Optimization of CRISPR/Cas9 Gene Editing System in Sheep (Ovis aries) Oocytes via Microinjection
Abstract
1. Introduction
2. Results
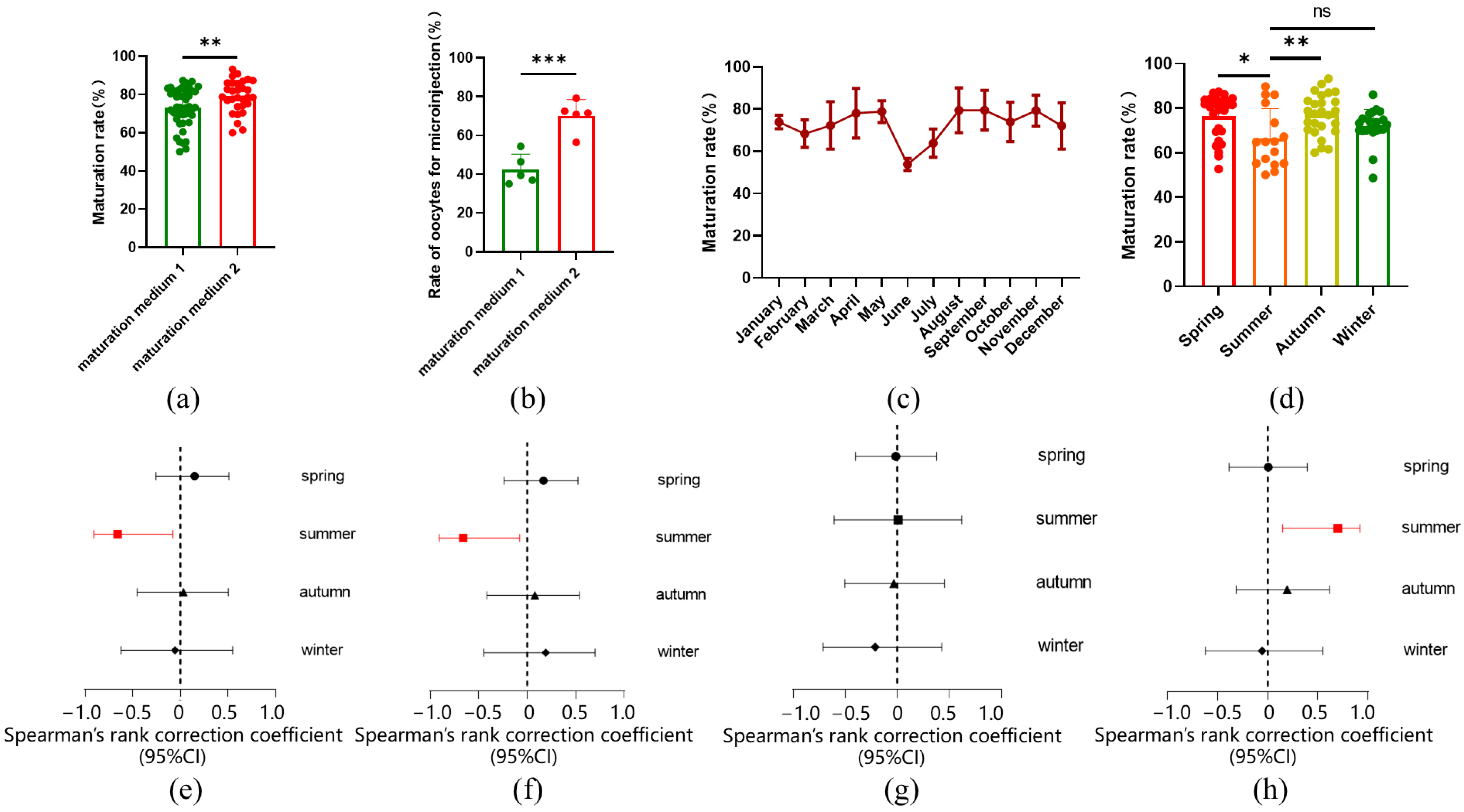
2.1. In Vitro Maturation Rate of Sheep Oocytes Influenced by Seasonal Variations
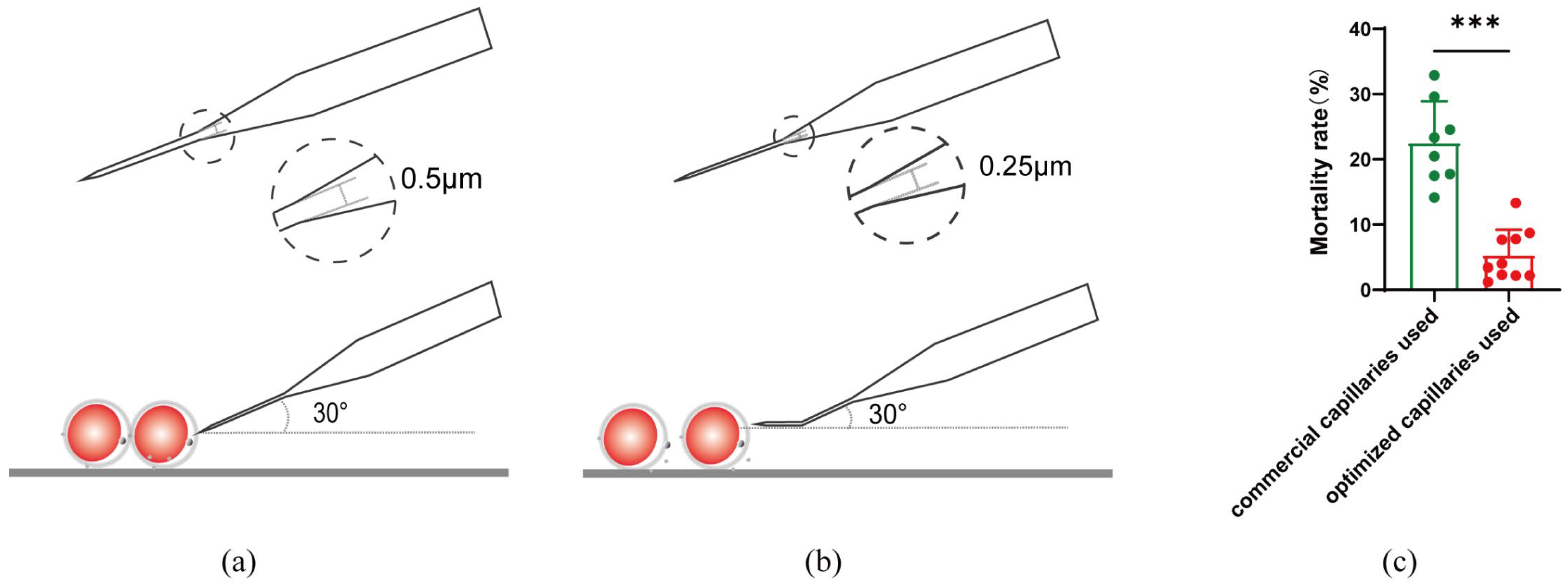
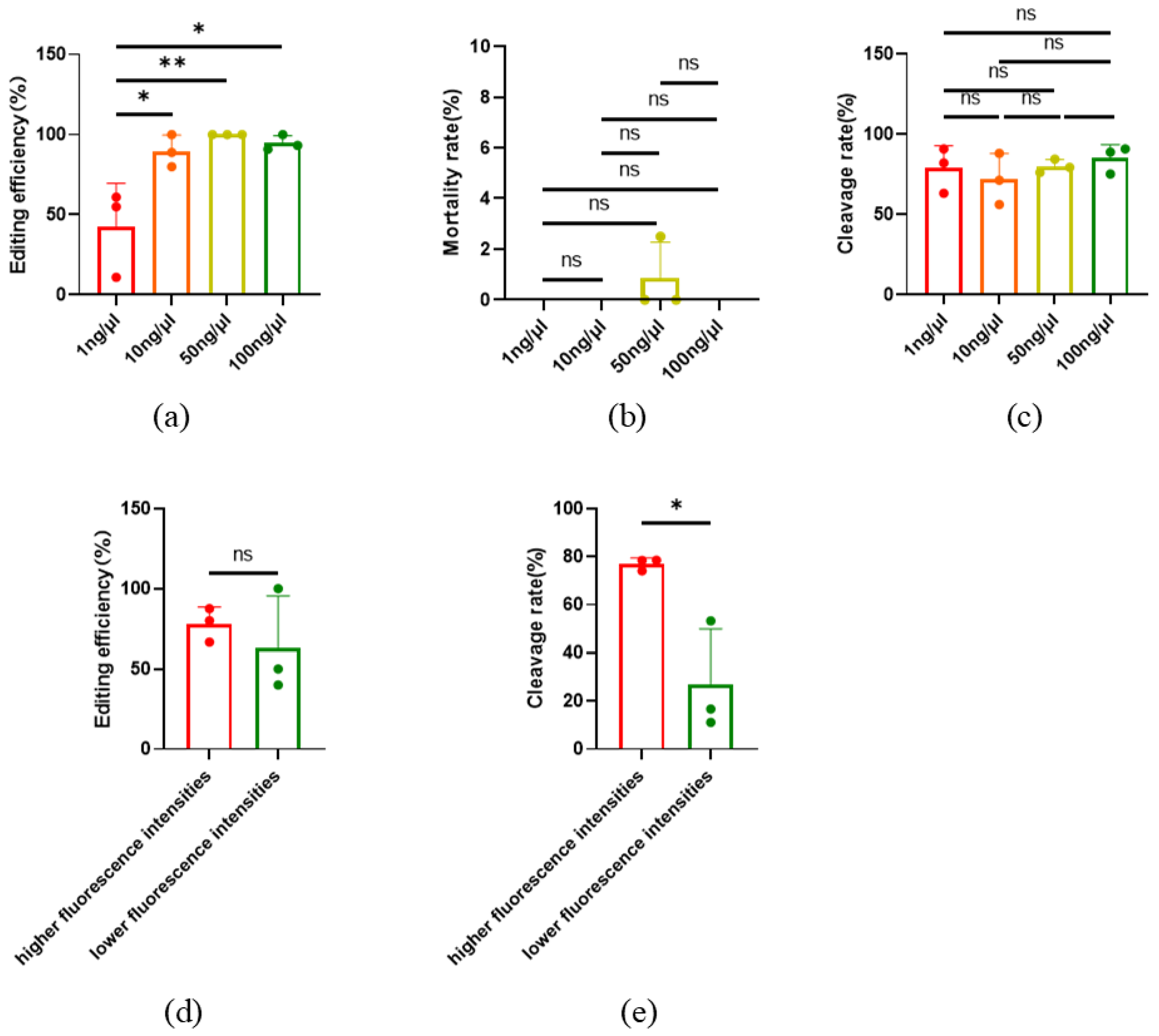
2.2. Optimization of CRISPR/Cas9 Editing Microinjection System in Oocytes
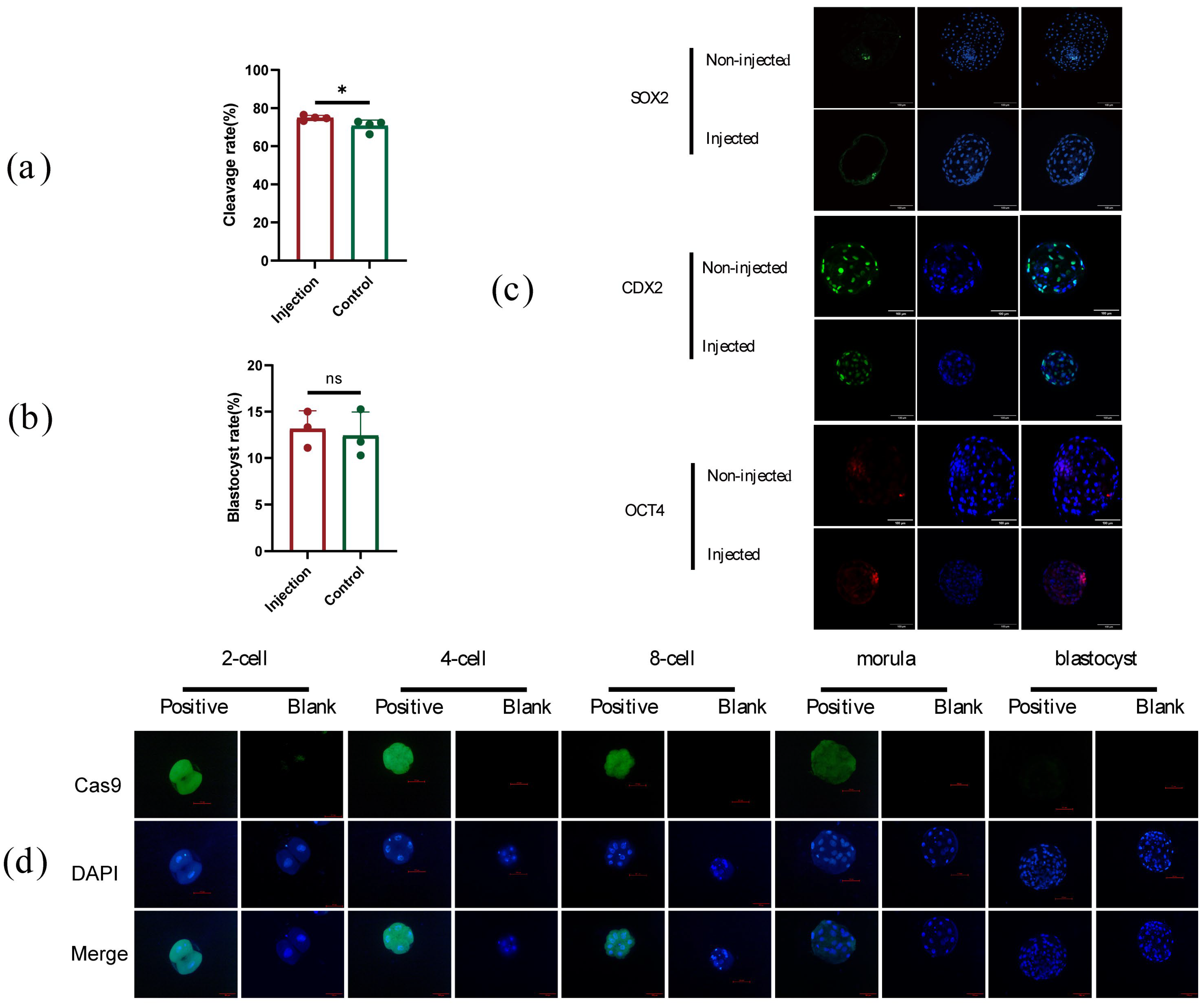
2.3. The Development of Early Embryos Was Not Influenced by Microinjection
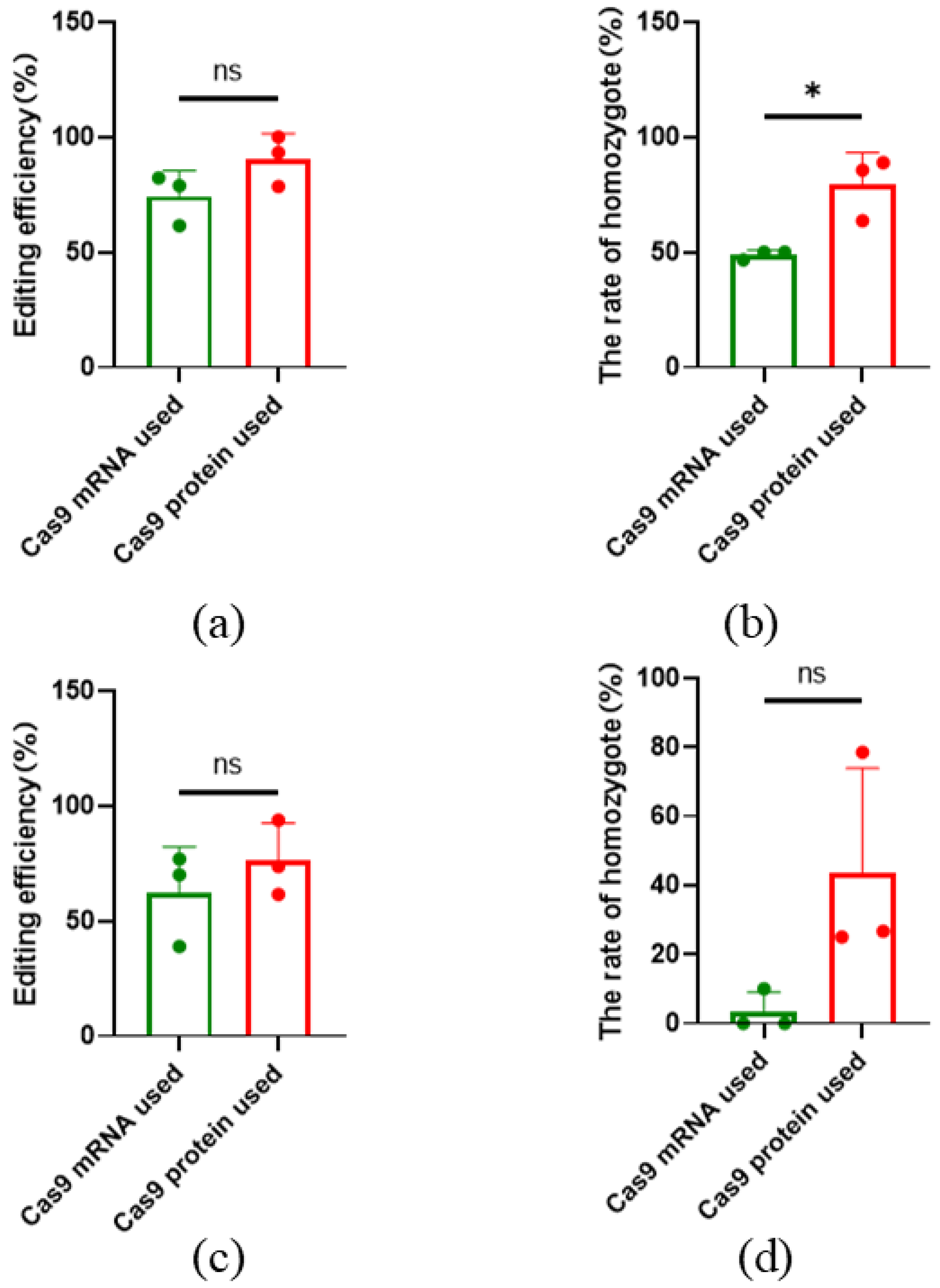
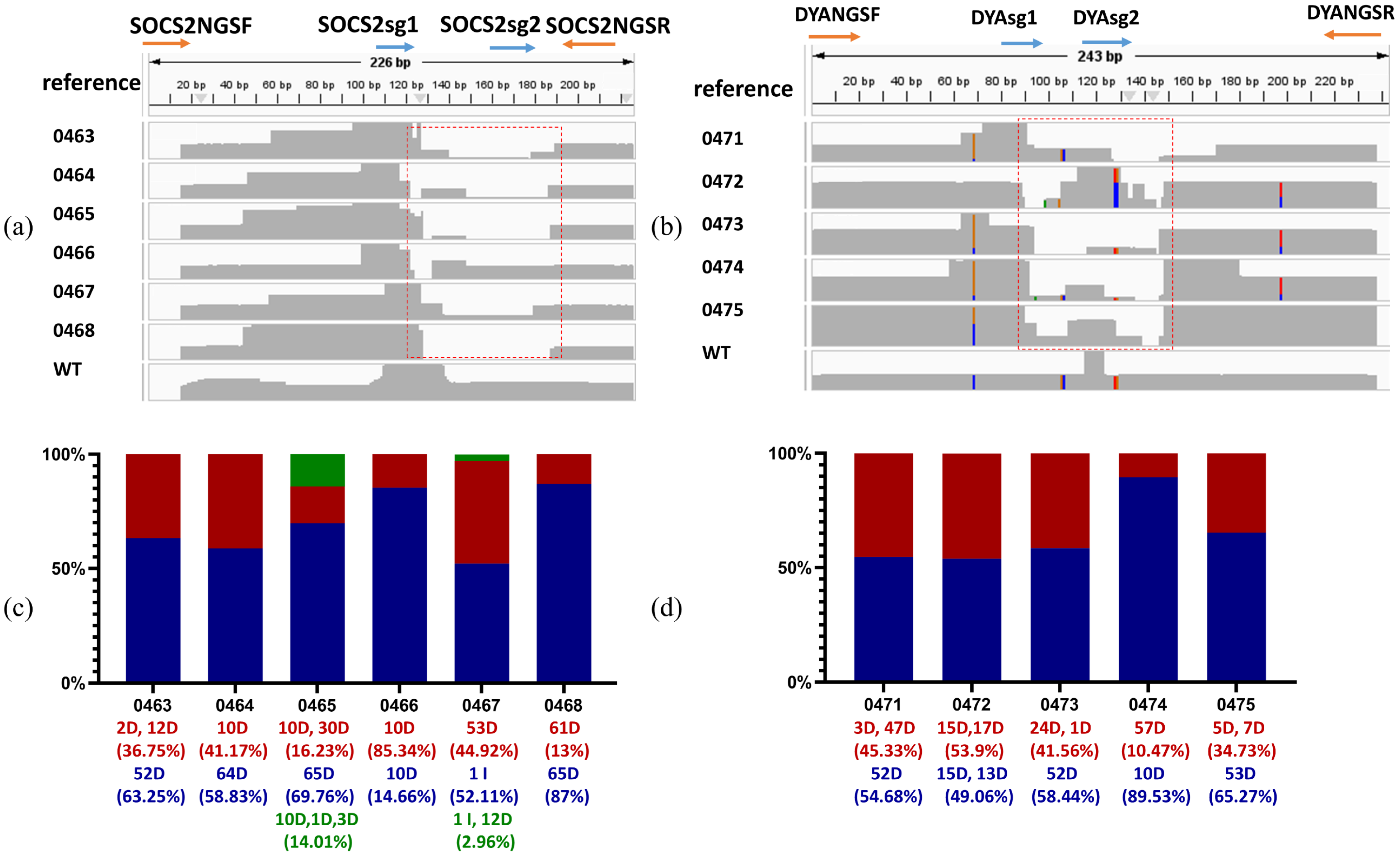
2.4. High-Efficiency of CRISPR/Cas9 Editing in Sheep by Microinjection
3. Discussion
4. Materials and Methods
4.1. Animals and Ethical Statement
4.2. sgRNA and Cas9 mRNA or Protein
4.3. Manufacture of Capillaries for Microinjection
4.4. In Vitro Maturation of Sheep Oocytes
4.5. Microinjection
4.6. Parthenogenetic Activation and In Vitro Development
4.7. Immunofluorescence
4.8. Statistical Analysis
4.9. Generation of Gene-Edited Sheep
4.10. Genotyping of Edited Sheep
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Jabbar, A.; Zulfiqar, F.; Mahnoor, M.; Mushtaq, N.; Zaman, M.H.; Din, A.S.U.; Khan, M.A.; Ahmad, H.I. Advances and Perspectives in the Application of CRISPR-Cas9 in Livestock. Mol. Biotechnol. 2021, 63, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Menchaca, A.; Dos Santos-Neto, P.C.; Mulet, A.P.; Crispo, M. CRISPR in livestock: From editing to printing. Theriogenology 2020, 150, 247–254. [Google Scholar] [CrossRef]
- Gao, F.; Li, P.; Yin, Y.; Du, X.; Cao, G.; Wu, S.; Zhao, Y. Molecular breeding of livestock for disease resistance. Virology 2023, 587, 109862. [Google Scholar] [CrossRef]
- Turnley, A.M. Role of SOCS2 in growth hormone actions. Trends Endocrinol. Metab. 2005, 16, 53–58. [Google Scholar] [CrossRef]
- Zhou, S.; Cai, B.; He, C.; Wang, Y.; Ding, Q.; Liu, J.; Liu, Y.; Ding, Y.; Zhao, X.; Li, G.; et al. Programmable Base Editing of the Sheep Genome Revealed No Genome-Wide Off-Target Mutations. Front. Genet. 2019, 10, 215. [Google Scholar] [CrossRef]
- Dukkipati, V.S.; Blair, H.T.; Garrick, D.J.; Murray, A. ’-Mhc’—Ovine major histocompatibility complex: Structure and gene polymorphisms. Genet. Mol. Res. 2006, 5, 581–608. [Google Scholar]
- Mendoza-de Gives, P.; López-Arellano, M.E.; Olmedo-Juárez, A.; Higuera-Pierdrahita, R.I.; von Son-de Fernex, E. Recent Advances in the Control of Endoparasites in Ruminants from a Sustainable Perspective. Pathogens 2023, 12, 1121. [Google Scholar] [CrossRef]
- Li, X.; He, S.G.; Li, W.R.; Luo, L.Y.; Yan, Z.; Mo, D.X.; Wan, X.; Lv, F.H.; Yang, J.; Xu, Y.X.; et al. Genomic analyses of wild argali, domestic sheep, and their hybrids provide insights into chromosome evolution, phenotypic variation, and germplasm innovation. Genome Res. 2022, 32, 1669–1684. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Zhang, W.; Zhao, G.; Zhang, X.; Bai, J.; Brosh, R.; Wudzinska, A.; Huang, E.; Ashe, H.; Ellis, G.; et al. On the genetic basis of tail-loss evolution in humans and apes. Nature 2024, 626, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Weng, Y.; Bai, D.; Jia, Y.; Liu, Y.; Zhang, Y.; Kou, X.; Zhao, Y.; Ruan, J.; Chen, J.; et al. Precise allele-specific genome editing by spatiotemporal control of CRISPR-Cas9 via pronuclear transplantation. Nat. Commun. 2020, 11, 4593. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Ma, Y.; Wang, T.; Lian, L.; Tian, X.; Rui, H.; Deng, S.; Li, K.; Wang, F.; Li, N.; et al. One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system. Front. Agr. Sci. Eng. 2014, 1, 2–5. [Google Scholar]
- Wang, X.; Niu, Y.; Zhou, J.; Yu, H.; Kou, Q.; Lei, A.; Zhao, X.; Yan, H.; Cai, B.; Shen, Q.; et al. Multiplex gene editing via CRISPR/Cas9 exhibits desirable muscle hypertrophy without detectable off-target effects in sheep. Sci. Rep. 2016, 6, 32271. [Google Scholar] [CrossRef]
- Hu, R.; Fan, Z.Y.; Wang, B.Y.; Deng, S.L.; Zhang, X.S.; Zhang, J.L.; Han, H.B.; Lian, Z.X. RAPID COMMUNICATION: Generation of FGF5 knockout sheep via the CRISPR/Cas9 system. J. Anim. Sci. 2017, 95, 2019–2024. [Google Scholar] [CrossRef]
- Zhang, X.; Li, W.; Liu, C.; Peng, X.; Lin, J.; He, S.; Li, X.; Han, B.; Zhang, N.; Wu, Y.; et al. Alteration of sheep coat color pattern by disruption of ASIP gene via CRISPR Cas9. Sci. Rep. 2016, 7, 8149. [Google Scholar] [CrossRef]
- Zhou, S.; Yu, H.; Zhao, X.; Cai, B.; Ding, Q.; Huang, Y.; Li, Y.; Li, Y.; Niu, Y.; Lei, A.; et al. Generation of gene-edited sheep with a defined Booroola fecundity gene (FecBB) mutation in bone morphogenetic protein receptor type 1B (BMPR1B) via clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) 9. Reprod. Fertil. Dev. 2018, 30, 1616–1621. [Google Scholar] [CrossRef]
- Zhou, S.; Ding, Y.; Liu, J.; Liu, Y.; Zhao, X.; Li, G.; Zhang, C.; Li, C.; Wang, Y.; Kalds, P.; et al. Highly efficient generation of sheep with a defined FecBB mutation via adenine base editing. Genet. Sel. Evol. 2020, 52, 35. [Google Scholar] [CrossRef]
- Zhou, S.; Kalds, P.; Luo, Q.; Sun, K.; Zhao, X.; Gao, Y.; Cai, B.; Huang, S.; Kou, Q.; Petersen, B.; et al. Optimized Cas9: sgRNA delivery efficiently generates biallelic MSTN knockout sheep without affecting meat quality. BMC Genom. 2022, 23, 348. [Google Scholar] [CrossRef]
- Zhou, S.; Lenk, L.J.; Gao, Y.; Wang, Y.; Zhao, X.; Pan, M.; Huang, S.; Sun, K.; Kalds, P.; Luo, Q.; et al. Generation of sheep with defined FecBB and TBXT mutations and porcine blastocysts with KCNJ5G151R/+ mutation using prime editing. BMC Genom. 2023, 24, 313. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Wang, H.; Meng, C.; Gui, H.; Li, Y.; Chen, F.; Zhang, C.; Zhang, H.; Ding, Q.; Zhang, J.; et al. Efficient and Specific Generation of MSTN-Edited Hu Sheep Using C-CRISPR. Genes 2023, 14, 1216. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.M.; Zhao, Y.; Yu, K.; Xu, X.L.; Zhang, X.S.; Zhang, J.L.; Wu, S.J.; Liu, Z.M.; Yuan, Y.M.; Guo, X.F.; et al. A MSTNDel73C mutation with FGF5 knockout sheep by CRISPR/Cas9 promotes skeletal muscle myofiber hyperplasia. eLife 2024, 12, RP86827. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Shen, J.; Li, D.; Cheng, Y. Strategies in the delivery of Cas9 ribonucleoprotein for CRISPR/Cas9 genome editing. Theranostics 2021, 11, 614–648. [Google Scholar] [CrossRef]
- Paffoni, A.; Brevini, T.A.; Somigliana, E.; Restelli, L.; Gandolfi, F.; Ragni, G. In vitro development of human oocytes after parthenogenetic activation or intracytoplasmic sperm injection. Fertil. Steril. 2007, 87, 77–82. [Google Scholar] [CrossRef]
- Da Silva, J.C.B.; Alves, M.B.R.; Bridi, A.; Bohrer, R.C.; Escobar, G.S.L.; de Carvalho, J.A.B.A.; Binotti, W.A.B.; Pugliesi, G.; Lemes, K.M.; Chello, D.; et al. Reproductive seasonality influences oocyte retrieval and embryonic competence but not uterine receptivity in buffaloes. Theriogenology 2021, 170, 77–84. [Google Scholar] [CrossRef]
- Lorca, T.; Cruzalegui, F.H.; Fesquet, D.; Cavadore, J.C.; Méry, J.; Means, A.; Dorée, M. Calmodulin-dependent protein kinase II mediates inactivation of MPF and CSF upon fertilization of Xenopus eggs. Nature 1993, 366, 270–273. [Google Scholar] [CrossRef]
- Quan, Z.J.; Li, S.A.; Yang, Z.X.; Zhao, J.J.; Li, G.H.; Zhang, F.; Wen, W.; Cheng, T.; Zhang, X.B. GREPore-seq: A Robust Workflow to Detect Changes After Gene Editing Through Long-range PCR and Nanopore Sequencing. Genom. Proteom. Bioinf. 2023, 21, 1221–1236. [Google Scholar] [CrossRef]
- Navarro-Serna, S.; Vilarino, M.; Park, I.; Gadea, J.; Ross, P.J. Livestock Gene Editing by One-step Embryo Manipulation. J. Equine Vet. Sci. 2020, 89, 103025. [Google Scholar] [CrossRef]


| Target Gene | Zygotes | Transplanted Zygotes | Recipient | Pregnant Recipients | Delivered Lambs | Edited Lambs | Ratio of Lambs with Detectable Gene Editing * |
|---|---|---|---|---|---|---|---|
| SOCS2 | 58 | 47 | 7 | 5 | 6 | 6 | 100% |
| DYA | 135 | 90 | 15 | 4 | 5 | 5 | 100% |
| Target Gene | Lambs | cDNA Sequences | Genotypes | Predicted Animo Acids Sequences | Animo Acids Number |
|---|---|---|---|---|---|
| SOCS2 | WT | ---GGGAATGGCGCGGAAGGG------CCAGAGGCGGCGCGTCTGGCG--------CAGGTATAA | / | ---GNGAEG---PEAARLA------QV* | 198aa |
| 0463a | ---GGGAATGGCGC---------------------CCAGAGGCGGCGCGTCTGGCG------ATATGA | −52 bp | ---GNGAQRRRVWR---I* | 35aa | |
| 0463b | ---GGGAATA-----------------------------------------------------------TCTGG----------ATATGA | −71 bp | ---GNIW--------------I* | 29aa | |
| 0464 | ---GGGAATC--------------------------------------------CGGCG--------------------------CAGGTATAA | −64 bp | ---GNPA------------------------QV* | 177aa | |
| 0465 | ---GGGAATGGCGCG------------------------------------------------TCTGGC------TGTTAA | −65 bp | ---GNGASG--------C* | 33aa | |
| 0466a | ---GGGAATGGCGCG------------------------------------------------TCTGGC------TGTTAA | −65 bp | ---GNGASG--------C* | 33aa | |
| 0466b | ---GGGAATGGCGC-----------------------CAGA------GACTGTTAA | −53 bp | ---GNGAR---------------DC* | 37aa | |
| 0467a | ---GGGAATGGCGC-----------------------CAGA------GACTGTTAA | −53 bp | ---GNGAR---------------DC* | 37aa | |
| 0467b | ---GGGAATGGCGCG------------------------------------------------TCTGGC------TGTTAA | −65 bp | ---GNGASG--------C* | 33aa | |
| 0468a | ---GGGAATGGCGCG------------------------------------------------TCTGGC------TGTTAA | −65 bp | ---GNGASG--------C* | 33aa | |
| 0468b | ---GGGAATGGCG-----------------GAC---------------------GCGTC------GAAATATGA | −61 bp | ---GNGGRV------EI* | 32aa | |
| DYA | WT | ATGAAGAAAGCTCTGATTCTG------ATCGTGGCGGAC----------------------AGGTGA | / | MKKALIL------IVAD----------R* | 288aa |
| 0471a | ATGAAGAAAGCTCTGATTC----------------------CG----------ACCTGA | −52 bp | MKKALIP----------T* | 42aa | |
| 0471b | ATGAAGAAAGCTCTGAT---GAGGGCTCTCGCTCTGGCCGCCATGATGAGCC--TGTG------C---GTTTGA | −43 bp | MKKALMRALALAAMMSLC----V* | 40aa | |
| 0472a | ATGAAGAAAGCTCT--------CGC--------TG------CAT---GGC------AGGTGA | −18 bp | MKKALA------------------R* | 282aa | |
| 0472b | ATGAAGAAAGCTCTGATTCTG--------------GCCGCCATGATGAGCCCCTGTGG--------------C------GTTTGA | −32 bp | MKKALILAAMMSPCG------V* | 37aa | |
| 0473 | ATGAAGAAAGCTCTGATTC----------------------CG----------ACCTGA | −52 bp | MKKALIP----------T* | 42aa | |
| 0474 | ATGAAGAAAGCTCTGAT--------------------------GGCG----------------AGGTGA | −57 bp | MKKALMA----------------R* | 269aa | |
| 0475a | ATGAAGAAAGCTCTGATTCTT--------------------TCT------AGGTGA | −49 bp | MKKALILS----------------T* | 43aa | |
| 0475b | ATGAAGAAAGCTCTG-----GAGGGCTCTCATTCTGGCCGCCATGATGAGCCCCTGTGGAGGTG-------C------AGGTGA | −12 bp | MKKALEGSHSGRHDEPLWRC----R* | 284aa |
| Target Gene | Transplanted Zygotes | Pregnant Recipients/Recipients | Edited Lambs/Delivered Lambs | Editing Efficiency (%) | Editing Tools |
|---|---|---|---|---|---|
| MSTN [14] | 213 | 31/55 | 2/35 | 5.7 | Cas9 |
| MSTN, ASIP and BCO2 [15] | 578 | 77/82 | 35/36 | 97.2 | Cas9 |
| FGF5 [16] | 100 | 14/53 | 3/18 | 16.7 | Cas9 |
| ASIP [17] | 92 | 6/60 | 5/6 | 83.3 | Cas9 |
| BMPR1B [18] | 279 | 16/39 | 7/21 | 33.3 | Cas9 |
| SOCS2 [8] | 53 | 3/8 | 3/4 | 75 | CBE (Cytosine base editor) |
| FecB [19] | 95 | 6/18 | 6/8 | 75 | ABE (adenine base editing) |
| MSTN [20] | 345 | 14/58 | 8/16 | 50 | Cas9 |
| TBXT [11] | 338 | 31/216 | 19/28 | 67.9 | Cas9&ssODN |
| FecB [21] | 122 | 15/45 | 5/7 | 71.4 | PE (prime editing) |
| TBXT [21] | 140 | 3/8 | 37.5 | ||
| MSTN [22] | 70 | 5/13 | 9/10 | 90 | Cas9 |
| MSTN/FGF5 [23] | 1201 | 78/236 | 12/64 | 18.8 | Cas9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Yang, H.; Li, T.; Chen, Y.; Chen, J.; Zhang, X.; Zhang, J.; Zhang, Y.; Zhang, N.; Ma, R.; et al. Optimization of CRISPR/Cas9 Gene Editing System in Sheep (Ovis aries) Oocytes via Microinjection. Int. J. Mol. Sci. 2025, 26, 1065. https://doi.org/10.3390/ijms26031065
Wang H, Yang H, Li T, Chen Y, Chen J, Zhang X, Zhang J, Zhang Y, Zhang N, Ma R, et al. Optimization of CRISPR/Cas9 Gene Editing System in Sheep (Ovis aries) Oocytes via Microinjection. International Journal of Molecular Sciences. 2025; 26(3):1065. https://doi.org/10.3390/ijms26031065
Chicago/Turabian StyleWang, Haitao, Hengqian Yang, Tingting Li, Yan Chen, Jieran Chen, Xiaosheng Zhang, Jinlong Zhang, Yuting Zhang, Na Zhang, Runlin Ma, and et al. 2025. "Optimization of CRISPR/Cas9 Gene Editing System in Sheep (Ovis aries) Oocytes via Microinjection" International Journal of Molecular Sciences 26, no. 3: 1065. https://doi.org/10.3390/ijms26031065
APA StyleWang, H., Yang, H., Li, T., Chen, Y., Chen, J., Zhang, X., Zhang, J., Zhang, Y., Zhang, N., Ma, R., Huang, X., & Liu, Q. (2025). Optimization of CRISPR/Cas9 Gene Editing System in Sheep (Ovis aries) Oocytes via Microinjection. International Journal of Molecular Sciences, 26(3), 1065. https://doi.org/10.3390/ijms26031065







