Deciphering Colorectal Cancer–Hepatocyte Interactions: A Multiomics Platform for Interrogation of Metabolic Crosstalk in the Liver–Tumor Microenvironment
Abstract
:1. Introduction
2. Results
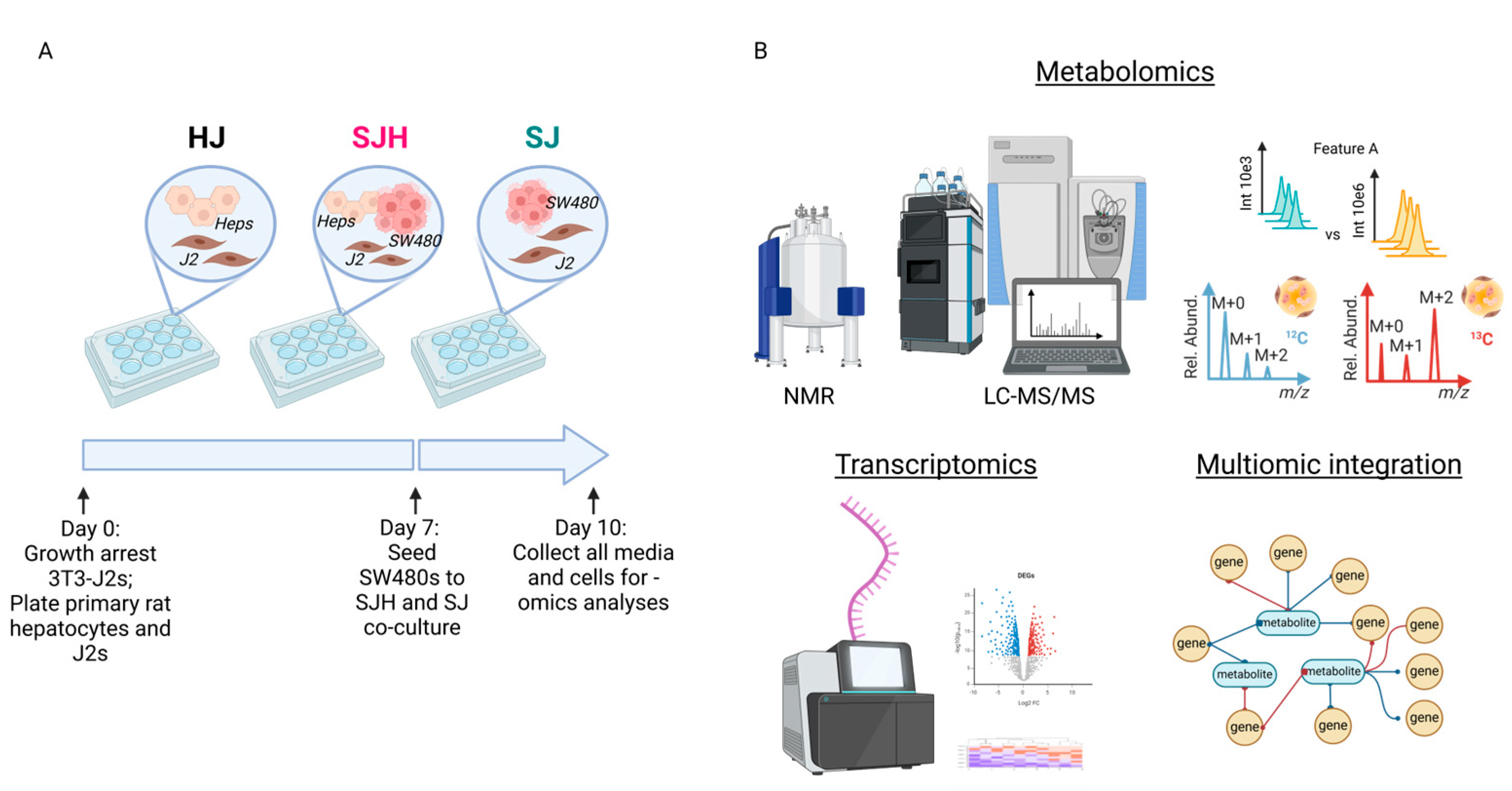
2.1. Co-Culture of SW480 Cells with Primary Hepatocytes Reprograms Metabolism
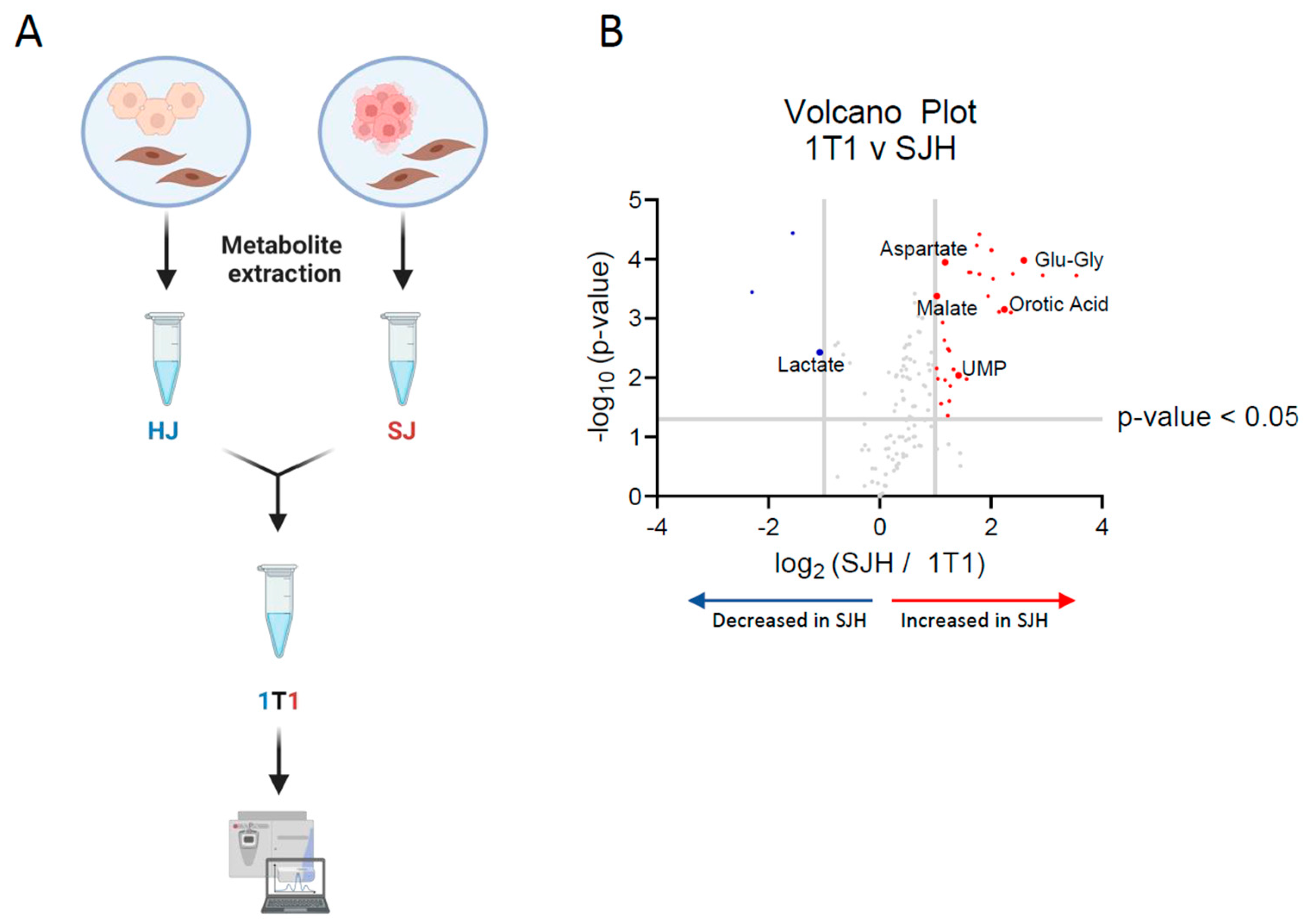
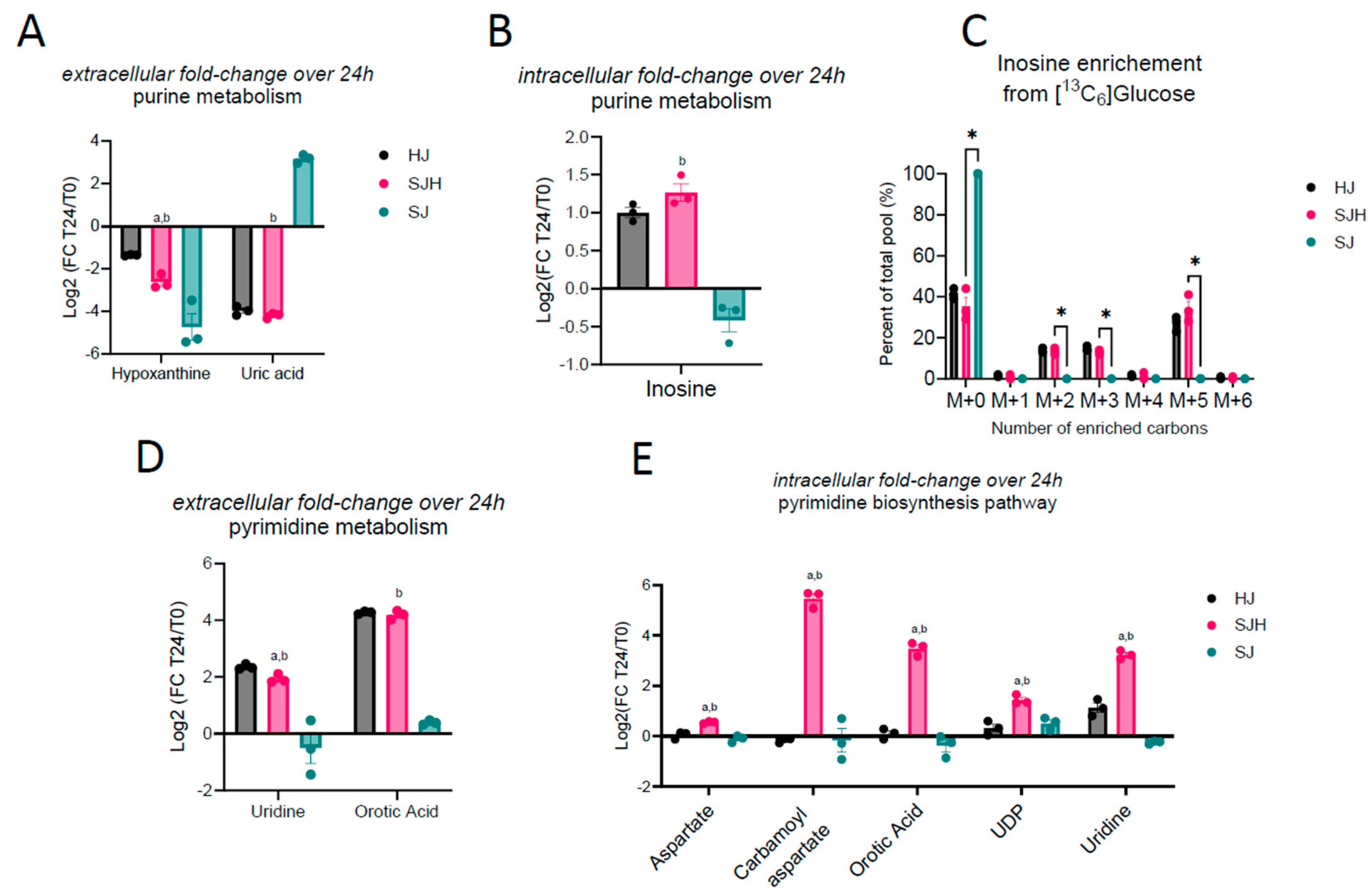
2.2. Metabolite Exchange of Nucleoside Intermediates
Pyrimidine Metabolism Is Increased in Hepatocytes by Co-Culture with SW480 Cells
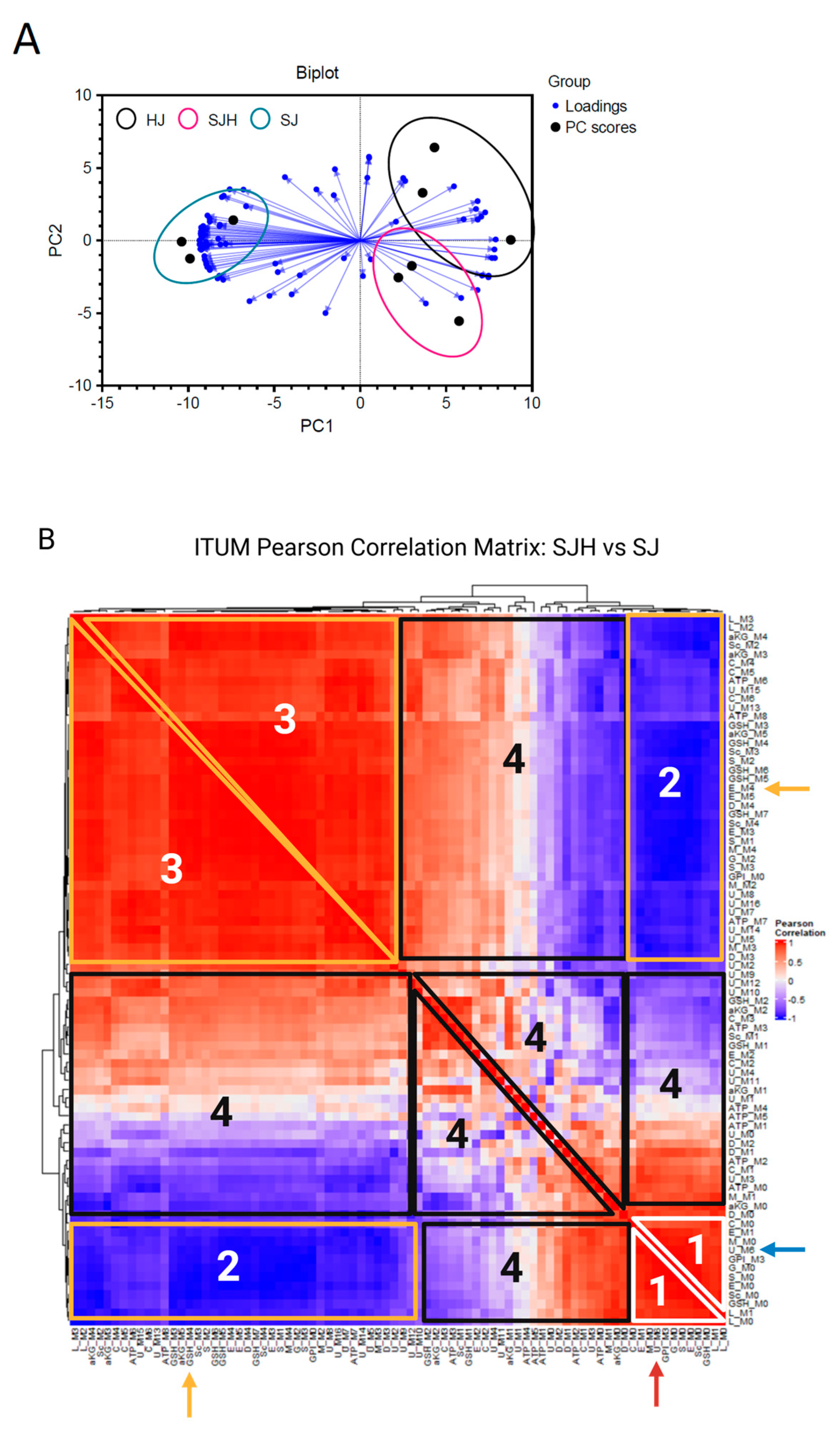
2.3. Discriminant 13C-ITUM Analysis of SJH Co-Cultures
2.4. Transcriptomic Analysis of 3-Dimensional Hepatocyte–SW480 Microtissue Organoids
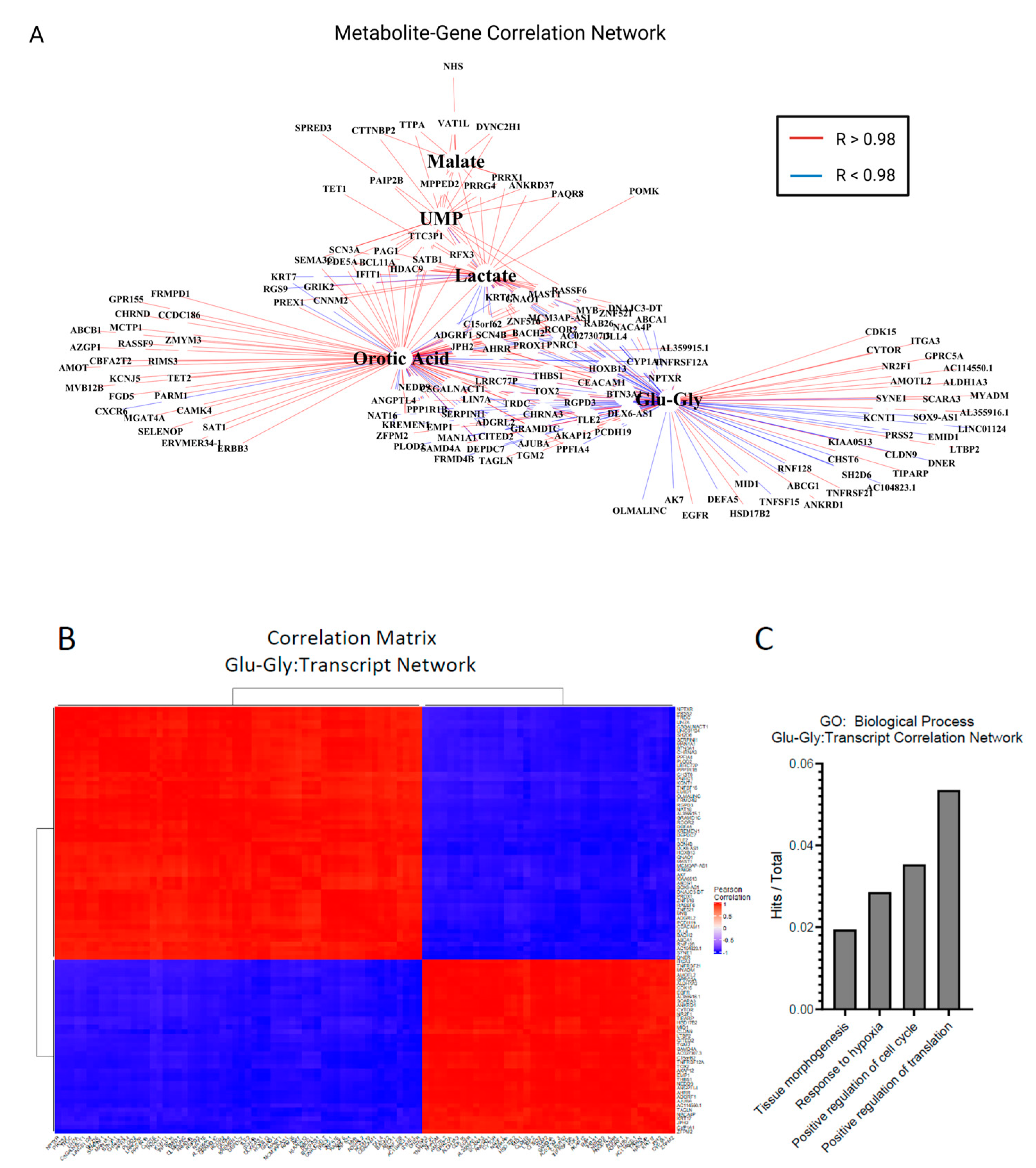
2.5. Multiomics Analysis of Differentially Expressed Genes and Static Metabolite Pools
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Two- and Three-Dimensional Culture Platform
4.3. Quantification of Glucose and Lactate via 1 H NMR
4.4. Untargeted Metabolomics and Isotope Tracing Untargeted Metabolomics Pipeline
4.4.1. Cell Collection for Metabolomics
4.4.2. Metabolite Extraction
4.4.3. Data Acquisition
4.4.4. Data Preparation
4.5. Quantification of Total Ketone Bodies
4.6. Bulk RNA Sequencing and Analysis
Bulk RNA-Seq Analysis
4.7. Statistics and Multiomics Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| UHPLC | ultra-high-pressure liquid chromatography |
| MS | mass spectrometry |
| MS/MS | tandem mass spectrometry (MS2) |
| HCC | hepatocellular carcinoma |
| m/z | mass-to-charge ratio |
| RT | retention time |
| ITUM | isotope tracing untargeted metabolomics |
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA A Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Horn, S.R.; Stoltzfus, K.C.; Lehrer, E.J.; Dawson, L.A.; Tchelebi, L.; Gusani, N.J.; Sharma, N.K.; Chen, H.; Trifiletti, D.M.; Zaorsky, N.G. Epidemiology of liver metastases. Cancer Epidemiol. 2020, 67, 101760. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg Robert, A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lyssiotis, C.A.; Kimmelman, A.C. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017, 27, 863–875. [Google Scholar] [CrossRef]
- Sullivan, M.R.; Danai, L.V.; Lewis, C.A.; Chan, S.H.; Gui, D.Y.; Kunchok, T.; Dennstedt, E.A.; Vander Heiden, M.G.; Muir, A. Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. eLife 2019, 8, e44235. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Rao, D.; Zhang, M.; Gao, Q. Metabolic reprogramming in the tumor microenvironment of liver cancer. J. Hematol. Oncol. 2024, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Fowle-Grider, R.; Rowles, J.L.; Shen, I.; Wang, Y.; Schwaiger-Haber, M.; Dunham, A.J.; Jayachandran, K.; Inkman, M.; Zahner, M.; Naser, F.J.; et al. Dietary fructose enhances tumour growth indirectly via interorgan lipid transfer. Nature 2024, 636, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Tan, Y.; Yin, P.; Ye, G.; Gao, P.; Lu, X.; Wang, H.; Xu, G. Metabolic Characterization of Hepatocellular Carcinoma Using Nontargeted Tissue Metabolomics. Cancer Res. 2013, 73, 4992–5002. [Google Scholar] [CrossRef] [PubMed]
- Nwosu, Z.C.; Megger, D.A.; Hammad, S.; Sitek, B.; Roessler, S.; Ebert, M.P.; Meyer, C.; Dooley, S. Identification of the Consistently Altered Metabolic Targets in Human Hepatocellular Carcinoma. Cell. Mol. Gastroenterol. Hepatol. 2017, 4, 303–323.e1. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, F.; Tabariès, S.; Andrzejewski, S.; Dong, Z.; Blagih, J.; Annis, M.G.; Omeroglu, A.; Gao, D.; Leung, S.; Amir, E.; et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell Metab. 2015, 22, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Bu, P.; Chen, K.-Y.; Xiang, K.; Johnson, C.; Crown, S.B.; Rakhilin, N.; Ai, Y.; Wang, L.; Xi, R.; Astapova, I.; et al. Aldolase B-Mediated Fructose Metabolism Drives Metabolic Reprogramming of Colon Cancer Liver Metastasis. Cell Metab. 2018, 27, 1249–1262.e4. [Google Scholar] [CrossRef] [PubMed]
- Loo, J.M.; Scherl, A.; Nguyen, A.; Man, F.Y.; Weinberg, E.; Zeng, Z.; Saltz, L.; Paty, P.B.; Tavazoie, S.F. Extracellular Metabolic Energetics Can Promote Cancer Progression. Cell 2015, 160, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Clish, C.B. Metabolomics: An emerging but powerful tool for precision medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nelson, A.B.; Chow, L.S.; Stagg, D.B.; Gillingham, J.R.; Evans, M.D.; Pan, M.; Hughey, C.C.; Myers, C.L.; Han, X.; Crawford, P.A.; et al. Acute aerobic exercise reveals FAHFAs distinguish the metabolomes of overweight and normal weight runners. JCI Insight 2022, 7, e158037. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.; Chen, L.; Rabinowitz, J.D. Metabolomics and Isotope Tracing. Cell 2018, 173, 822–837. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Puchalska, P.; Crawford, P.A. Application of Stable Isotope Labels for Metabolomics in Studies in Fatty Liver Disease. Methods Mol. Biol. 2019, 1996, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Stagg, D.B.; Gillingham, J.R.; Nelson, A.B.; Lengfeld, J.E.; D’avignon, D.A.; Puchalska, P.; Crawford, P.A. Diminished ketone interconversion, hepatic TCA cycle flux, and glucose production in D-β-hydroxybutyrate dehydrogenase hepatocyte-deficient mice. Mol. Metab. 2021, 53, 101269. [Google Scholar] [CrossRef] [PubMed]
- Puchalska, P.; Huang, X.; Martin, S.E.; Han, X.; Patti, G.J.; Crawford, P.A. Isotope Tracing Untargeted Metabolomics Reveals Macrophage Polarization-State-Specific Metabolic Coordination across Intracellular Compartments. iScience 2018, 9, 298–313. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Buescher, J.M.; Antoniewicz, M.R.; Boros, L.G.; Burgess, S.C.; Brunengraber, H.; Clish, C.B.; DeBerardinis, R.J.; Feron, O.; Frezza, C.; Ghesquiere, B.; et al. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 2015, 34, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Muir, A.; Danai, L.V.; Vander Heiden, M.G. Microenvironmental regulation of cancer cell metabolism: Implications for experimental design and translational studies. Dis. Models Mech. 2018, 11, dmm035758. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.; Du, Y.; Meng, G.; Yi, L.S.; Sun, S.; Song, N.; Zhang, X.; Xiao, Y.; Wang, J.; Yi, Z.; et al. Long-term functional maintenance of primary human hepatocytes in vitro. Science 2019, 364, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Zhang, G.; Su, X.; Jin, C.; Yu, B.; Yu, X.; Lv, Z.; Ma, H.; Zhang, M.; Wei, W.; et al. Maintenance of Primary Hepatocyte Functions In Vitro by Inhibiting Mechanical Tension-Induced YAP Activation. Cell Rep. 2019, 29, 3212–3222.e4. [Google Scholar] [CrossRef]
- Kukla, D.A.; Crampton, A.L.; Wood, D.K.; Khetani, S.R. Microscale Collagen and Fibroblast Interactions Enhance Primary Human Hepatocyte Functions in Three-Dimensional Models. Gene Expr. 2020, 20, 1–18. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- De Berardinis, R.J.; Chandel, N.S. We need to talk about the Warburg effect. Nat. Metab. 2020, 2, 127–129. [Google Scholar] [CrossRef]
- Warburg, O. The metabolism of carcinoma cells. J. Cancer Res. 1925, 9, 148–163. [Google Scholar] [CrossRef]
- Ma, E.H.; Verway, M.J.; Johnson, R.M.; Roy, D.G.; Steadman, M.; Hayes, S.; Williams, K.S.; Sheldon, R.D.; Samborska, B.; Kosinski, P.A.; et al. Metabolic Profiling Using Stable Isotope Tracing Reveals Distinct Patterns of Glucose Utilization by Physiologically Activated CD8+ T Cells. Immunity 2019, 51, 856–870.e5. [Google Scholar] [CrossRef] [PubMed]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Deng, Y.; Wang, Z.V.; Gordillo, R.; An, Y.; Zhang, C.; Liang, Q.; Yoshino, J.; Cautivo, K.M.; De Brabander, J.; Elmquist, J.K.; et al. An adipo-biliary-uridine axis that regulates energy homeostasis. Science 2017, 355, 6330. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Puchalska, P.; Crawford, P.A. Metabolic and Signaling Roles of Ketone Bodies in Health and Disease. In Annual Review of Nutrition; Stover, P.J., Balling, R., Eds.; Annual Reviews: San Mateo, CA, USA, 2021; Volume 41, pp. 49–77. [Google Scholar]
- Camarero, N.; Mascaró, C.; Mayordomo, C.; Vilardell, F.; Haro, D.; Marrero, P.F. Ketogenic HMGCS2 Is a c-Myc target gene expressed in differentiated cells of human colonic epithelium and down-regulated in colon cancer. Mol. Cancer Res. 2006, 4, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Liu, C.L.; Chiu, W.C.; Twu, Y.C.; Liao, Y.J. HMGCS2 Mediates Ketone Production and Regulates the Proliferation and Metastasis of Hepatocellular Carcinoma. Cancers 2019, 11, 1876. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zou, K.; Hu, Y.; Li, M.; Wang, H.; Zhang, Y.; Huang, L.; Xie, Y.; Li, S.; Dai, X.; Xu, W.; et al. Potential Role of HMGCS2 in Tumor Angiogenesis in Colorectal Cancer and Its Potential Use as a Diagnostic Marker. Can. J. Gastroenterol. Hepatol. 2019, 2019, 8348967. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, J.; Jia, P.-P.; Liu, Q.-L.; Cong, M.-H.; Gao, Y.; Shi, H.-P.; Yu, W.-N.; Miao, M.-Y. Low ketolytic enzyme levels in tumors predict ketogenic diet responses in cancer cell lines in vitro and in vivo. J. Lipid Res. 2018, 59, 625–634. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shukla, S.K.; Gebregiworgis, T.; Purohit, V.; Chaika, N.V.; Gunda, V.; Radhakrishnan, P.; Mehla, K.; Pipinos, I.I.; Powers, R.; Yu, F.; et al. Metabolic reprogramming induced by ketone bodies diminishes pancreatic cancer cachexia. Cancer Metab. 2014, 2, 18. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sperry, J.; Condro, M.C.; Guo, L.; Braas, D.; Vanderveer-Harris, N.; Kim, K.K.; Pope, W.B.; Divakaruni, A.S.; Lai, A.; Christofk, H.; et al. Glioblastoma Utilizes Fatty Acids and Ketone Bodies for Growth Allowing Progression during Ketogenic Diet Therapy. iScience 2020, 23, 101453. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xia, S.; Lin, R.; Jin, L.; Zhao, L.; Kang, H.-B.; Pan, Y.; Liu, S.; Qian, G.; Qian, Z.; Konstantakou, E.; et al. Prevention of Dietary-Fat-Fueled Ketogenesis Attenuates BRAF V600E Tumor Growth. Cell Metab. 2017, 25, 358–373. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xiong, L.; Yang, X.; Liu, H.; Wu, X.; Cai, T.; Yuan, M.; Huang, L.; Zhou, C.; Zheng, X.; Li, W.; et al. Glutamic-pyruvic transaminase 1 deficiency-mediated metabolic reprogramming facilitates colorectal adenoma-carcinoma progression. Sci. Transl. Med. 2025, 17, eadp9805. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Hilakivi-Clarke, L.; Shaha, A.; Wang, Y.; Wang, X.; Deng, Y.; Lai, J.; Kang, N. Metabolic reprogramming and its clinical implication for liver cancer. Hepatology 2023, 78, 1602–1624. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shi, M.; Yang, Y.; Huang, N.; Zeng, D.; Mo, Z.; Wang, J.; Zhang, X.; Liu, R.; Wang, C.; Rong, X.; et al. Genetic and microenvironmental evolution of colorectal liver metastases under chemotherapy. Cell Rep. Med. 2024, 5, 101838. [Google Scholar] [CrossRef] [PubMed]
- Gunji, D.; Abe, Y.; Muraoka, S.; Narumi, R.; Isoyama, J.; Ikemoto, N.; Ishida, M.; Shinkura, A.; Tomonaga, T.; Nagayama, S.; et al. Longitudinal phosphoproteomics reveals the PI3K-PAK1 axis as a potential target for recurrent colorectal liver metastases. Cell Rep. 2024, 43, 115061. [Google Scholar] [CrossRef]
- Lima, W.G.; Martins-Santos, M.E.; Chaves, V.E. Uric acid as a modulator of glucose and lipid metabolism. Biochimie 2015, 116, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Yiu, A.; Van Hemelrijck, M.; Garmo, H.; Holmberg, L.; Malmström, H.; Lambe, M.; Hammar, N.; Walldius, G.; Jungner, I.; Wulaningsih, W. Circulating uric acid levels and subsequent development of cancer in 493,281 individuals: Findings from the AMORIS Study. Oncotarget 2017, 8, 42332–42342. [Google Scholar] [CrossRef] [PubMed]
- Fini, M.A.; Elias, A.; Johnson, R.J.; Wright, R.M. Contribution of uric acid to cancer risk, recurrence, and mortality. Clin. Transl. Med. 2012, 1, 16. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nwosu, Z.C.; Ward, M.H.; Sajjakulnukit, P.; Poudel, P.; Ragulan, C.; Kasperek, S.; Radyk, M.; Sutton, D.; Menjivar, R.E.; Andren, A.; et al. Uridine-derived ribose fuels glucose-restricted pancreatic cancer. Nature 2023, 618, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Skinner, O.S.; Blanco-Fernández, J.; Goodman, R.P.; Kawakami, A.; Shen, H.; Kemény, L.V.; Joesch-Cohen, L.; Rees, M.G.; Roth, J.A.; Fisher, D.E.; et al. Salvage of ribose from uridine or RNA supports glycolysis in nutrient-limited conditions. Nat. Metab. 2023, 5, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Barcena, A.J.R.; Owens, T.C.; Melancon, S.; Workeneh, I.; Cao, H.S.T.; Vauthey, J.-N.; Huang, S.Y. Current Perspectives and Progress in Preoperative Portal Vein Embolization with Stem Cell Augmentation (PVESA). Stem Cell Rev. Rep. 2024, 20, 1236–1251. [Google Scholar] [CrossRef]
- Dong, Y.; Tu, R.; Liu, H.; Qing, G. Regulation of cancer cell metabolism: Oncogenic MYC in the driver’s seat. Signal Transduct. Target. Ther. 2020, 5, 124. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Chen, J.; He, L.; Stiles, B.L. PTEN: Tumor Suppressor and Metabolic Regulator. Front. Endocrinol. 2018, 9, 338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, H.; Li, Y.; Xia, D.; Yang, L.; Ma, Y.; Li, H. The role of YAP/TAZ activity in cancer metabolic reprogramming. Mol. Cancer 2018, 17, 134. [Google Scholar] [CrossRef]
- Affo, S.; Nair, A.; Brundu, F.; Ravichandra, A.; Bhattacharjee, S.; Matsuda, M.; Chin, L.; Filliol, A.; Wen, W.; Song, X.; et al. Promotion of cholangiocarcinoma growth by diverse cancer-associated fibroblast subpopulations. Cancer Cell 2021, 39, 866–882.e11. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhu, C.-X.; Yan, K.; Chen, L.; Huang, R.-R.; Bian, Z.-H.; Wei, H.-R.; Gu, X.-M.; Zhao, Y.-Y.; Liu, M.-C.; Suo, C.-X.; et al. Targeting OXCT1-mediated ketone metabolism reprograms macrophages to promote antitumor immunity via CD8+ T cells in hepatocellular carcinoma. J. Hepatol. 2024, 81, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, S.; Yuan, R.; Huang, H.-L.; Zhang, W.; Chiu, D.K.-C.; Kim, H.; Cha, S.L.; Tolentino, L.; Lowitz, J.; Liu, Y.; et al. The acid-sensing receptor GPR65 on tumor macrophages drives tumor growth in obesity. Sci. Immunol. 2024, 9, eadg6453. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, L.; Xu, J.; Ma, J. The Proteomic Landscape of Monocytes in Response to Colorectal Cancer Cells. J. Proteome Res. 2024, 23, 4067–4081. [Google Scholar] [CrossRef] [PubMed]
- Ivanisevic, J.; Zhu, Z.-J.; Plate, L.; Tautenhahn, R.; Chen, S.; O’brien, P.J.; Johnson, C.H.; Marletta, M.A.; Patti, G.J.; Siuzdak, G. Toward ’omic scale metabolite profiling: A dual separation-mass spectrometry approach for coverage of lipid and central carbon metabolism. Anal. Chem. 2013, 85, 6876–6884. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Koelmel, J.P.; Kroeger, N.M.; Gill, E.L.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Yost, R.A.; Garrett, T.J. Expanding Lipidome Coverage Using LC-MS/MS Data-Dependent Acquisition with Automated Exclusion List Generation. J. Am. Soc. Mass. Spectrom. 2017, 28, 908–917. [Google Scholar] [CrossRef]
- Silva, L.P.; Lorenzi, P.L.; Purwaha, P.; Yong, V.; Hawke, D.H.; Weinstein, J.N. Measurement of DNA Concentration as a Normalization Strategy for Metabolomic Data from Adherent Cell Lines. Anal. Chem. 2013, 85, 9536–9542. [Google Scholar] [CrossRef]
- Puchalska, P.; Nelson, A.B.; Stagg, D.B.; Crawford, P.A. Determination of ketone bodies in biological samples via rapid UPLC-MS/MS. Talanta 2021, 225, 122048. [Google Scholar] [CrossRef]
- Queathem, E.D.; Nelson, A.B.; Puchalska, P. Quantification of Total Ketone Bodies in Biological Matrices Using Stable Isotope-Labeled Internal Standards and RP-UHPLC-MS/MS. In Clinical Metabolomics: Methods and Protocols; Giera, M., Sánchez-López, E., Eds.; Springer: New York, NY, USA, 2025; pp. 117–131. [Google Scholar]
- Baller, J.; Kono, T.; Herman, A.; Zhang, Y. CHURP: A Lightweight CLI Framework to Enable Novice Users to Analyze Sequencing Datasets in Parallel. In Practice and Experience in Advanced Research Computing 2019: Rise of the Machines (Learning); Association for Computing Machinery: Chicago, IL, USA, 2019; p. 96. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2018, 47, D330–D338. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ewald, J.; Zhou, G.; Lu, Y.; Xia, J. Using ExpressAnalyst for Comprehensive Gene Expression Analysis in Model and Non-Model Organisms. Curr. Protoc. 2023, 3, e922. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Xia, J. Using MetaboAnalyst 4.0 for Metabolomics Data Analysis, Interpretation, and Integration with Other Omics Data. Methods Mol. Biol. 2020, 2104, 337–360. [Google Scholar] [CrossRef] [PubMed]
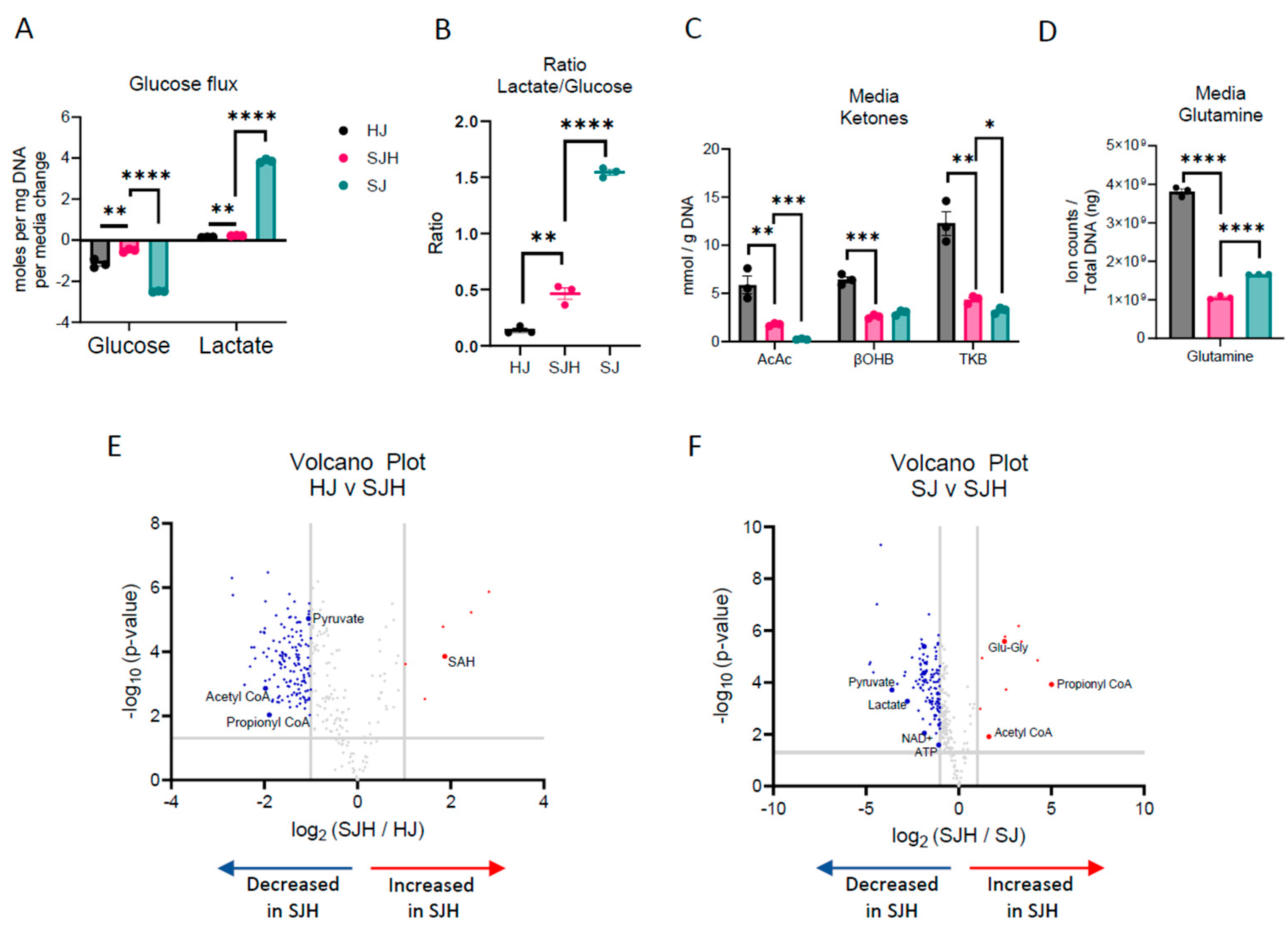

| Putative Metabolite | Log2 (FC: SJH/ctrl) | Adjusted p-Value | Direction Relative to [HJ] or [SJ] |
|---|---|---|---|
| S-adenosylhomocysteine | 1.87 | 0.0004 | ↑; HJ |
| Pyruvate | −1.05 | 0.0001 | ↓; HJ |
| Acetyl-CoA | −1.98 | 0.002 | ↓; HJ |
| Propionyl-CoA | −1.89 | 0.012 | ↓; HJ |
| Propionyl-CoA | 5.01 | 0.0004 | ↑; SJ |
| Glutamyl-glycine | 2.47 | 0.00007 | ↑; SJ |
| Acetyl-CoA | 1.63 | 0.016 | ↑; SJ |
| Pyruvate | −3.60 | 0.0006 | ↓; SJ |
| Lactate | −2.78 | 0.0012 | ↓; SJ |
| NAD+ | −1.85 | 0.012 | ↓; SJ |
| ATP | −1.09 | 0.03 | ↓; SJ |
| Putative Metabolite | Log2 (FC: SJH/HJ) | Adjusted p-Value | Direction Relative to 1T1 |
|---|---|---|---|
| Glutamyl-glycine | 2.59 | 0.0018 | ↑ |
| Uracil | 2.24 | 0.0044 | ↑ |
| Uridine diphosphate | 1.42 | 0.021 | ↑ |
| Aspartate | 1.17 | 0.0018 | ↑ |
| Malate | 1.03 | 0.0029 | ↑ |
| Log2 (FC: T24/T0) [adj. p-Value] | |||
|---|---|---|---|
| Putative Metabolite (E/I) | HJ | SJH | Direction Relative to T0 |
| Hypoxanthine (E) | −1.36 [<0.0001] | −2.62 [<0.0001] | ↓; ↓; ↓ |
| Uric acid (E) | −3.97 [<0.001] | −4.20 [<0.001] | ↓; ↓; ↑ |
| Inosine (I) | 1.00 [0.002] | 1.27 [0.003] | ↑; ↑; NC |
| Uridine (E) | 2.36 [<0.0001] | 1.94 [<0.001] | ↑; ↑; NC |
| Orotic acid (E) | 4.27 [<0.00001] | 4.19 [<0.001] | ↑; ↑; ↑ |
| Aspartate (I) | 0.04 [0.87] | 0.55 [0.02] | NC; ↑; NC |
| Carbamoyl aspartate (I) | −0.16 [0.19] | 5.47 [0.036] | NC; ↑; NC |
| UDP (I) | 0.32 [0.62] | 1.44 [0.017] | NC; ↑; NC |
| Uridine (I) | 1.13 [0.10] | 3.23 [0.005] | NC; ↑; NC |
| Orotic acid (I) | 0.09 [0.62] | 3.48 [0.029] | NC; ↑; NC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nelson, A.B.; Reese, L.E.; Rono, E.; Queathem, E.D.; Qiu, Y.; McCluskey, B.M.; Crampton, A.; Conniff, E.; Cummins, K.; Boytim, E.; et al. Deciphering Colorectal Cancer–Hepatocyte Interactions: A Multiomics Platform for Interrogation of Metabolic Crosstalk in the Liver–Tumor Microenvironment. Int. J. Mol. Sci. 2025, 26, 1976. https://doi.org/10.3390/ijms26051976
Nelson AB, Reese LE, Rono E, Queathem ED, Qiu Y, McCluskey BM, Crampton A, Conniff E, Cummins K, Boytim E, et al. Deciphering Colorectal Cancer–Hepatocyte Interactions: A Multiomics Platform for Interrogation of Metabolic Crosstalk in the Liver–Tumor Microenvironment. International Journal of Molecular Sciences. 2025; 26(5):1976. https://doi.org/10.3390/ijms26051976
Chicago/Turabian StyleNelson, Alisa B., Lyndsay E. Reese, Elizabeth Rono, Eric D. Queathem, Yinjie Qiu, Braedan M. McCluskey, Alexandra Crampton, Eric Conniff, Katherine Cummins, Ella Boytim, and et al. 2025. "Deciphering Colorectal Cancer–Hepatocyte Interactions: A Multiomics Platform for Interrogation of Metabolic Crosstalk in the Liver–Tumor Microenvironment" International Journal of Molecular Sciences 26, no. 5: 1976. https://doi.org/10.3390/ijms26051976
APA StyleNelson, A. B., Reese, L. E., Rono, E., Queathem, E. D., Qiu, Y., McCluskey, B. M., Crampton, A., Conniff, E., Cummins, K., Boytim, E., Dansou, S., Hwang, J., Safo, S. E., Puchalska, P., Wood, D. K., Schwertfeger, K. L., & Crawford, P. A. (2025). Deciphering Colorectal Cancer–Hepatocyte Interactions: A Multiomics Platform for Interrogation of Metabolic Crosstalk in the Liver–Tumor Microenvironment. International Journal of Molecular Sciences, 26(5), 1976. https://doi.org/10.3390/ijms26051976







