Genome-Wide Investigation of MADS-Box Genes in Flower Development and Environmental Acclimation of Lumnitzera littorea (Jack) Voigt
Abstract
1. Introduction
2. Results
2.1. Identification of MADS-Box Genes in L. littorea
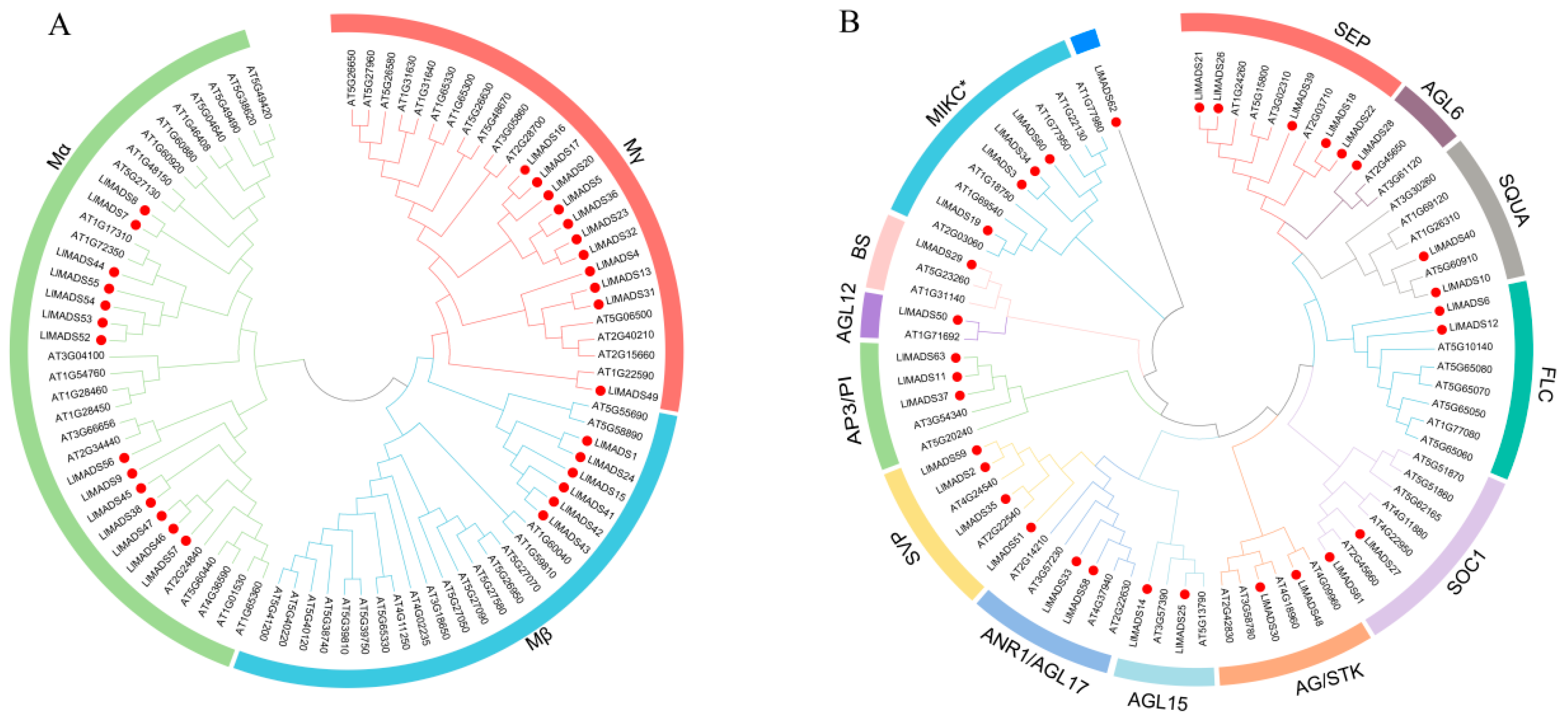
2.2. Classification and Phylogenetic Analysis
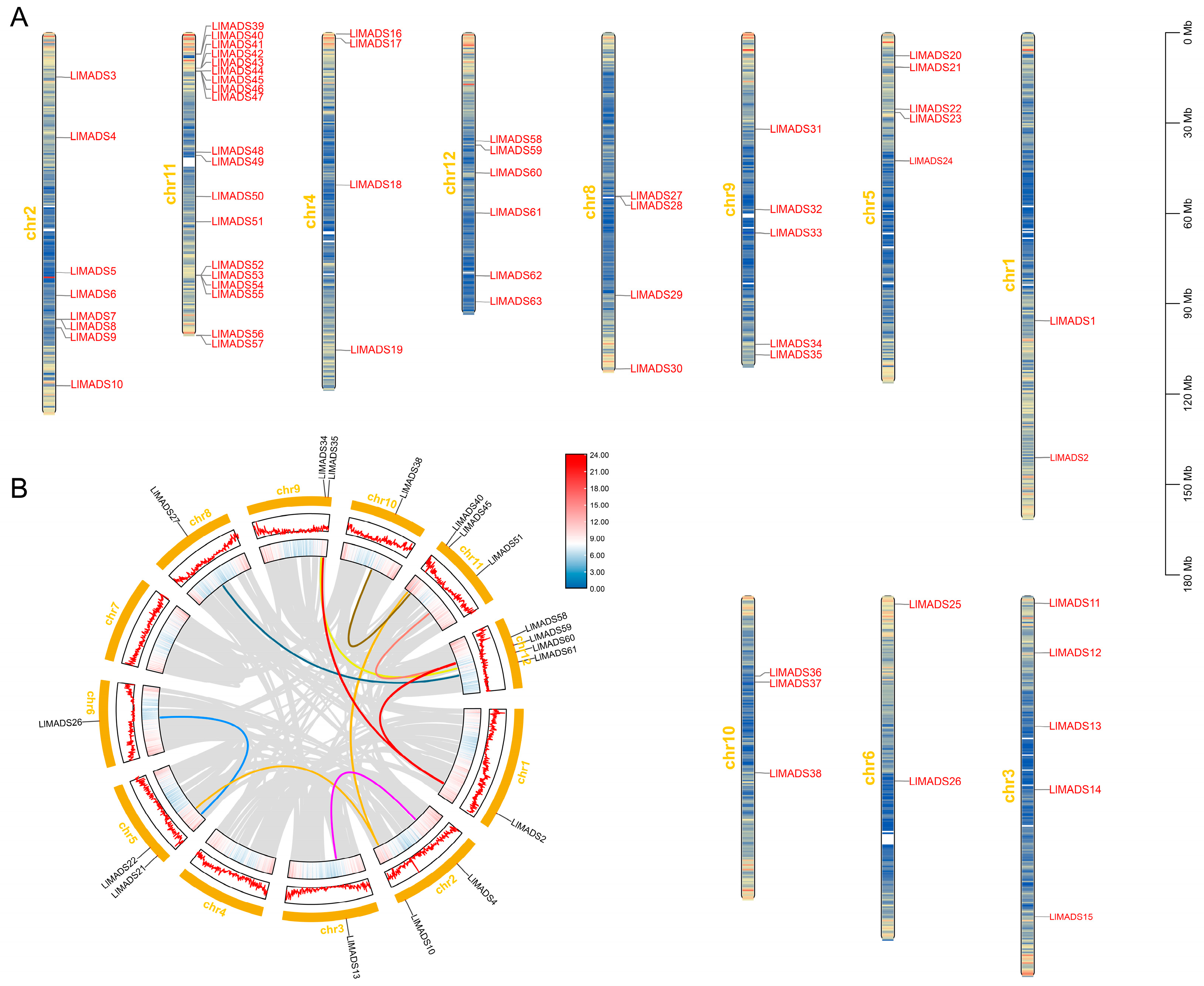
2.3. Chromosomal Locations and Collinearity Analysis
2.4. Conserved Motif and Gene Structure Analysis
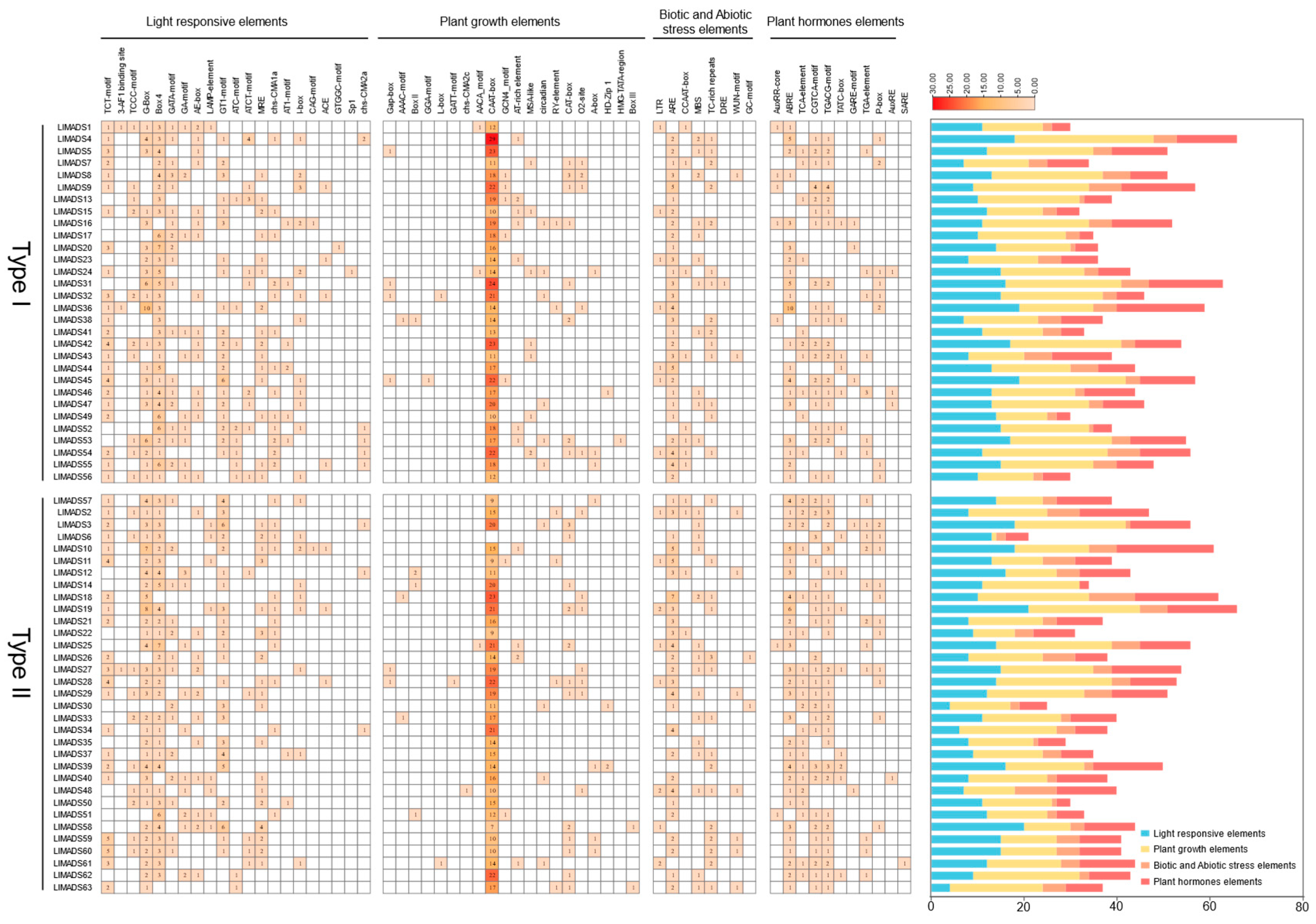
2.5. Cis-Regulatory Element Prediction
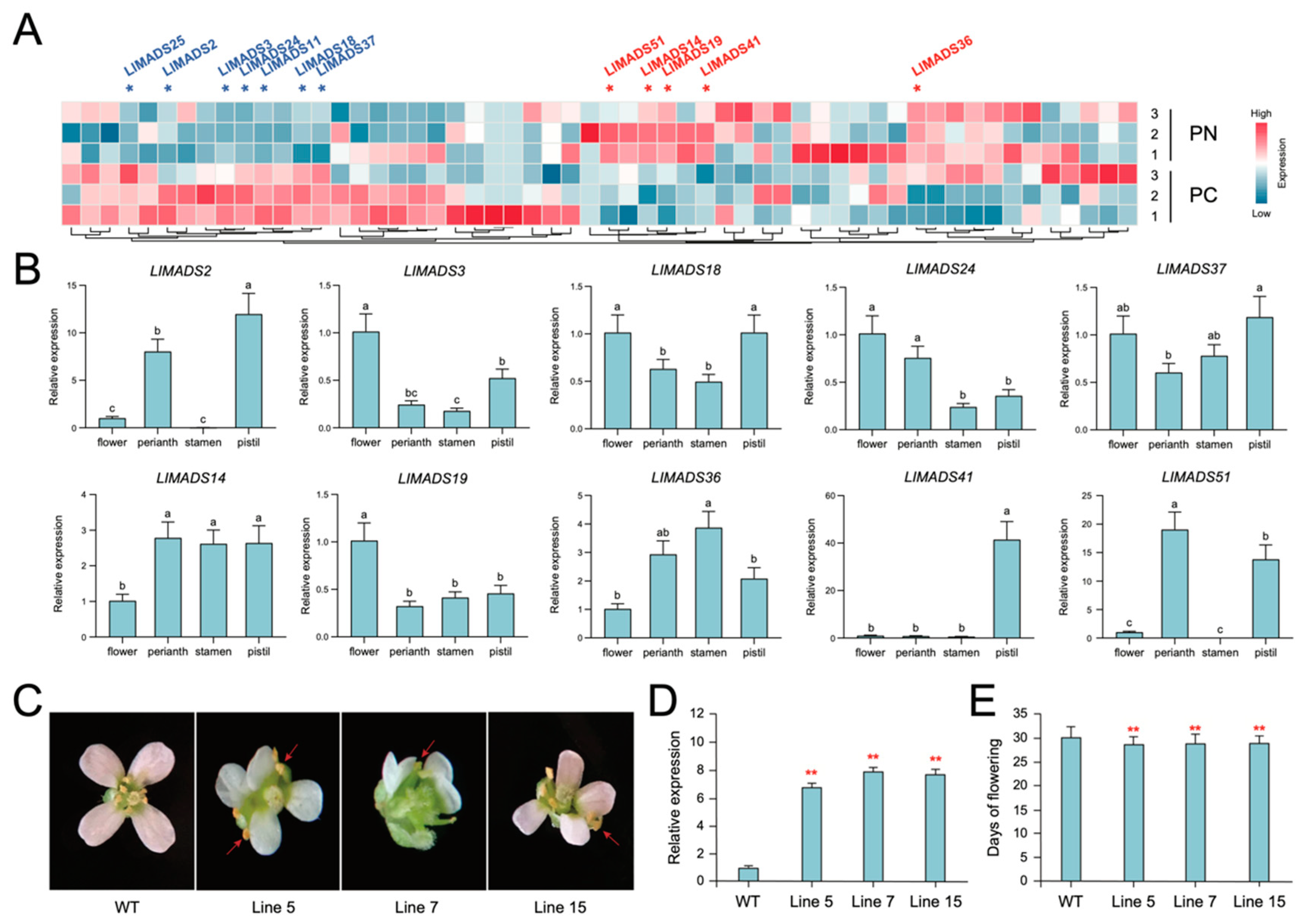
2.6. Expression Profiles of LlMADS Genes Involved in Flower Development
2.7. Expression Profiles of LlMADS Genes Under Cold and Salt Stress
3. Discussion
4. Materials and Methods
4.1. Identification of LlMADS Genes, Chromosomal Location, and Phylogenetic, Gene Structure, and Conserved Motif Analysis, as Well as Cis-Regulatory Element Prediction
4.2. Expression Characterization of LlMADS Genes in L. littorea Flowers and Functional Validation
4.3. Comparative Transcriptome Analysis of LlMADS Gene in Leaves in Response to Salt and Cold Stresses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, W.; Banerjee, A.K.; Wu, H.; Ng, W.L.; Feng, H.; Qiao, S.; Liu, Y.; Huang, Y. Contrasting Phylogeographic Patterns in Lumnitzera Mangroves Across the Indo-West Pacific. Front. Plant Sci. 2021, 12, 637009. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Chen, Y.; Wu, W.; Zhou, R.; Zhang, Y. The complete chloroplast genome sequence of an endangered mangrove tree Lumnitzera littorea (Combretaceae). Conserv. Genet. Resour. 2017, 10, 911–913. [Google Scholar] [CrossRef]
- He, Z.; Feng, X.; Chen, Q.; Li, L.; Li, S.; Han, K.; Guo, Z.; Wang, J.; Liu, M.; Shi, C.; et al. Evolution of coastal forests based on a full set of mangrove genomes. Nat. Ecol. Evol. 2022, 6, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Cecilia, M.; Almahasheer, H.; Duarte, C.M. Mangrove forests as traps for marine litter. Environ. Pollut. 2019, 247, 499–508. [Google Scholar]
- Sandilyan, S.; Kathiresan, K. Mangrove conservation: A global perspective. Biodivers. Conserv. 2012, 21, 3523–3542. [Google Scholar] [CrossRef]
- Giesen, W.; Wulffraat, S.; Zieren, M.; Scholten, L. Mangrove Guidebook for Southeast Asia; Rap Publication: Bangkok, Thailand, 2007. [Google Scholar]
- Nguyen, T.T.; Pham, T.T.; Hansen, P.E.; Nguyen, P.K. In vitro α-glucosidase inhibitory activity of compounds isolated from mangrove Lumnitzera littorea leaves. Sci. Technol. Dev. J. 2019, 22, 106–113. [Google Scholar] [CrossRef]
- Manurung, J.; Kappen, J.; Schnitzler, J.; Frolov, A.; Wessjohann, L.A.; Agusta, A.; Muellner-Riehl, A.N.; Franke, K. Analysis of Unusual Sulfated Constituents and Anti-infective Properties of Two Indonesian Mangroves, Lumnitzera littorea and Lumnitzera racemosa (Combretaceae). Separations 2021, 8, 82. [Google Scholar] [CrossRef]
- García-Niño, W.R.; Zazueta, C. Ellagic acid: Pharmacological activities and molecular mechanisms involved in liver protection. Pharmacol. Res. 2015, 97, 84–103. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zou, L.; Lei, X.; Su, J.; Yang, R.; Xie, W.; Li, W.; Chen, G. OSMAC strategy integrated with molecular networking discovery peniciacetals A−I, nine new meroterpenoids from the mangrove-derived fungus Penicillium sp. HLLG-122. Bioorganic Chem. 2023, 130, 106271. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, W.; Xu, Z.; Bai, Q.; Zhou, X.; Zheng, C.; Bai, M.; Chen, G. Biological Secondary Metabolites from the Lumnitzera littorea-Derived Fungus Penicillium oxalicum HLLG-13. Mar. Drugs 2023, 21, 22. [Google Scholar] [CrossRef]
- Vattem, D.A.; Shetty, K. Biological Functionality of Ellagic Acid: A Review. J. Food Biochem. 2005, 29, 234–266. [Google Scholar] [CrossRef]
- Zhong, C.; Li, S.; Guan, W.; Li, H.; Lin, X.; Liao, B. Current distributions of three endangered mangrove species in China. Ecol. Sci. 2011, 30, 431–435. [Google Scholar]
- Zhang, Y.; Zhong, C.; Li, S.; Yan, T.; Guan, W. Endangered species of mangrove plants: Lumnitzera littore. For. Resour. Manag. 2013, 5, 103–107. [Google Scholar]
- Zhang, Y.; Zhong, C.; Yang, Y.; Zhong, H.; Zeng, Z.; Zhang, J.; Zhang, S. Rescue of germplasm resources of endangered mangrove plant Lumnitzera littorea. Mol. Plant Breed. 2018, 16, 4112–4118. [Google Scholar]
- Aerts, N.; de Bruijn, S.; van Mourik, H.; Angenent, G.C.; van Dijk, A.D.J. Comparative analysis of binding patterns of MADS-domain proteins in Arabidopsis thaliana. BMC Plant Biol. 2018, 18, 131. [Google Scholar] [CrossRef]
- Heisler, M.G.; Jönsson, H.; Wenkel, S.; Kaufmann, K. Context-specific functions of transcription factors controlling plant development: From leaves to flowers. Curr. Opin. Plant Biol. 2022, 69, 102262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Zhou, Y.; Zhang, J.; Bai, H.; Zheng, C. Comparative Transcriptome Reveals the Genes’ Adaption to Herkogamy of Lumnitzera littorea (Jack) Voigt. Front. Genet. 2020, 11, 584817. [Google Scholar] [CrossRef]
- Patil, R.V.; Hadawale, K.N.; Ramli, A.N.M.; Wadkar, S.S.; Bhuyar, P. An Overview of Molecular Basis and Genetic Modification of Floral Organs Genes: Impact of Next-Generation Sequencing. Mol. Biotechnol. 2022, 65, 833–848. [Google Scholar] [CrossRef]
- Gramzow, L.; Ritz, M.S.; Theißen, G. On the origin of MADS-domain transcription factors. Trends Genet. 2010, 26, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Bey, M.; Stüber, K.; Fellenberg, K.; Schwarz-Sommer, Z.; Sommer, H.; Saedler, H.; Zachgo, S. Characterization of Antirrhinum Petal Development and Identification of Target Genes of the Class B MADS Box GeneDEFICIENS [W]. Plant Cell 2004, 16, 3197–3215. [Google Scholar] [CrossRef] [PubMed]
- Treisman, R.; Ammerer, G. The SRF and MCM1 transcription factors. Curr. Opin. Genet. Dev. 1992, 2, 221. [Google Scholar] [CrossRef] [PubMed]
- Shore, P.; Sharrocks, A.D. The MADS-box family of transcription factors. Eur. J. Biochem. 1995, 229, 1. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.G.H.; Muiño, J.M.; Blanvillain, R.; Busscher, M.; Busscher-Lange, J.; Dinh, Q.D.; Liu, S.; Westphal, A.H.; Boeren, S.; et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 2012, 109, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Vega-Léon, R.; Hugouvieux, V.; Blanc-Mathieu, R.; van der Wal, F.; Lucas, J.; Silva, C.S.; Jourdain, A.; Muino, J.M.; Nanao, M.H.; et al. The intervening domain is required for DNA-binding and functional identity of plant MADS transcription factors. Nat. Commun. 2021, 12, 4760. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.; Yanofsky, M.F. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001, 2, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Pařenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and Phylogenetic Analyses of the Complete MADS-Box Transcription Factor Family in Arabidopsis. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Theißen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenetics Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.H.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef]
- Jetha, K.; Theißen, G.; Melzer, R. Arabidopsis SEPALLATA proteins differ in cooperative DNA-binding during the formation of floral quartet-like complexes. Nucleic Acids Res. 2014, 42, 10927–10942. [Google Scholar] [CrossRef]
- Murai, K. Homeotic Genes and the ABCDE Model for Floral Organ Formation in Wheat. Plants 2013, 2, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Irish, V. The ABC model of floral development. Curr. Biol. 2017, 27, R887–R890. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Moyroud, E. Reflections on the ABC model of flower development. Plant Cell 2024, 36, 1334–1357. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, S.H.; Yasui, Y.; Ohmori, S.; Tanaka, W.; Hirano, H.Y. Rice Flower Development Revisited: Regulation of Carpel Specification and Flower Meristem Determinacy. Plant Cell Physiol. 2019, 60, 1284–1295. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Yadav, R.K. C-terminal domain of APETALA1 is essential for its functional divergence from CAULIFLOWER in Arabidopsis. J. Plant Biochem. Biotechnol. 2020, 29, 824–831. [Google Scholar] [CrossRef]
- Irish, V.F.; Sussex, L.M. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 1990, 2, 741–753. [Google Scholar]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Castelán-Muñoz, N.; Herrera, J.; Cajero-Sánchez, W.; Arrizubieta, M.; Trejo, C.; García-Ponce, B.; Sánchez, M.d.l.P.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. MADS-Box Genes Are Key Components of Genetic Regulatory Networks Involved in Abiotic Stress and Plastic Developmental Responses in Plants. Front. Plant Sci. 2019, 10, 853. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhou, Z.; Zhu, L.; Gu, Y.; Guo, B.; Lv, C.; Zhu, J.; Xu, R. Genome-wide analysis of the MADS-box gene family involved in salt and waterlogging tolerance in barley (Hordeum vulgare L.). Front. Plant Sci. 2023, 14, 1178065. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 242. [Google Scholar] [CrossRef] [PubMed]
- Duke, N.C. Mangrove floristics and biogeography revisited:Further deductions from biodiversity hot spots, ancestral discontinuities, and common evolutionary processes. In Mangrove Ecosystems: A Global Biogeographic Perspective; Springer International Publishing: Cham, Switzerland, 2017; pp. 17–53. [Google Scholar]
- Chen, W.; Tong, Y.Y.; Feng, Y.; Hao, L.L.; Zhang, H.Y.; Yue, D.F.; Zhang, Y.; Zheng, C.F. Effects of salt stress in chloroplast ultrastructure and photosynthetic fluorescence characteristics of Lumnizerea littorea (Jack) Voigt seedlings. Chin. J. Ecol. 2024, 43, 716–723. [Google Scholar]
- Xia, X.; Hao, L.; Sun, Y.; Lv, Y.; Wang, Y.; Wu, H.; Jiang, Z.; Li, X.; Yan, Y.; Chen, X.; et al. Unravelling chilling-stress resistance mechanisms in endangered Mangrove plant Lumnitzera littorea (Jack) Voigt. Mar. Environ. Res. 2023, 192, 106210. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Cho, L.H.; Jung, K.H. Hierarchical Structures and Dissected Functions of MADS-Box Transcription Factors in Rice Development. J. Plant Biol. 2022, 65, 99–109. [Google Scholar] [CrossRef]
- Raza, Q.; Riaz, A.; Atif, R.M.; Hussain, B.; Rana, I.A.; Ali, Z.; Budak, H.; Alaraidh, I.A. Genome-Wide Diversity of MADS-Box Genes in Bread Wheat is Associated with its Rapid Global Adaptability. Front. Genet. 2022, 12, 818880. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Z.; Li, S.; Hou, M.; Zhou, Y.; Zhao, Y.; Li, G.; Zhao, H.; Ma, H. Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A. BMC Genom. 2018, 19, 726. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Sun, W.; Tsai, W.; Xiang, S.; Lai, X.; Chen, D.; Liu, X.; Wang, Y.; Le, Y.; Chen, S.; et al. Chromosome-scale assembly of the Kandelia obovata genome. Hortic. Res. 2020, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-Wide Analysis of the MADS-Box Transcription Factor Family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef] [PubMed]
- Leseberg, C.H.; Li, A.Z.; Kang, H.; Duvall, M.; Mao, L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 2006, 378, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Wang, J.; Yu, J.; Huang, X.; Gu, X. Evolution of alternative splicing after gene duplication. Genome Res. 2006, 16, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Shao, S.; Feng, X.; Li, S.; Zhang, L.; Wu, W.; Liu, M.; Tracy, M.; Zhong, C.; Guo, Z.; et al. Adaptation in Unstable Environments and Global Gene Losses: Small but Stable Gene Networks by the May–Wigner Theory. Mol. Biol. Evol. 2024, 41, msae059. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; de Pouplana, L.R.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Wuest, S.E.; O’Maoileidigh, D.S.; Rae, L.; Kwasniewska, K.; Raganelli, A.; Hanczaryk, K.; Lohan, A.J.; Loftus, B.; Graciet, E.; Wellmer, F. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 2012, 109, 13452–13457. [Google Scholar] [CrossRef]
- Hernández-Hernández, T.; Martínez-Castilla, L.P.; Alvarez-Buylla, E.R. Functional Diversification of B MADS-Box Homeotic Regulators of Flower Development: Adaptive Evolution in Protein–Protein Interaction Domains after Major Gene Duplication Events. Mol. Biol. Evol. 2007, 24, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Yoo, M.J.; Albert, V.A.; Farris, J.S.; Soltis, P.S.; Soltis, D.E. Phylogeny and diversification of B-function MADS-box genes in angiosperms: Evolutionary and functional implications of a 260-million-year-old duplication. Am. J. Bot. 2004, 91, 2102–2118. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Lu, L.; Guan, J.; Yan, M.; Liu, Z.; Wan, Y.; Zhou, G. Gene co-expression network analysis in areca floral organ and the potential role of the AcMADS17 and AcMADS23 in transgenic Arabidopsis. Plant Sci. 2024, 342, 112049. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; He, H.; Li, Y.; Wang, L.; Liu, Y.; Luan, X.; Wang, J.; Liu, H.; Liu, S.; Zhang, J.; et al. MADS-Box Subfamily Gene GmAP3 from Glycine max Regulates Early Flowering and Flower Development. Int. J. Mol. Sci. 2023, 24, 2751. [Google Scholar] [CrossRef]
- Marand, A.P.; Eveland, A.L.; Kaufmann, K.; Springer, N.M. cis-Regulatory Elements in Plant Development, Adaptation, and Evolution. Annu. Rev. Plant Biol. 2023, 74, 111–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ruan, J.; Ho, T.D.; You, Y.; Yu, T.; Quatrano, R.S. Cis-regulatory element based targeted gene finding: Genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics 2005, 21, 3074–3081. [Google Scholar] [CrossRef] [PubMed]
- Heidari, P.; Ahmadizadeh, M.; Najafi-Zarrini, H. In silico analysis of Cis-regulatory elements on co-expressed genes. J. Biol. Environ. Sci. 2015, 9, 1–9. [Google Scholar]
- Kavi Kishor, P.B.; Tiozon, R.N.; Fernie, A.R.; Sreenivasulu, N. Abscisic acid and its role in the modulation of plant growth, development, and yield stability. Trends Plant Sci. 2022, 27, 1283–1295. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Zhang, D. Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 2015, 27, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Negin, B.; Yaaran, A.; Kelly, G.; Zait, Y.; Moshelion, M. Mesophyll Abscisic Acid Restrains Early Growth and Flowering But Does Not Directly Suppress Photosynthesis. Plant Physiol. 2019, 180, 910–925. [Google Scholar] [CrossRef] [PubMed]
- Castro-Camba, R.; Sánchez, C.; Vidal, N.; Vielba, J. Interactions of Gibberellins with Phytohormones and Their Role in Stress Responses. Horticulturae 2022, 8, 241. [Google Scholar] [CrossRef]
- He, H.; Yang, M.; Li, S.; Zhang, G.; Ding, Z.; Zhang, L.; Shi, G.; Li, Y. Mechanisms and biotechnological applications of transcription factors. Synth. Syst. Biotechnol. 2023, 8, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cui, S.; Wu, F.; Yan, S.; Lin, X.; Du, X.; Chong, K.; Schilling, S.; Theißen, G.; Meng, Z. Functional Conservation of MIKC*-Type MADS Box Genes in Arabidopsis and Rice Pollen Maturation. Plant Cell 2013, 25, 1288–1303. [Google Scholar] [CrossRef]
- Qiu, Y.; Köhler, C. Endosperm Evolution by Duplicated and Neofunctionalized Type I MADS-Box Transcription Factors. Mol. Biol. Evol. 2022, 39, msab355. [Google Scholar] [CrossRef] [PubMed]
- Masiero, S.; Colombo, L.; Grini, P.E.; Schnittger, A.; Kater, M.M. The Emerging Importance of Type I MADS Box Transcription Factors for Plant Reproduction. Plant Cell 2011, 23, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Guo, Z.; Wang, J.; He, Z.; Li, Y.; Feng, X.; Zhong, C.; Shi, S. Evolution of woody plants to the land-sea interface-The atypical genomic features of mangroves with atypical phenotypic adaptation. Mol. Ecol. 2022, 32, 1351–1365. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Ren, L.; Zhu, B.Q.; Zhang, Y.B.; Wang, H.Y.; Ba, C.F. The research of applying primer premier 5.0 to design pcr primer. J. Jinzhou Med. Coll. 2004, 1, 270. [Google Scholar]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Wang, L.; Han, Y.; He, Q. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Yang, Y.; Zhang, Y.; Yang, F. Genome-Wide Investigation of MADS-Box Genes in Flower Development and Environmental Acclimation of Lumnitzera littorea (Jack) Voigt. Int. J. Mol. Sci. 2025, 26, 1680. https://doi.org/10.3390/ijms26041680
Zhang L, Yang Y, Zhang Y, Yang F. Genome-Wide Investigation of MADS-Box Genes in Flower Development and Environmental Acclimation of Lumnitzera littorea (Jack) Voigt. International Journal of Molecular Sciences. 2025; 26(4):1680. https://doi.org/10.3390/ijms26041680
Chicago/Turabian StyleZhang, Linbi, Yuchen Yang, Ying Zhang, and Fusun Yang. 2025. "Genome-Wide Investigation of MADS-Box Genes in Flower Development and Environmental Acclimation of Lumnitzera littorea (Jack) Voigt" International Journal of Molecular Sciences 26, no. 4: 1680. https://doi.org/10.3390/ijms26041680
APA StyleZhang, L., Yang, Y., Zhang, Y., & Yang, F. (2025). Genome-Wide Investigation of MADS-Box Genes in Flower Development and Environmental Acclimation of Lumnitzera littorea (Jack) Voigt. International Journal of Molecular Sciences, 26(4), 1680. https://doi.org/10.3390/ijms26041680






