Graph and Multi-Level Sequence Fusion Learning for Predicting the Molecular Activity of BACE-1 Inhibitors
Abstract
1. Introduction
2. Results and Discussion
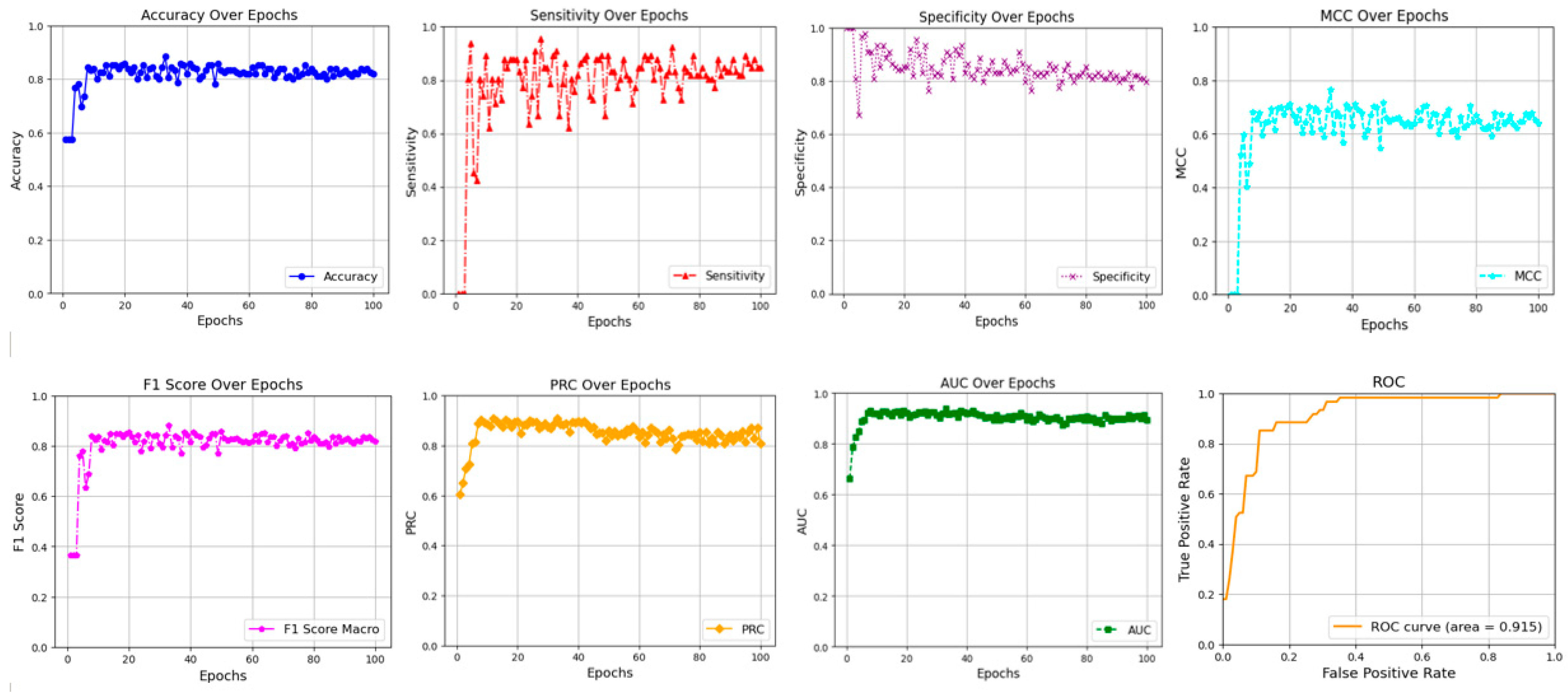
2.1. Hyperparameter Tuning
2.2. Comparison Results and Discussion
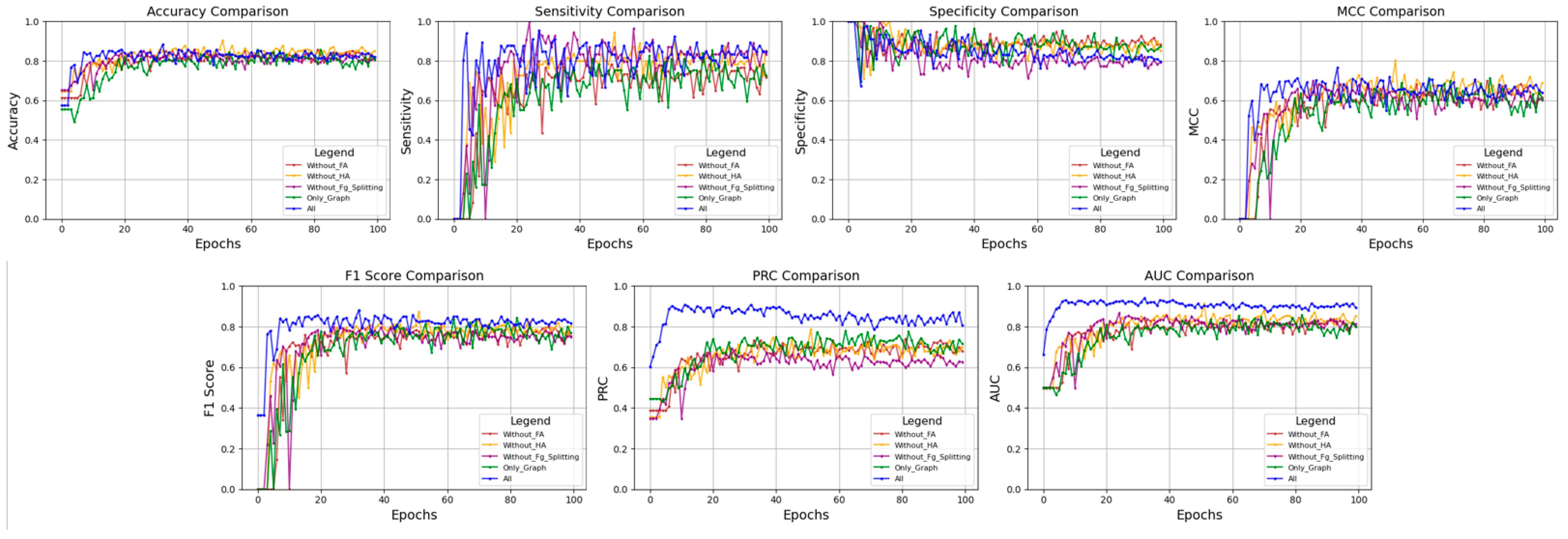
2.3. Ablation and Discussion
3. Materials and Methods
3.1. Dataset
3.2. Methods
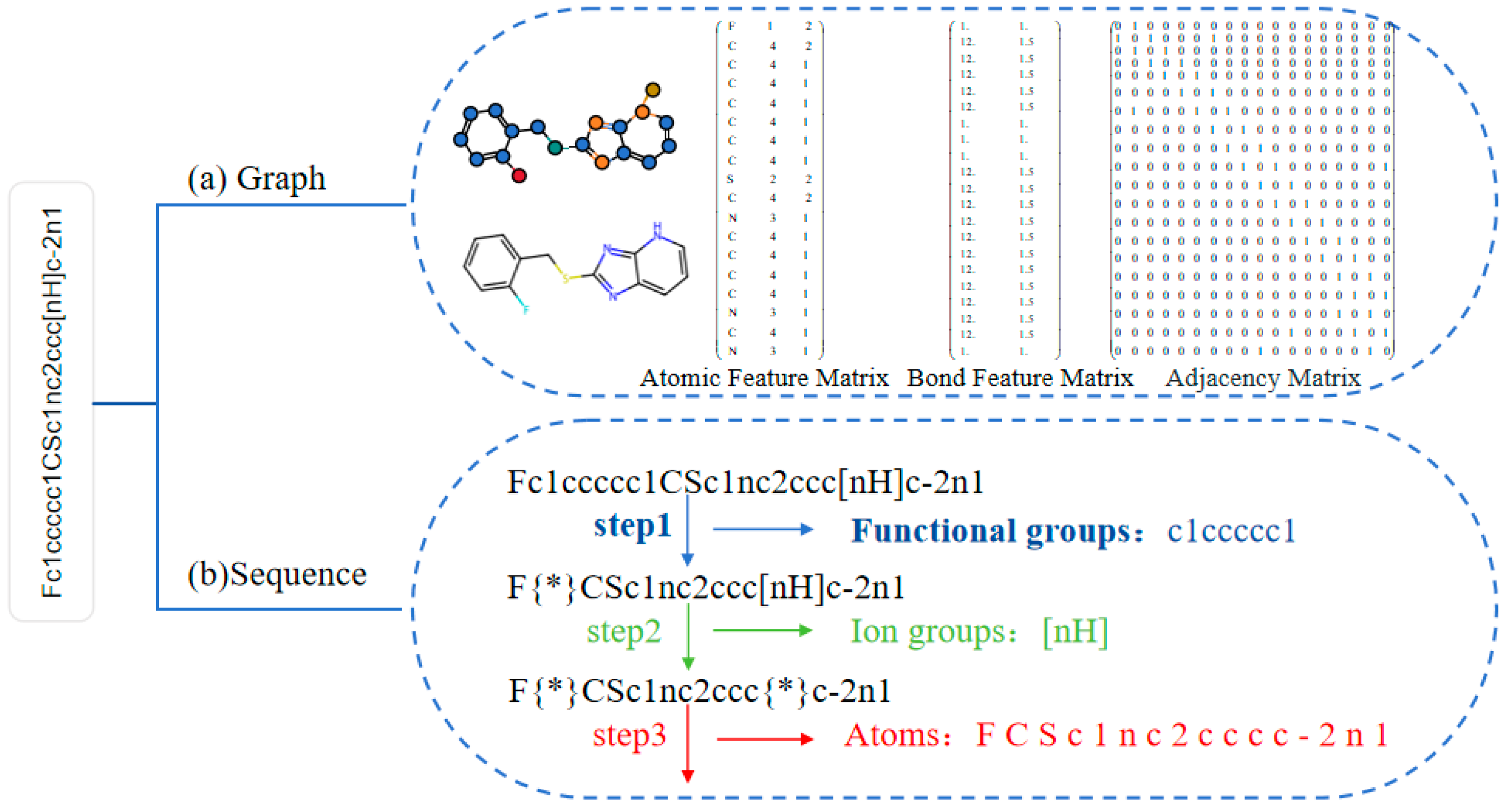
3.2.1. Data Pre-Processing
- (1).
- Molecular Graph Construction
- (2).
- Multi-level substring sequences extraction
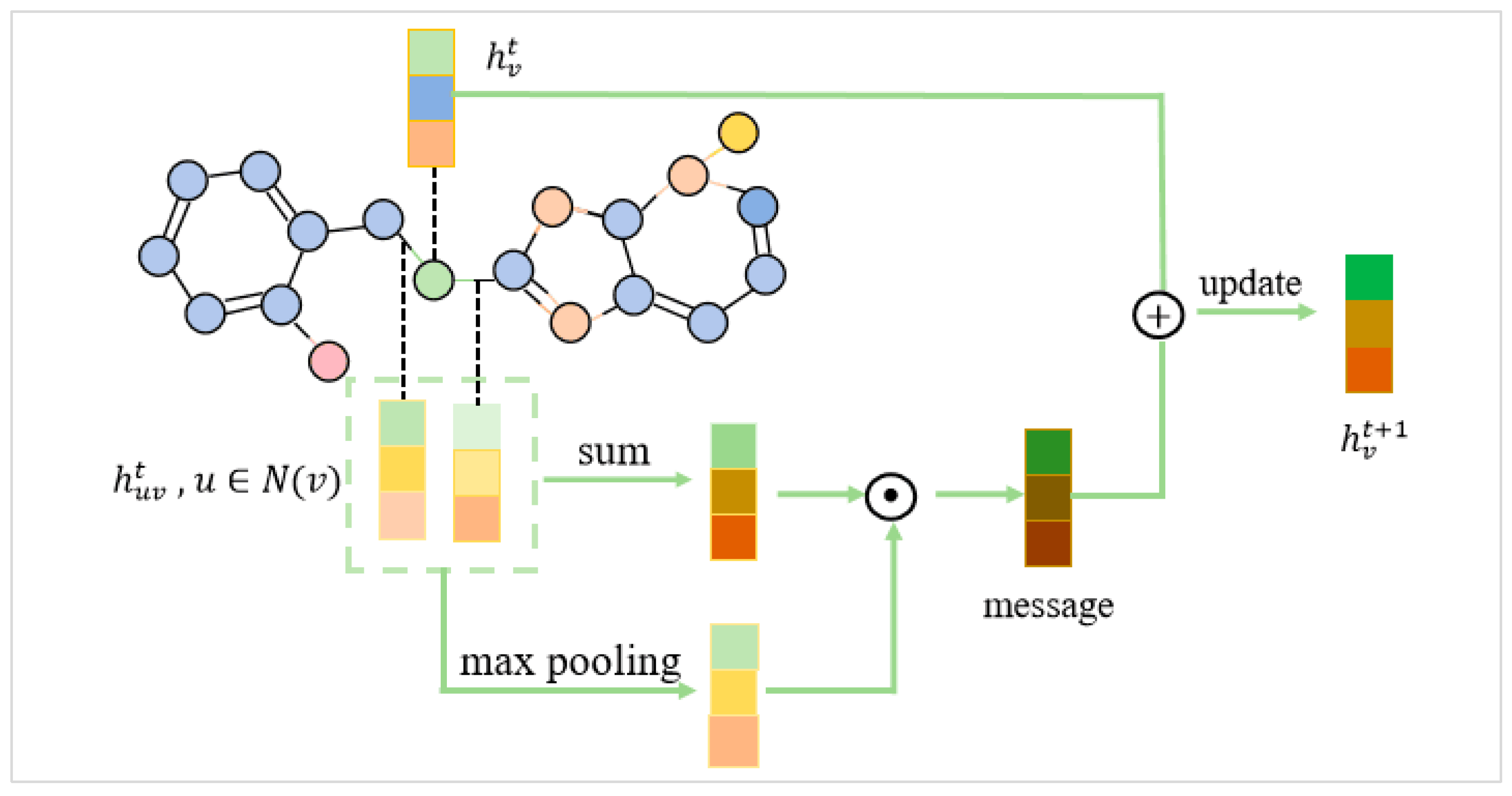
3.2.2. Graph Encoding Module
- (1).
- Message Passing with an Atomic-Level Feature Attention Encoder
- (1)
- Aggregation:where was the message generated by the node.
- (2)
- Communication:where was the hidden representation of the node.
- (3)
- Edge representation update:where represented the hidden representation of the node, and represented the hidden representation of the edge. Here, was a learnable weight matrix, and the activation function was denoted as . After iterations, the following operations were performed to obtain the final message and node representation.
- (4)
- Final message aggregation:
- (5)
- Final node representation update:
- (2).
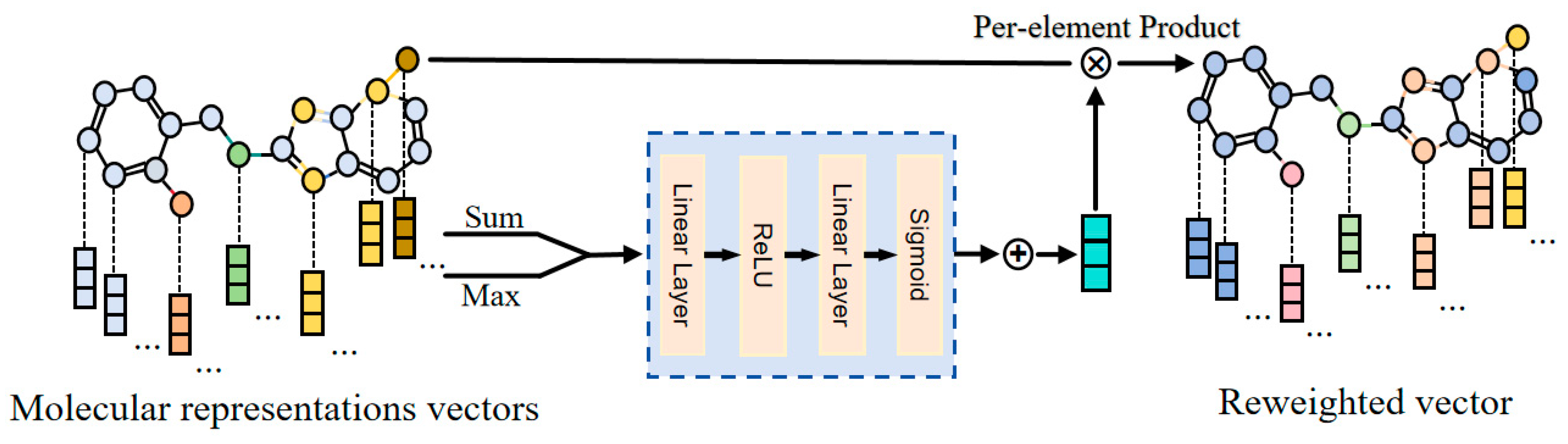
- Feature Attention Module
3.2.3. Multi-Level Substring Sequence Encoding Module
- (1).
- SMILES Embedding
- (2).
- Hierarchical Attention
- (3).
- BiLSTM and Transformer Coding Module
3.2.4. Weight Fusion Learning Module
3.2.5. Loss Function
3.3. Metrics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levey, A.I. Progress with Treatments for Alzheimer’s Disease. N. Engl. J. Med. 2021, 384, 1762–1763. [Google Scholar] [CrossRef]
- Behl, T.; Kaur, I.; Fratila, O.; Brata, R.; Bungau, S. Exploring the Potential of Therapeutic Agents Targeted towards Mitigating the Events Associated with Amyloid-β Cascade in Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 7443. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; He, S.; Chen, Y.; Feng, F.; Qu, W.; Sun, H. Donepezil-based Multi-functional Cholinesterase Inhibitors for Treatment of Alzheimer’s Disease. Eur. J. Med. Chem. 2018, 158, 463–477. [Google Scholar] [CrossRef]
- Sun, X.; Chen, W.D.; Wang, Y.D. β-Amyloid: The Key Peptide in the Pathogenesis of Alzheimer’s Disease. Front. Pharmacol. 2015, 6, 221. [Google Scholar] [CrossRef]
- Cervellati, C.; Valacchi, G.; Zuliani, G. BACE1 Role in Alzheimer’s Disease and Other Dementias: From Theory to Practice. Neural Regen. Res. 2021, 16, 2407–2408. [Google Scholar] [CrossRef]
- Ponzoni, I.; Sebastián-Pérez, V.; Martínez, M.J.; Roca, C.; De la Cruz Pérez, C.; Cravero, F.; Vazquez, G.E.; Páez, J.A.; Díaz, M.F.; Campillo, N.E. QSAR Classification Models for Predicting the Activity of Inhibitors of Beta-Secretase (BACE1) Associated with Alzheimer’s Disease. Sci. Rep. 2019, 9, 9102. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Baecker, D.; Mai Dung, D.T.; Phuong Nhung, N.; Thi Thuan, N.; Nguyen, P.L.; Phuong Dung, P.T.; Huong, T.T.L.; Rasulev, B.; Casanola-Martin, G.M.; et al. Development of Activity Rules and Chemical Fragment Design for In Silico Discovery of AChE and BACE1 Dual Inhibitors against Alzheimer’s Disease. Molecules 2023, 28, 3588. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Liu, Y.; Shi, B. Comprehensive 3D-QSAR and Binding Mode of BACE-1 Inhibitors Using R-group Search and Molecular Docking. J. Mol. Graph. Modell. 2013, 45, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Aswathy, L.; Jisha, R.S.; Masand, V.H.; Gajbhiye, J.M.; Shibi, I.G. Design of Novel Amyloid β Aggregation Inhibitors Using QSAR, Pharmacophore Modeling, Molecular Docking and ADME Prediction. In Silico Pharmacol. 2018, 6, 12. [Google Scholar] [CrossRef]
- Singh, R.; Ganeshpurkar, A.; Ghosh, P.; Pokle, A.V.; Kumar, D.; Singh, R.B.; Singh, S.K.; Kumar, A. Classification of Beta-site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitors by Using Machine Learning Methods. Chem. Biol. Drug Des. 2021, 98, 1079–1097. [Google Scholar] [CrossRef]
- Rong, Y.; Bian, Y.; Xu, T.; Xie, W.; Wei, Y.; Huang, W.; Huang, J. Self-supervised Graph Transformer on Large-scale Molecular Data. Adv. Neural Inf. Process. Syst. 2020, 33, 12559–12571. [Google Scholar]
- Deng, D.; Chen, X.; Zhang, R.; Lei, Z.; Wang, X.; Zhou, F. XGraphBoost: Extracting Graph Neural Network-Based Features for a Better Prediction of Molecular Properties. J. Chem. Inf. Model. 2021, 61, 2697–2705. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Guan, J.; Zhou, S. FraGAT: A Fragment-Oriented Multi-Scale Graph Attention Model for Molecular Property Prediction. Bioinformatics 2021, 37, 2981–2987. [Google Scholar] [CrossRef]
- Choo, H.Y.; Wee, J.; Shen, C.; Xia, K. Fingerprint-Enhanced Graph Attention Network (FinGAT) Model for Antibiotic Discovery. J. Chem. Inf. Model. 2023, 63, 2928–2935. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Fu, H.; Wu, Y.; Zhao, G.; Song, Z.; Huang, F.; Zhang, Z.; Liu, S.; Zhang, W. HimGNN: A Novel Hierarchical Molecular Graph Representation Learning Framework for Property Prediction. Brief. Bioinform. 2023, 24, bbad305. [Google Scholar] [CrossRef] [PubMed]
- Bongini, P.; Pancino, N.; Bendjeddou, A.; Scarselli, F.; Maggini, M.; Bianchini, M. Composite Graph Neural Networks for Molecular Property Prediction. Int. J. Mol. Sci. 2024, 25, 6583. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, S.; Zhu, F.; Huang, J. Seq2seq Fingerprint: An Unsupervised Deep Molecular Embedding for Drug Discovery. In Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics (BCB), Seattle, WA, USA, 2–5 October 2017; pp. 285–294. [Google Scholar]
- Honda, S.; Shi, S.; Ueda, H.R. SMILES Transformer: Pre-trained Molecular Fingerprint for Low Data Drug Discovery. arXiv 2019, arXiv:1911.04738. [Google Scholar]
- Wang, S.; Guo, Y.; Wang, Y.; Sun, H.; Huang, J. SMILES-BERT: Large Scale Unsupervised Pre-training for Molecular Property Prediction. In Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics (BCB), Niagara Falls, NY, USA, 22–25 September 2019; pp. 429–436. [Google Scholar]
- Kim, H.; Lee, J.; Ahn, S.; Lee, J.R. A Merged Molecular Representation Learning for Molecular Properties Prediction with a Web-Based Service. Sci. Rep. 2021, 11, 11028. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.; Ekenna, C. Molecular Descriptors Property Prediction Using Transformer-Based Approach. Int. J. Mol. Sci. 2023, 24, 11948. [Google Scholar] [CrossRef]
- Zheng, X.; Tomiura, Y. A BERT-Based Pretraining Model for Extracting Molecular Structural Information from a SMILES Sequence. J. Cheminform. 2024, 16, 71. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Ba, L.J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference for Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015; pp. 1–13. [Google Scholar]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- Kunnakkattu, I.R.; Choudhary, P.; Pravda, L.; Nadzirin, N.; Smart, O.S.; Yuan, Q.; Anyango, S.; Nair, S.; Varadi, M.; Velankar, S. PDBe CCDUtils: An RDKit-based Toolkit for Handling and Analysing Small Molecules in the Protein Data Bank. J. Cheminform. 2023, 15, 117. [Google Scholar] [CrossRef] [PubMed]
- Fey, M.; Lenssen, J.E. In Fast Graph Representation Learning with PyTorch Geometric. In Proceedings of the 6th International Conference on Learning Representations, RLGM Workshop, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Ren, T.; Zhang, H.; Shi, Y.; Luo, X.; Zhou, S. SuHAN: Substructural hierarchical attention network for molecular representation. J. Mol. Graph. Model. 2023, 119, 108401. [Google Scholar] [CrossRef]
- Li, B.; Lin, M.; Chen, T.; Wang, L. FG-BERT: A Generalized and Self-Supervised Functional Group-based Molecular Representation Learning Framework for Properties Prediction. Brief. Bioinform. 2023, 24, bbad398. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zheng, S.; Niu, Z.; Fu, Z.H.; Lu, Y.; Yang, Y. Communicative Representation Learning on Attributed Molecular Graphs. In Proceedings of the 29th International Joint Conference on Artificial Intelligence, Yokohama, Japan, 11–17 July 2020; pp. 2831–2838. [Google Scholar]
- Chen, J.; Zheng, S.; Song, Y.; Rao, J.; Yang, Y. Learning Attributed Graph Representations with Communicative Message Passing Transformer. arXiv 2021, arXiv:2107.08773. [Google Scholar]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-Excitation Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 2011–2023. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is All You Need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NeurIPS), Long Beach, CA, USA, 4–9 December 2017; pp. 6000–6010. [Google Scholar]


| Methods | Accuracy ↑ | SE ↑ | SP ↑ | MCC ↑ | F1-Score ↑ | PRC ↑ | AUC ↑ | |
|---|---|---|---|---|---|---|---|---|
| Training | Test | |||||||
| SVM | 0.993 | 0.810 | 0.694 | 0.887 | 0.598 | 0.745 | 0.680 | 0.790 |
| KNN | 0.994 | 0.813 | 0.758 | 0.849 | 0.609 | 0.764 | 0.681 | 0.804 |
| XGBoost | 0.961 | 0.816 | 0.734 | 0.871 | 0.613 | 0.762 | 0.687 | 0.802 |
| CNN | 0.797 | 0.797 | 0.815 | 0.785 | 0.590 | 0.762 | 0.658 | 0.799 |
| RF [7] | 0.850 | 0.830 | 0.850 | null | 0.660 | 0.800 | null | 0.880 |
| XGraphBoost [11] | 0.839 | 0.834 | 0.814 | 0.855 | 0.667 | 0.813 | 0.857 | 0.891 |
| FraGAT [13] | 0.895 | 0.857 | 0.825 | 0.876 | 0.696 | 0.810 | 0.833 | 0.911 |
| Ours | 0.941 | 0.877 | 0.852 | 0.894 | 0.744 | 0.872 | 0.869 | 0.915 |
| Metrics | Accuracy ↑ | SE ↑ | SP ↑ | MCC ↑ | F1-Score ↑ | PRC ↑ | AUC ↑ |
|---|---|---|---|---|---|---|---|
| Without FA: Graph + Sequence + HA + Fg_Spliting | 0.858 | 0.783 | 0.905 | 0.698 | 0.810 | 0.741 | 0.899 |
| Without HA: Graph + Sequence + FA + Fg_Spliting | 0.871 | 0.800 | 0.910 | 0.716 | 0.814 | 0.735 | 0.895 |
| Without Fg_Spliting: Graph + Sequence + HA + FA | 0.858 | 0.778 | 0.901 | 0.685 | 0.792 | 0.706 | 0.881 |
| Only Graph model: Graph + FA | 0.858 | 0.826 | 0.884 | 0.712 | 0.838 | 0.780 | 0.909 |
| All | 0.877 | 0.852 | 0.894 | 0.744 | 0.872 | 0.869 | 0.915 |
| Functional Groups | Ion Groups | Atoms |
|---|---|---|
| C(=O)N, OCC, [N+](=O)[O−], clccccc1 S(=O), O=c, NC(=O), NS(=O)(=O), OC, S(=O)(=O), C=O, O=C, [N+](=O)[O−] | [C@H], [C@@H], [C@@], [nH], [C@], [S@+], [S+], [N+], [S@@+], [N+](=O)[O–], [O] | C, c, O, N, S, F, Cl, Br, I, =, –, #, /, \, 1, 2, 3, 4, 5, 6 |
| SMILES | Substructures | ||
|---|---|---|---|
| Functional Groups | Ion Groups | Atoms | |
| CC(C)CNC(=O)[C@H](C)[C@H](O)[C@H](CC(C)C)NC(=O)[C@H]C(Cc1ccccc1)NC(=O)c1ccccc1 | NC(=O), NC(=O) Cc1ccccc1, NC(=O), c1ccccc1 | [C@H], [C@H] [C@H], [C@H] | C, C, (, C, ), C, (, C, ), (, O, ), (, C, C, (, C, ), C, ), C, (, C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S.; Zhang, C.; Chen, Y.; Chen, M. Graph and Multi-Level Sequence Fusion Learning for Predicting the Molecular Activity of BACE-1 Inhibitors. Int. J. Mol. Sci. 2025, 26, 1681. https://doi.org/10.3390/ijms26041681
Zheng S, Zhang C, Chen Y, Chen M. Graph and Multi-Level Sequence Fusion Learning for Predicting the Molecular Activity of BACE-1 Inhibitors. International Journal of Molecular Sciences. 2025; 26(4):1681. https://doi.org/10.3390/ijms26041681
Chicago/Turabian StyleZheng, Shaohua, Changwang Zhang, Youjia Chen, and Meimei Chen. 2025. "Graph and Multi-Level Sequence Fusion Learning for Predicting the Molecular Activity of BACE-1 Inhibitors" International Journal of Molecular Sciences 26, no. 4: 1681. https://doi.org/10.3390/ijms26041681
APA StyleZheng, S., Zhang, C., Chen, Y., & Chen, M. (2025). Graph and Multi-Level Sequence Fusion Learning for Predicting the Molecular Activity of BACE-1 Inhibitors. International Journal of Molecular Sciences, 26(4), 1681. https://doi.org/10.3390/ijms26041681






