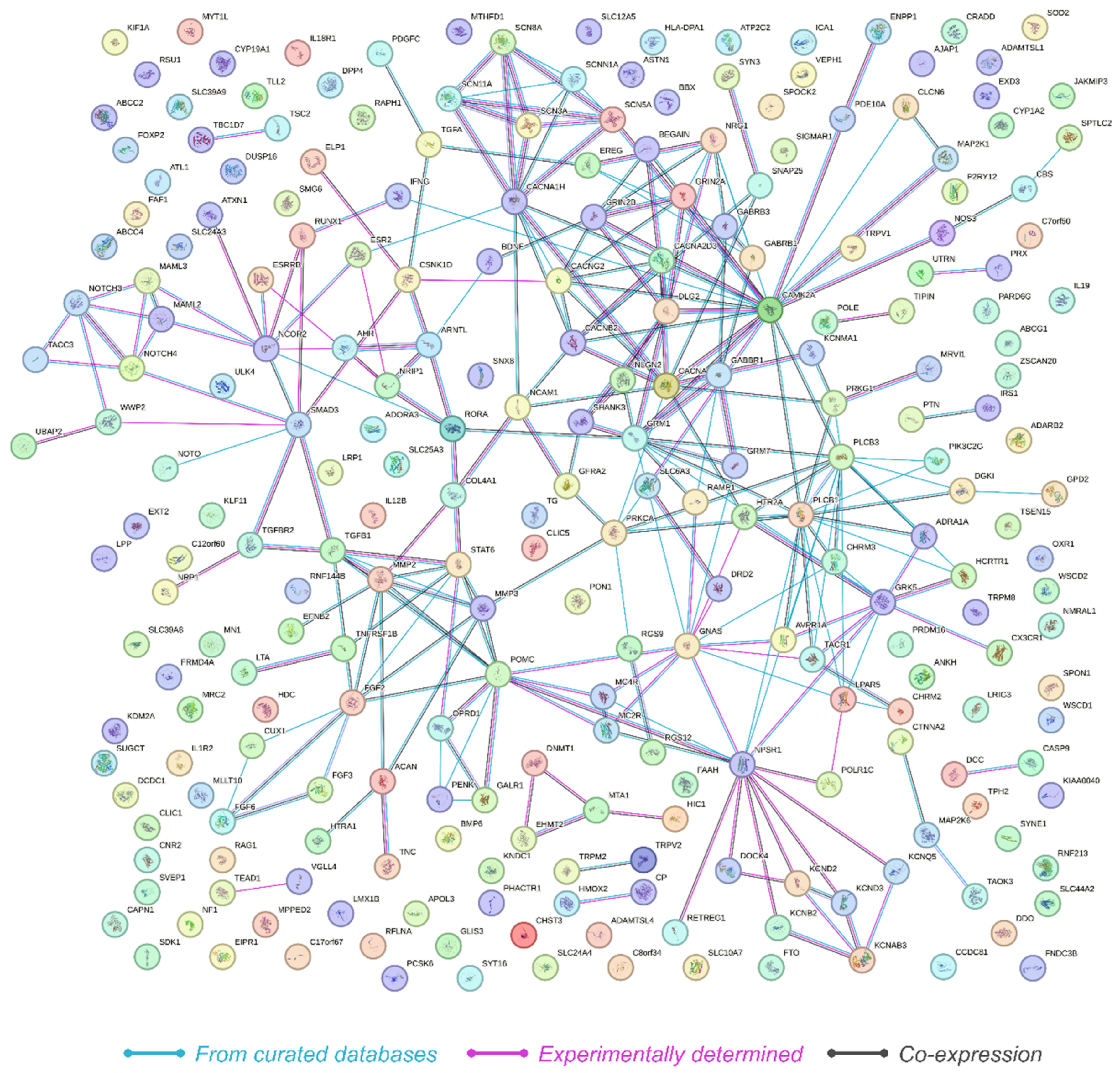
2.3. Network Analysis
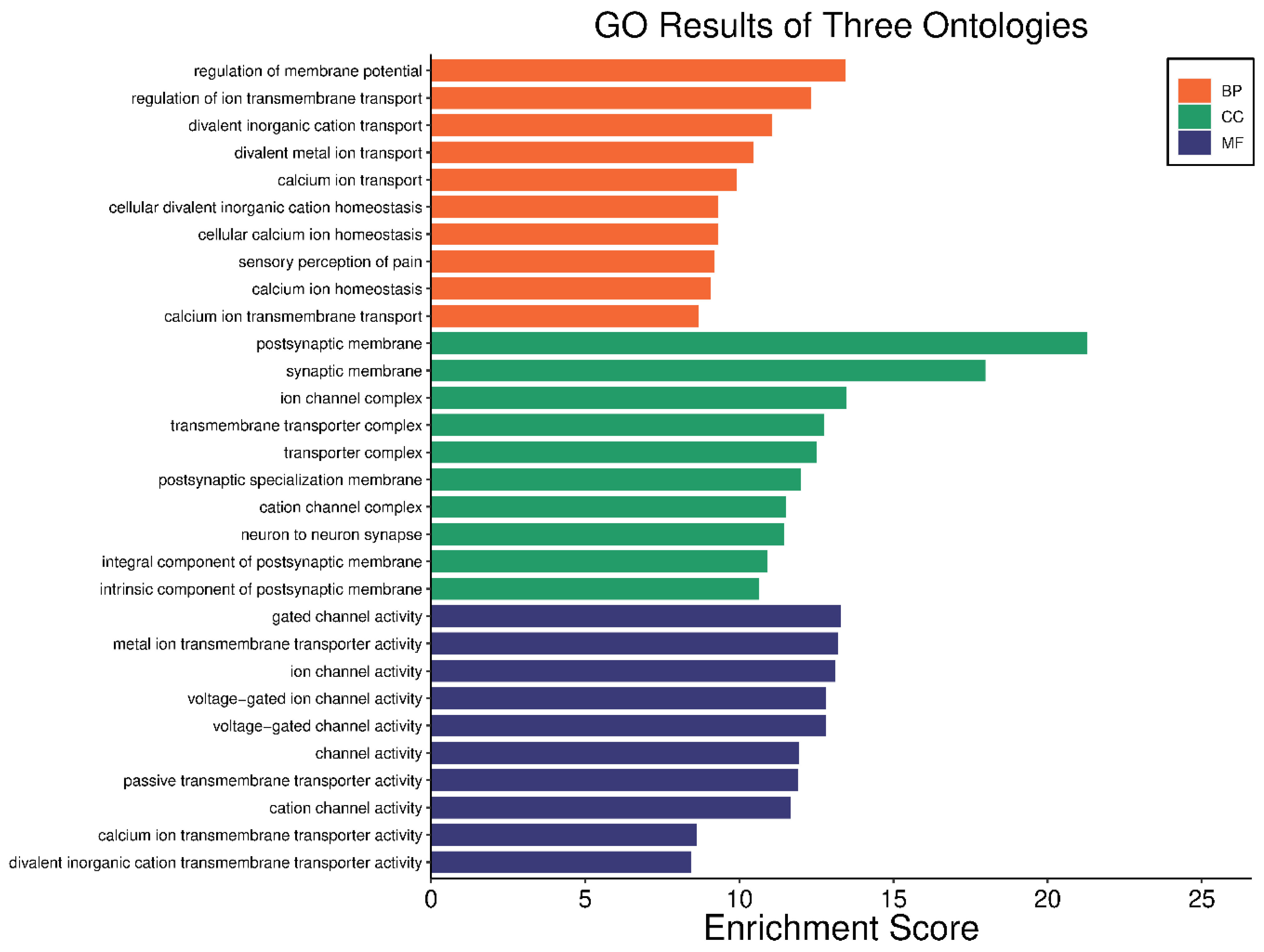
2.4. Gene Ontology (GO) and KEGG Pathway Analysis
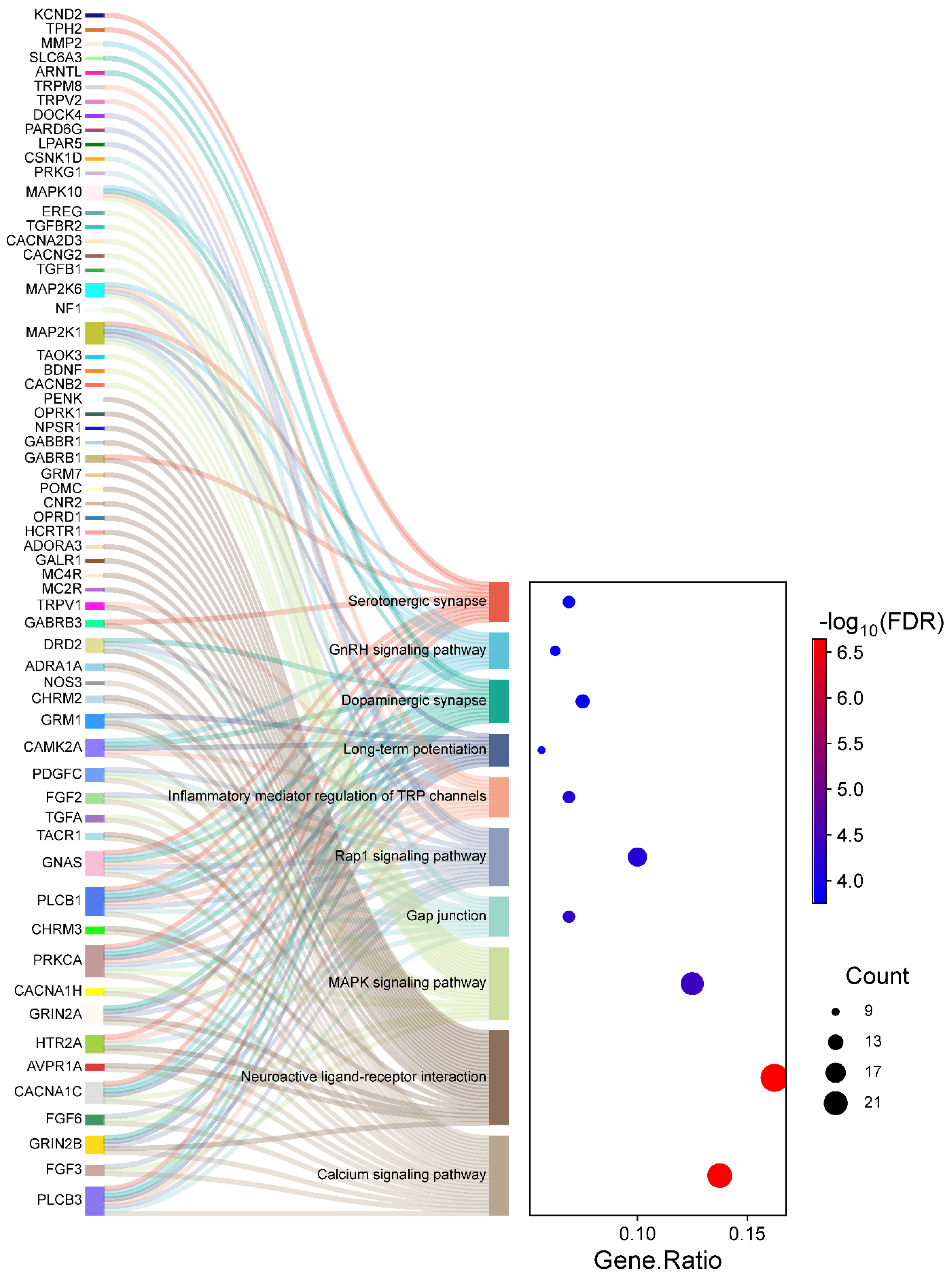
2.5. KEGG Pathway Analysis
3. Discussion
4. Materials and Methods
Hidradenitis Suppurativa Sample Selection
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crowley, J.J.; Mekkes, J.R.; Zouboulis, C.C.; Scheinfeld, N.; Kimball, A.; Sundaram, M.; Gu, Y.; Okun, M.M.; Kerdel, F. Association of hidradenitis suppurativa disease severity with increased risk for systemic comorbidities. Br. J. Dermatol. 2014, 171, 1561–1565. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T. Hidradenitis Suppurativa as a Potential Subtype of Autoinflammatory Keratinization Disease. Front. Immunol. 2020, 11, 847. [Google Scholar] [CrossRef] [PubMed]
- Metpally, R.P.; Vishweswaraiah, S.; Krishnamurthy, S.; Saiyed, N.; Stahl, R.C.; Golden, A.; Denisenko, A.; Staples, J.; Gonzaga-Jauregui, C.; Carey, D.J.; et al. Identification of Novel Genetic Risk Variants Associated with Hidradenitis Suppurativa in an Exome Sequencing Cohort of 92,455 Individuals. Dermatology 2024, 240, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Ratnamala, U.; Jain, N.K.; Jhala, D.D.; Prasad, P.V.S.; Saiyed, N.; Nair, S.; Radhakrishna, U. An Updated Mutation Spectrum of the gamma-Secretase Complex: Novel NCSTN Gene Mutation in an Indian Family with Hidradenitis Suppurativa and Acne Conglobata. Indian. J. Dermatol. 2023, 68, 141–147. [Google Scholar] [CrossRef]
- Radhakrishna, U.; Ratnamala, U.; Jhala, D.D.; Vadsaria, N.; Patel, M.; Uppala, L.V.; Vishweswaraiah, S.; Vedangi, A.; Saiyed, N.; Damiani, G.; et al. Methylated miRNAs may serve as potential biomarkers and therapeutic targets for hidradenitis suppurativa. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 2199–2213. [Google Scholar] [CrossRef]
- Radhakrishna, U.; Ratnamala, U.; Jhala, D.D.; Uppala, L.V.; Vedangi, A.; Saiyed, N.; Patel, M.; Shah, S.R.; Rawal, R.M.; Jemec, G.B.E.; et al. Deregulated Long Non-Coding RNAs (lncRNA) as Promising Biomarkers in Hidradenitis Suppurativa. J. Clin. Med. 2024, 13, 3016. [Google Scholar] [CrossRef]
- Ratnamala, U.; Jhala, D.; Jain, N.K.; Saiyed, N.M.; Raveendrababu, M.; Rao, M.V.; Mehta, T.Y.; Al-Ali, F.M.; Raval, K.; Nair, S.; et al. Expanding the spectrum of gamma-secretase gene mutation-associated phenotypes: Two novel mutations segregating with familial hidradenitis suppurativa (acne inversa) and acne conglobata. Exp. Dermatol. 2016, 25, 314–316. [Google Scholar] [CrossRef]
- Saunte, D.M.; Boer, J.; Stratigos, A.; Szepietowski, J.C.; Hamzavi, I.; Kim, K.H.; Zarchi, K.; Antoniou, C.; Matusiak, L.; Lim, H.W.; et al. Diagnostic delay in hidradenitis suppurativa is a global problem. Br. J. Dermatol. 2015, 173, 1546–1549. [Google Scholar] [CrossRef]
- Pelekanou, A.; Kanni, T.; Savva, A.; Mouktaroudi, M.; Raftogiannis, M.; Kotsaki, A.; Giamarellos-Bourboulis, E.J. Long-term efficacy of etanercept in hidradenitis suppurativa: Results from an open-label phase II prospective trial. Exp. Dermatol. 2010, 19, 538–540. [Google Scholar] [CrossRef]
- Vinkel, C.; Thomsen, S.F. Hidradenitis Suppurativa: Causes, Features, and Current Treatments. J. Clin. Aesthet. Dermatol. 2018, 11, 17–23. [Google Scholar]
- Okun, M.M.; Flamm, A.; Werley, E.B.; Kirby, J.S. Hidradenitis Suppurativa: Diagnosis and Management in the Emergency Department. J. Emerg. Med. 2022, 63, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Alavi, A.; Farzanfar, D.; Lee, R.K.; Almutairi, D. The Contribution of Malodour in Quality of Life of Patients With Hidradenitis Suppurativa. J. Cutan. Med. Surg. 2018, 22, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Savage, K.T.; Singh, V.; Patel, Z.S.; Yannuzzi, C.A.; McKenzie-Brown, A.M.; Lowes, M.A.; Orenstein, L.A.V. Pain management in hidradenitis suppurativa and a proposed treatment algorithm. J. Am. Acad. Dermatol. 2021, 85, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Ballard, K.; Shuman, V.L. Hidradenitis Suppurativa. In StatPearls; Treasure Island (FL) Ineligible Companies: Treasure Island, FL, USA, 2023. [Google Scholar]
- Kaaz, K.; Szepietowski, J.C.; Matusiak, L. Influence of Itch and Pain on Sleep Quality in Patients with Hidradenitis Suppurativa. Acta Derm. Venereol. 2018, 98, 757–761. [Google Scholar] [CrossRef]
- Alotaibi, H.M. Incidence, Risk Factors, and Prognosis of Hidradenitis Suppurativa Across the Globe: Insights from the Literature. Clin. Cosmet. Investig. Dermatol. 2023, 16, 545–552. [Google Scholar] [CrossRef]
- Amtmann, D.; Askew, R.L.; Kim, J.; Chung, H.; Ehde, D.M.; Bombardier, C.H.; Kraft, G.H.; Jones, S.M.; Johnson, K.L. Pain affects depression through anxiety, fatigue, and sleep in multiple sclerosis. Rehabil. Psychol. 2015, 60, 81–90. [Google Scholar] [CrossRef]
- Brasure, M.; Nelson, V.A.; Scheiner, S.; Forte, M.L.; Butler, M.; Nagarkar, S.; Saha, J.; Wilt, T.J. Treatment for Acute Pain: An Evidence Map; Agency for Healthcare Research and Quality: Rockville, MD, USA, 2019.
- Patel, Z.S.; Hoffman, L.K.; Buse, D.C.; Grinberg, A.S.; Afifi, L.; Cohen, S.R.; Lowes, M.A.; Seng, E.K. Pain, Psychological Comorbidities, Disability, and Impaired Quality of Life in Hidradenitis Suppurativa [corrected]. Curr. Pain. Headache Rep. 2017, 21, 49. [Google Scholar] [CrossRef]
- Matusiak, L. Profound consequences of hidradenitis suppurativa: A review. Br. J. Dermatol. 2020, 183, e171–e177. [Google Scholar] [CrossRef]
- Moller Johansen, L.; Gerra, M.C.; Arendt-Nielsen, L. Time course of DNA methylation in pain conditions: From experimental models to humans. Eur. J. Pain. 2021, 25, 296–312. [Google Scholar] [CrossRef]
- Krajewski, P.K.; Matusiak, L.; von Stebut, E.; Schultheis, M.; Kirschner, U.; Nikolakis, G.; Szepietowski, J.C. Pain in Hidradenitis Suppurativa: A Cross-sectional Study of 1795 Patients. Acta Derm. Venereol. 2021, 101, adv00364. [Google Scholar]
- Whitley, S.K. Elucidation of Pain Mechanisms in Hidradenitis Suppurativa. JAMA Dermatol. 2023, 159, 1047–1049. [Google Scholar] [CrossRef] [PubMed]
- Horváth, B.; Janse, I.C.; Sibbald, G.R. Pain management in patients with hidradenitis suppurativa. J. Am. Acad. Dermatol. 2015, 73, S47–S51. [Google Scholar] [CrossRef] [PubMed]
- Amat-Samaranch, V.; Agut-Busquet, E.; Vilarrasa, E.; Puig, L. New perspectives on the treatment of hidradenitis suppurativa. Ther. Adv. Chronic Dis. 2021, 12, 20406223211055920. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yao, J.; Rong, M. Editorial: Role of Ion Channels in Pain. Front. Pharmacol. 2022, 13, 884665. [Google Scholar] [CrossRef]
- Alles, S.R.A.; Gomez, K.; Moutal, A.; Khanna, R. Putative roles of SLC7A5 (LAT1) transporter in pain. Neurobiol. Pain. 2020, 8, 100050. [Google Scholar] [CrossRef]
- Radhakrishna, U.; Ratnamala, U.; Jhala, D.D.; Uppala, L.V.; Vedangi, A.; Saiyed, N.; Patel, M.; Vadsaria, N.; Shah, S.R.; Rawal, R.M.; et al. Hidradenitis suppurativa associated telomere-methylome dysregulations in blood. J. Eur. Acad. Dermatol. Venereol. 2024, 38, 393–403. [Google Scholar] [CrossRef]
- Jha, M.K.; Song, G.J.; Lee, M.G.; Jeoung, N.H.; Go, Y.; Harris, R.A.; Park, D.H.; Kook, H.; Lee, I.K.; Suk, K. Metabolic Connection of Inflammatory Pain: Pivotal Role of a Pyruvate Dehydrogenase Kinase-Pyruvate Dehydrogenase-Lactic Acid Axis. J. Neurosci. 2015, 35, 14353–14369. [Google Scholar] [CrossRef]
- Zhang, J.M.; An, J. Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef]
- Radhakrishna, U.; Ratnamala, U.; Jhala, D.D.; Uppala, L.V.; Vedangi, A.; Patel, M.; Vadsaria, N.; Shah, S.; Saiyed, N.; Rawal, R.M.; et al. Hidradenitis suppurativa presents a methylome dysregulation capable to explain the pro-inflammatory microenvironment: Are these DNA methylations potential therapeutic targets? J. Eur. Acad. Dermatol. Venereol. 2023, 37, 2109–2123. [Google Scholar] [CrossRef]
- Bumgarner, J.R.; McCray, E.W.; Nelson, R.J. The disruptive relationship among circadian rhythms, pain, and opioids. Front. Neurosci. 2023, 17, 1109480. [Google Scholar] [CrossRef]
- Stavropoulou, E.; Pircalabioru, G.G.; Bezirtzoglou, E. The Role of Cytochromes P450 in Infection. Front. Immunol. 2018, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishna, U.; Ratnamala, U.; Jhala, D.D.; Vadsaria, N.; Patel, M.; Uppala, L.V.; Vedangi, A.; Saiyed, N.; Rawal, R.M.; Damiani, G.; et al. Cytochrome P450 Genes Mediated by DNA Methylation Are Involved in the Resistance to Hidradenitis Suppurativa. J. Investig. Dermatol. 2023, 143, 670–673.e19. [Google Scholar] [CrossRef] [PubMed]
- Goodin, B.R.; Overstreet, D.S.; Penn, T.M.; Bakshi, R.; Quinn, T.L.; Sims, A.; Ptacek, T.; Jackson, P.; Long, D.L.; Aroke, E.N. Epigenome-wide DNA methylation profiling of conditioned pain modulation in individuals with non-specific chronic low back pain. Clin. Epigenet. 2022, 14, 45. [Google Scholar] [CrossRef] [PubMed]
- Tajerian, M.; Alvarado, S.; Millecamps, M.; Dashwood, T.; Anderson, K.M.; Haglund, L.; Ouellet, J.; Szyf, M.; Stone, L.S. DNA methylation of SPARC and chronic low back pain. Mol. Pain. 2011, 7, 65. [Google Scholar] [CrossRef]
- Garriga, J.; Laumet, G.; Chen, S.R.; Zhang, Y.; Madzo, J.; Issa, J.J.; Pan, H.L.; Jelinek, J. Nerve Injury-Induced Chronic Pain Is Associated with Persistent DNA Methylation Reprogramming in Dorsal Root Ganglion. J. Neurosci. 2018, 38, 6090–6101. [Google Scholar] [CrossRef]
- Zheng, G.; Ren, J.; Shang, L.; Bao, Y. Role of autophagy in the pathogenesis and regulation of pain. Eur. J. Pharmacol. 2023, 955, 175859. [Google Scholar] [CrossRef]
- Merighi, A. Brain-Derived Neurotrophic Factor, Nociception, and Pain. Biomolecules 2024, 14, 539. [Google Scholar] [CrossRef]
- Kozera, E.K.; Lowes, M.A.; Hsiao, J.L.; Frew, J.W. Clinical considerations in the management of hidradenitis suppurativa in women. Int. J. Womens Dermatol. 2021, 7 5Pt B, 664–671. [Google Scholar] [CrossRef]
- Yamamoto, S.; Kashimoto, R.; Furukawa, S.; Ohashi, A.; Satoh, A. Lmx1b activation in axolotl limb regeneration. Dev. Dyn. 2022, 251, 1509–1523. [Google Scholar] [CrossRef]
- Warner, S.C.; van Meurs, J.B.; Schiphof, D.; Bierma-Zeinstra, S.M.; Hofman, A.; Uitterlinden, A.G.; Richardson, H.; Jenkins, W.; Doherty, M.; Valdes, A.M. Genome-wide association scan of neuropathic pain symptoms post total joint replacement highlights a variant in the protein-kinase C gene. Eur. J. Hum. Genet. 2017, 25, 446–451. [Google Scholar] [CrossRef]
- Lazarus, M.B.; Novotny, C.J.; Shokat, K.M. Structure of the human autophagy initiating kinase ULK1 in complex with potent inhibitors. ACS Chem. Biol. 2015, 10, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Steward, A.M.; Morgan, J.D.; Espinosa, J.P.; Turk, D.C.; Patel, K.V. Chronic Pain and Telomere Length in Community-Dwelling Adults: Findings From the 1999 to 2002 National Health and Nutrition Examination Survey. J. Pain 2017, 18, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.; Monteiro, C.; Cardoso-Cruz, H.; Galhardo, V. Altered Brain Expression of DNA Methylation and Hydroxymethylation Epigenetic Enzymes in a Rat Model of Neuropathic Pain. Int. J. Mol. Sci. 2023, 24, 7305. [Google Scholar] [CrossRef]
- Coppede, F.; Stoccoro, A.; Tannorella, P.; Migliore, L. Plasma Homocysteine and Polymorphisms of Genes Involved in Folate Metabolism Correlate with DNMT1 Gene Methylation Levels. Metabolites 2019, 9, 298. [Google Scholar] [CrossRef] [PubMed]
- Buesing, S.; Costa, M.; Schilling, J.M.; Moeller-Bertram, T. Vitamin B12 as a Treatment for Pain. Pain. Physician 2019, 22, E45–E52. [Google Scholar] [CrossRef]
- Sutherland, H.G.; Albury, C.L.; Griffiths, L.R. Advances in genetics of migraine. J. Headache Pain. 2019, 20, 72. [Google Scholar] [CrossRef]
- Deng, Z.; Fan, T.; Xiao, C.; Tian, H.; Zheng, Y.; Li, C.; He, J. TGF-beta signaling in health, disease, and therapeutics. Signal Transduct. Target. Ther. 2024, 9, 61. [Google Scholar] [CrossRef]
- Griesius, S.; O’Donnell, C.; Waldron, S.; Thomas, K.L.; Dwyer, D.M.; Wilkinson, L.S.; Hall, J.; Robinson, E.S.J.; Mellor, J.R. Reduced expression of the psychiatric risk gene DLG2 (PSD93) impairs hippocampal synaptic integration and plasticity. Neuropsychopharmacology 2022, 47, 1367–1378. [Google Scholar] [CrossRef]
- Daguet, I.; Raverot, V.; Bouhassira, D.; Gronfier, C. Circadian rhythmicity of pain sensitivity in humans. Brain 2022, 145, 3225–3235. [Google Scholar] [CrossRef]
- Radhakrishna, U.; Ratnamala, U.; Jhala, D.D.; Uppala, L.V.; Vedangi, A.; Saiyed, N.; Shah, S.R.; Patel, M.; Rawal, R.M.; Mazza, T.; et al. DNA methylation patterns of circadian and ultradian genes are altered in the peripheral blood of patients with hidradenitis suppurativa. Front. Immunol. 2024, 15, 1475424. [Google Scholar] [CrossRef]
- Callaghan, B.; Feldman, E. The metabolic syndrome and neuropathy: Therapeutic challenges and opportunities. Ann. Neurol. 2013, 74, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Baik, J.H. Dopamine signaling in food addiction: Role of dopamine D2 receptors. BMB Rep. 2013, 46, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Fairchild, T.J.; Vo, L.; Drummond, P.D. High Blood Glucose and Excess Body fat Enhance Pain Sensitivity and Weaken Pain Inhibition in Healthy Adults: A Single-blind Cross-over Randomized Controlled Trial. J. Pain 2023, 24, 128–144. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Suarez, V.J.; Beltran-Velasco, A.I.; Redondo-Florez, L.; Martin-Rodriguez, A.; Yanez-Sepulveda, R.; Tornero-Aguilera, J.F. Neuro-Vulnerability in Energy Metabolism Regulation: A Comprehensive Narrative Review. Nutrients 2023, 15, 3106. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.Y.; Jones, B.L.; Haas, G.L. Suicidal ideation and aggression in childhood, genetic variation and young adult depression. J. Affect. Disord. 2020, 276, 954–962. [Google Scholar] [CrossRef]
- Saez, E.; Erkoreka, L.; Moreno-Calle, T.; Berjano, B.; Gonzalez-Pinto, A.; Basterreche, N.; Arrue, A. Genetic variables of the glutamatergic system associated with treatment-resistant depression: A review of the literature. World J. Psychiatry 2022, 12, 884–896. [Google Scholar] [CrossRef]
- Rafiei, D.; Kolla, N.J. Elevated Brain Fatty Acid Amide Hydrolase Induces Depressive-Like Phenotypes in Rodent Models: A Review. Int. J. Mol. Sci. 2021, 22, 1047. [Google Scholar] [CrossRef]
- Lopez, J.F.; Palkovits, M.; Arato, M.; Mansour, A.; Akil, H.; Watson, S.J. Localization and quantification of pro-opiomelanocortin mRNA and glucocorticoid receptor mRNA in pituitaries of suicide victims. Neuroendocrinology 1992, 56, 491–501. [Google Scholar] [CrossRef]
- Gonzalez-Castro, T.B.; Genis-Mendoza, A.D.; Tovilla-Zarate, C.A.; Juarez-Rojop, I.E.; Lopez-Narvaez, M.L.; Perez-Hernandez, N.; Rodriguez-Perez, J.M.; Martinez-Magana, J.J. Association between polymorphisms of NOS1, NOS2 and NOS3 genes and suicide behavior: A systematic review and meta-analysis. Metab. Brain Dis. 2019, 34, 967–977. [Google Scholar] [CrossRef]
- Lee, H.Y.; Kim, Y.K. Transforming growth factor-beta1 and major depressive disorder with and without attempted suicide: Preliminary study. Psychiatry Res. 2010, 178, 92–96. [Google Scholar] [CrossRef]
- Bonsignore, G.; Martinotti, S.; Ranzato, E. Wound Repair and Ca2+ Signalling Interplay: The Role of Ca2+ Channels in Skin. Cells 2024, 13, 491. [Google Scholar] [CrossRef] [PubMed]
- Andrade, A.; Brennecke, A.; Mallat, S.; Brown, J.; Gomez-Rivadeneira, J.; Czepiel, N.; Londrigan, L. Genetic Associations between Voltage-Gated Calcium Channels and Psychiatric Disorders. Int. J. Mol. Sci. 2019, 20, 3537. [Google Scholar] [CrossRef] [PubMed]
- Nissenbaum, J. From mouse to humans: Discovery of the CACNG2 pain susceptibility gene. Clin. Genet. 2012, 82, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Bellessort, B.; Bachelot, A.; Grouthier, V.; De Lombares, C.; Narboux-Neme, N.; Garagnani, P.; Pirazzini, C.; Astigiano, S.; Mastracci, L.; Fontaine, A.; et al. Comparative analysis of molecular signatures suggests the use of gabapentin for the management of endometriosis-associated pain. J. Pain. Res. 2018, 11, 715–725. [Google Scholar] [CrossRef]
- Duan, J.; Grando, C.; Liu, S.; Chernyavsky, A.; Chen, J.K.; Andersen, B.; Grando, S.A. The M3 Muscarinic Acetylcholine Receptor Promotes Epidermal Differentiation. J. Investig. Dermatol. 2022, 142, 3211–3221.e2. [Google Scholar] [CrossRef]
- Kowalska, M.; Kapelusiak-Pielok, M.; Grzelak, T.; Wypasek, E.; Kozubski, W.; Dorszewska, J. The New *G29A and G1222A of HCRTR1, 5-HTTLPR of SLC6A4 Polymorphisms and Hypocretin-1, Serotonin Concentrations in Migraine Patients. Front. Mol. Neurosci. 2018, 11, 191. [Google Scholar] [CrossRef]
- Bardin, L. The complex role of serotonin and 5-HT receptors in chronic pain. Behav. Pharmacol. 2011, 22, 390–404. [Google Scholar] [CrossRef]
- Echeverria, F.; Gonzalez-Sanabria, N.; Alvarado-Sanchez, R.; Fernandez, M.; Castillo, K.; Latorre, R. Large conductance voltage-and calcium-activated K+ (BK) channel in health and disease. Front. Pharmacol. 2024, 15, 1373507. [Google Scholar] [CrossRef]
- Wulff, H.; Castle, N.A.; Pardo, L.A. Voltage-gated potassium channels as therapeutic targets. Nat. Rev. Drug Discov. 2009, 8, 982–1001. [Google Scholar] [CrossRef]
- Du, X.; Gao, H.; Jaffe, D.; Zhang, H.; Gamper, N. M-type K+ channels in peripheral nociceptive pathways. Br. J. Pharmacol. 2018, 175, 2158–2172. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Wen, J.; Yang, W.; Wang, C.; Gao, L.; Zheng, L.H.; Wang, T.; Ran, K.; Li, Y.; Li, X.; et al. Gain-of-function mutations in SCN11A cause familial episodic pain. Am. J. Hum. Genet. 2013, 93, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Leipold, E.; Hanson-Kahn, A.; Frick, M.; Gong, P.; Bernstein, J.A.; Voigt, M.; Katona, I.; Oliver Goral, R.; Altmuller, J.; Nurnberg, P.; et al. Cold-aggravated pain in humans caused by a hyperactive NaV1.9 channel mutant. Nat. Commun. 2015, 6, 10049. [Google Scholar] [CrossRef] [PubMed]
- Vanoye, C.G.; Gurnett, C.A.; Holland, K.D.; George, A.L., Jr.; Kearney, J.A. Novel SCN3A variants associated with focal epilepsy in children. Neurobiol. Dis. 2014, 62, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Veerman, C.C.; Wilde, A.A.; Lodder, E.M. The cardiac sodium channel gene SCN5A and its gene product NaV1.5: Role in physiology and pathophysiology. Gene 2015, 573, 177–187. [Google Scholar] [CrossRef]
- O’Brien, J.E.; Meisler, M.H. Sodium channel SCN8A (Nav1.6): Properties and de novo mutations in epileptic encephalopathy and intellectual disability. Front. Genet. 2013, 4, 213. [Google Scholar] [CrossRef]
- Duan, Y.; Gong, K.; Xu, S.; Zhang, F.; Meng, X.; Han, J. Regulation of cholesterol homeostasis in health and diseases: From mechanisms to targeted therapeutics. Signal Transduct. Target. Ther. 2022, 7, 265. [Google Scholar] [CrossRef]
- Tang, B.L. The Expanding Therapeutic Potential of Neuronal KCC2. Cells 2020, 9, 240. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Yue, G.; Zhao, Y. Energy metabolism disturbance in migraine: From a mitochondrial point of view. Front. Physiol. 2023, 14, 1133528. [Google Scholar] [CrossRef]
- Barker, J.M.; Taylor, J.R.; De Vries, T.J.; Peters, J. Brain-derived neurotrophic factor and addiction: Pathological versus therapeutic effects on drug seeking. Brain Res. 2015, 1628 Pt A, 68–81. [Google Scholar] [CrossRef]
- Blum, K.; Oscar-Berman, M.; Barh, D.; Giordano, J.; Gold, M. Dopamine Genetics and Function in Food and Substance Abuse. J. Genet. Syndr. Gene Ther. 2013, 4, 1000121. [Google Scholar]
- Navarrete, F.; Garcia-Gutierrez, M.S.; Gasparyan, A.; Navarro, D.; Lopez-Picon, F.; Morcuende, A.; Femenia, T.; Manzanares, J. Biomarkers of the Endocannabinoid System in Substance Use Disorders. Biomolecules 2022, 12, 396. [Google Scholar] [CrossRef] [PubMed]
- Panlilio, L.V.; Justinova, Z.; Goldberg, S.R. Inhibition of FAAH and activation of PPAR: New approaches to the treatment of cognitive dysfunction and drug addiction. Pharmacol. Ther. 2013, 138, 84–102. [Google Scholar] [CrossRef] [PubMed]
- Nudmamud-Thanoi, S.; Veerasakul, S.; Thanoi, S. Pharmacogenetics of drug dependence: Polymorphisms of genes involved in GABA neurotransmission. Neurosci. Lett. 2020, 726, 134463. [Google Scholar] [CrossRef] [PubMed]
- Popescu, A.; Marian, M.; Dragoi, A.M.; Costea, R.V. Understanding the genetics and neurobiological pathways behind addiction (Review). Exp. Ther. Med. 2021, 21, 544. [Google Scholar] [CrossRef]
- Thakkar, B.; Acevedo, E.O. BDNF as a biomarker for neuropathic pain: Consideration of mechanisms of action and associated measurement challenges. Brain Behav. 2023, 13, e2903. [Google Scholar] [CrossRef]
- Anthoni, M.; Fyhrquist-Vanni, N.; Wolff, H.; Alenius, H.; Lauerma, A. Transforming growth factor-beta/Smad3 signalling regulates inflammatory responses in a murine model of contact hypersensitivity. Br. J. Dermatol. 2008, 159, 546–554. [Google Scholar]
- Eve, M.; Gandawijaya, J.; Yang, L.; Oguro-Ando, A. Neuronal Cell Adhesion Molecules May Mediate Neuroinflammation in Autism Spectrum Disorder. Front. Psychiatry 2022, 13, 842755. [Google Scholar] [CrossRef]
- Iemmolo, M.; Ghersi, G.; Bivona, G. The Cytokine CX3CL1 and ADAMs/MMPs in Concerted Cross-Talk Influencing Neurodegenerative Diseases. Int. J. Mol. Sci. 2023, 24, 8026. [Google Scholar] [CrossRef]
- Harno, E.; Gali Ramamoorthy, T.; Coll, A.P.; White, A. POMC: The Physiological Power of Hormone Processing. Physiol. Rev. 2018, 98, 2381–2430. [Google Scholar] [CrossRef]
- Cohen, B.; Cadesky, A.; Jaggi, S. Dermatologic manifestations of thyroid disease: A literature review. Front. Endocrinol. 2023, 14, 1167890. [Google Scholar] [CrossRef]
- Lightman, S.L.; Birnie, M.T.; Conway-Campbell, B.L. Dynamics of ACTH and Cortisol Secretion and Implications for Disease. Endocrine Reviews 2020, 41, 470–490. [Google Scholar] [CrossRef] [PubMed]
- Kramer, P.R.; Rao, M.; Stinson, C.; Bellinger, L.L.; Kinchington, P.R.; Yee, M.B. Aromatase Derived Estradiol Within the Thalamus Modulates Pain Induced by Varicella Zoster Virus. Front. Integr. Neurosci. 2018, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Husby, A. On the Use of Blood Samples for Measuring DNA Methylation in Ecological Epigenetic Studies. Integr. Comp. Biol. 2020, 60, 1558–1566. [Google Scholar] [CrossRef] [PubMed]
- Cazzaniga, S.; Naldi, L.; Damiani, G.; Atzori, L.; Patta, F.; Guidarelli, G.; Bettoli, V. Validation of a visual-aided questionnaire for the self-assessment of hidradenitits suppurativa. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 1993–1998. [Google Scholar] [CrossRef]
- Lipsker, D.; Severac, F.; Freysz, M.; Sauleau, E.; Boer, J.; Emtestam, L.; Matusiak, L.; Prens, E.; Velter, C.; Lenormand, C.; et al. The ABC of Hidradenitis Suppurativa: A Validated Glossary on how to Name Lesions. Dermatology 2016, 232, 137–142. [Google Scholar] [CrossRef]
- Hurley, H.J. Axillary hyperhidrosis, apocrine bromhidrosis, hidradenitis suppurativa, and familial benign pemphigus: Surgical approach. In Roenigk and Roenigk’s Dermatologic Surgery, Principles and Practice, 2nd ed.; Roenigk, R.K., Roenigk, H.H., Jr., Eds.; Marcel Dekker: New York, NY, USA, 1996; pp. 623–645. [Google Scholar]
- Damiani, G.; Della Valle, V.; Iannone, M.; Dini, V.; Marzano, A.V. Autoinflammatory Disease Damage Index (ADDI): A possible newborn also in hidradenitis suppurativa daily practice. Ann. Rheum. Dis. 2017, 76, e25. [Google Scholar] [CrossRef]
- Radhakrishna, U.; Vishweswaraiah, S.; Uppala, L.V.; Szymanska, M.; Macknis, J.; Kumar, S.; Saleem-Rasheed, F.; Aydas, B.; Forray, A.; Muvvala, S.B.; et al. Placental DNA methylation profiles in opioid-exposed pregnancies and associations with the neonatal opioid withdrawal syndrome. Genomics 2021, 113, 1127–1135. [Google Scholar] [CrossRef]
- Radhakrishna, U.; Albayrak, S.; Zafra, R.; Baraa, A.; Vishweswaraiah, S.; Veerappa, A.M.; Mahishi, D.; Saiyed, N.; Mishra, N.K.; Guda, C.; et al. Placental epigenetics for evaluation of fetal congenital heart defects: Ventricular Septal Defect (VSD). PLoS ONE 2019, 14, e0200229. [Google Scholar] [CrossRef]
- Parisien, M.; Khoury, S.; Chabot-Dore, A.J.; Sotocinal, S.G.; Slade, G.D.; Smith, S.B.; Fillingim, R.B.; Ohrbach, R.; Greenspan, J.D.; Maixner, W.; et al. Effect of Human Genetic Variability on Gene Expression in Dorsal Root Ganglia and Association with Pain Phenotypes. Cell Rep. 2017, 19, 1940–1952. [Google Scholar] [CrossRef]
- James, S. Human pain and genetics: Some basics. Br. J. Pain. 2013, 7, 171–178. [Google Scholar] [CrossRef]
- Bai, G.; Ren, K.; Dubner, R. Epigenetic regulation of persistent pain. Transl. Res. 2015, 165, 177–199. [Google Scholar] [CrossRef] [PubMed]
- Linnstaedt, S.D.; Rueckeis, C.A.; Riker, K.D.; Pan, Y.; Wu, A.; Yu, S.; Wanstrath, B.; Gonzalez, M.; Harmon, E.; Green, P.; et al. MicroRNA-19b predicts widespread pain and posttraumatic stress symptom risk in a sex-dependent manner following trauma exposure. Pain 2020, 161, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Zorina-Lichtenwalter, K.; Meloto, C.B.; Khoury, S.; Diatchenko, L. Genetic predictors of human chronic pain conditions. Neuroscience 2016, 338, 36–62. [Google Scholar] [CrossRef] [PubMed]
- Wistrom, E.; Chase, R.; Smith, P.R.; Campbell, Z.T. A compendium of validated pain genes. WIREs Mech. Dis. 2022, 14, e1570. [Google Scholar] [CrossRef]
- Sexton, J.E.; Cox, J.J.; Zhao, J.; Wood, J.N. The Genetics of Pain: Implications for Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 123–142. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Gu, Z. Complex heatmap visualization. Imeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]


| Target ID | Genes | Location | p-Val | FDR p-Val | % Methylation | AUC | CI | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Control | Difference | Lower | Upper | ||||||
| cg27280396 | RORA | 15q22.2 | 3.05 × 10−39 | 2.64 × 10−33 | 27.29 | 15.67 | 11.61 | 0.89 | 0.80 | 0.99 |
| cg09633973 | HIC1 | 17p13.3 | 6.69 × 10−38 | 5.79 × 10−32 | 19.45 | 11.02 | 8.42 | 0.84 | 0.73 | 0.96 |
| cg09820084 | RNF144B | 6p22.3 | 2.98 × 10−37 | 2.58 × 10−31 | 12.15 | 5.36 | 6.80 | 0.79 | 0.66 | 0.92 |
| cg16553796 | PARD6G | 18q23 | 6.72 × 10−29 | 5.82 × 10−23 | 23.84 | 40.87 | −17.03 | 0.82 | 0.70 | 0.94 |
| cg03466780 | EXD3 | 9q34.3 | 3.32 × 10−28 | 2.88 × 10−22 | 38.93 | 62.16 | −23.23 | 0.77 | 0.63 | 0.90 |
| cg14984943 | CACNA2D3 | 3p21.1-p14.3 | 1.59 × 10−24 | 1.38 × 10−18 | 63.36 | 77.64 | −14.28 | 0.92 | 0.84 | 1.00 |
| cg01364755 | ADARB2 | 10p15.3 | 3.05 × 10−24 | 2.64 × 10−18 | 91.33 | 97.30 | −5.97 | 0.85 | 0.73 | 0.96 |
| cg14311251 | COL4A1 | 13q34 | 4.25 × 10−22 | 3.68 × 10−16 | 51.37 | 66.76 | −15.40 | 0.92 | 0.84 | 1.00 |
| cg22572258 | KCND2 | 7q31.31 | 2.18 × 10−21 | 1.88 × 10−15 | 48.31 | 63.73 | −15.42 | 0.92 | 0.84 | 1.00 |
| cg03666597 | CUX1 | 7q22.1 | 5.58 × 10−21 | 4.82 × 10−15 | 38.87 | 54.45 | −15.58 | 0.88 | 0.77 | 0.98 |
| cg07913781 | NRP1 | 10p11.22 | 1.59 × 10−20 | 1.38 × 10−14 | 86.04 | 93.37 | −7.33 | 0.79 | 0.66 | 0.92 |
| cg17978764 | CLIC5 | 6p21.1 | 1.87 × 10−20 | 1.62 × 10−14 | 50.32 | 65.24 | −14.93 | 0.98 | 0.93 | 1.00 |
| cg13050884 | FNDC3B | 3q26.31 | 2.55 × 10−20 | 2.20 × 10−14 | 59.93 | 73.61 | −13.68 | 0.89 | 0.79 | 0.98 |
| cg11615758 | PHACTR1 | 6p24.1 | 3.30 × 10−20 | 2.86 × 10−14 | 59.06 | 72.84 | −13.78 | 0.94 | 0.87 | 1.00 |
| cg16201095 | ABCC2 | 10q24.2 | 9.14 × 10−19 | 7.90 × 10−14 | 88.21 | 94.72 | −6.51 | 0.89 | 0.79 | 0.98 |
| cg21752295 | BMP6 | 6p24.3 | 2.02 × 10−19 | 1.75 × 10−13 | 63.54 | 76.32 | −12.79 | 0.91 | 0.83 | 1.00 |
| cg09601770 | DPP4 | 2q24.2 | 4.93 × 10−19 | 4.26 × 10−13 | 10.64 | 20.92 | −10.28 | 0.78 | 0.65 | 0.91 |
| cg05707781 | LPP | 3q27.3-q28 | 9.41 × 10−19 | 8.14 × 10−13 | 65.84 | 77.99 | −12.15 | 0.96 | 0.90 | 1.00 |
| cg22018565 | ADORA3 | 1p13.2 | 1.75 × 10−18 | 1.51 × 10−12 | 70.46 | 81.58 | −11.12 | 0.76 | 0.62 | 0.90 |
| cg16873130 | CHRM2 | 7q33 | 2.58 × 10−18 | 2.23 × 10−12 | 48.61 | 62.88 | −14.27 | 0.91 | 0.82 | 1.00 |
| cg18356276 | AHR | 7p21.1 | 2.59 × 10−18 | 2.24 × 10−12 | 66.70 | 78.55 | −11.85 | 0.98 | 0.93 | 1.00 |
| cg11995490 | C7orf50 | 7p22.3 | 2.70 × 10−17 | 2.34 × 10−11 | 71.69 | 82.21 | −10.52 | 0.78 | 0.65 | 0.91 |
| cg07035454 | OXR1 | 8q23.1 | 3.96 × 10−17 | 3.43 × 10−11 | 57.42 | 70.37 | −12.95 | 0.88 | 0.77 | 0.98 |
| cg21189939 | SMG6 | 17p13.3 | 1.14 × 10−16 | 9.83 × 10−11 | 78.66 | 87.36 | −8.70 | 0.77 | 0.64 | 0.91 |
| cg21671386 | DRD2 | 11q23.2 | 1.24 × 10−16 | 1.07 × 10−10 | 61.12 | 73.38 | −12.26 | 0.97 | 0.92 | 1.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radhakrishna, U.; Kuracha, M.R.; Hamzavi, I.; Saiyed, N.; Prajapati, J.; Rawal, R.M.; Uppala, L.V.; Damiani, G.; Ratnamala, U.; Nath, S.K. Impaired Molecular Mechanisms Contributing to Chronic Pain in Patients with Hidradenitis Suppurativa: Exploring Potential Biomarkers and Therapeutic Targets. Int. J. Mol. Sci. 2025, 26, 1039. https://doi.org/10.3390/ijms26031039
Radhakrishna U, Kuracha MR, Hamzavi I, Saiyed N, Prajapati J, Rawal RM, Uppala LV, Damiani G, Ratnamala U, Nath SK. Impaired Molecular Mechanisms Contributing to Chronic Pain in Patients with Hidradenitis Suppurativa: Exploring Potential Biomarkers and Therapeutic Targets. International Journal of Molecular Sciences. 2025; 26(3):1039. https://doi.org/10.3390/ijms26031039
Chicago/Turabian StyleRadhakrishna, Uppala, Murali R. Kuracha, Iltefat Hamzavi, Nazia Saiyed, Jignesh Prajapati, Rakesh M. Rawal, Lavanya V. Uppala, Giovanni Damiani, Uppala Ratnamala, and Swapan K. Nath. 2025. "Impaired Molecular Mechanisms Contributing to Chronic Pain in Patients with Hidradenitis Suppurativa: Exploring Potential Biomarkers and Therapeutic Targets" International Journal of Molecular Sciences 26, no. 3: 1039. https://doi.org/10.3390/ijms26031039
APA StyleRadhakrishna, U., Kuracha, M. R., Hamzavi, I., Saiyed, N., Prajapati, J., Rawal, R. M., Uppala, L. V., Damiani, G., Ratnamala, U., & Nath, S. K. (2025). Impaired Molecular Mechanisms Contributing to Chronic Pain in Patients with Hidradenitis Suppurativa: Exploring Potential Biomarkers and Therapeutic Targets. International Journal of Molecular Sciences, 26(3), 1039. https://doi.org/10.3390/ijms26031039








