Evaluation of Salt-Tolerant Germplasms and Identification of Salt Tolerance-Related Proteins in Upland Cotton at the Seedling Stage
Abstract
:1. Introduction
2. Results
2.1. Classification of Salt Damage Symptoms at Seedling Stage in 430 Upland Cotton Populations
2.2. Variation Analysis of Salt Tolerance Coefficient-Related Traits in Ionomics at Seedling Stage of 430 Upland Cotton Populations
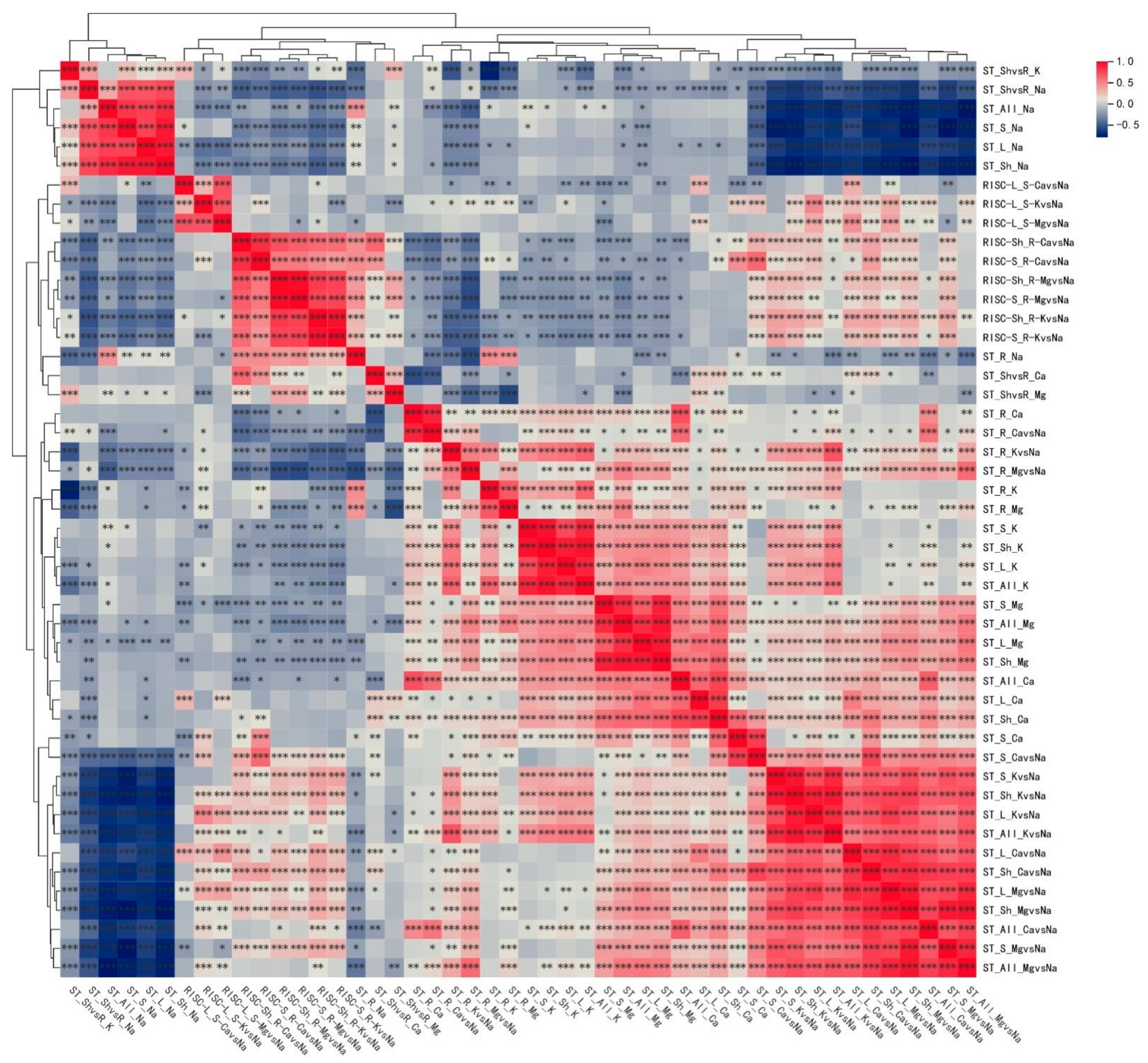
2.3. Correlation Analysis of 430 Traits Under Salt Stress at Seedling Stage of Upland Cotton
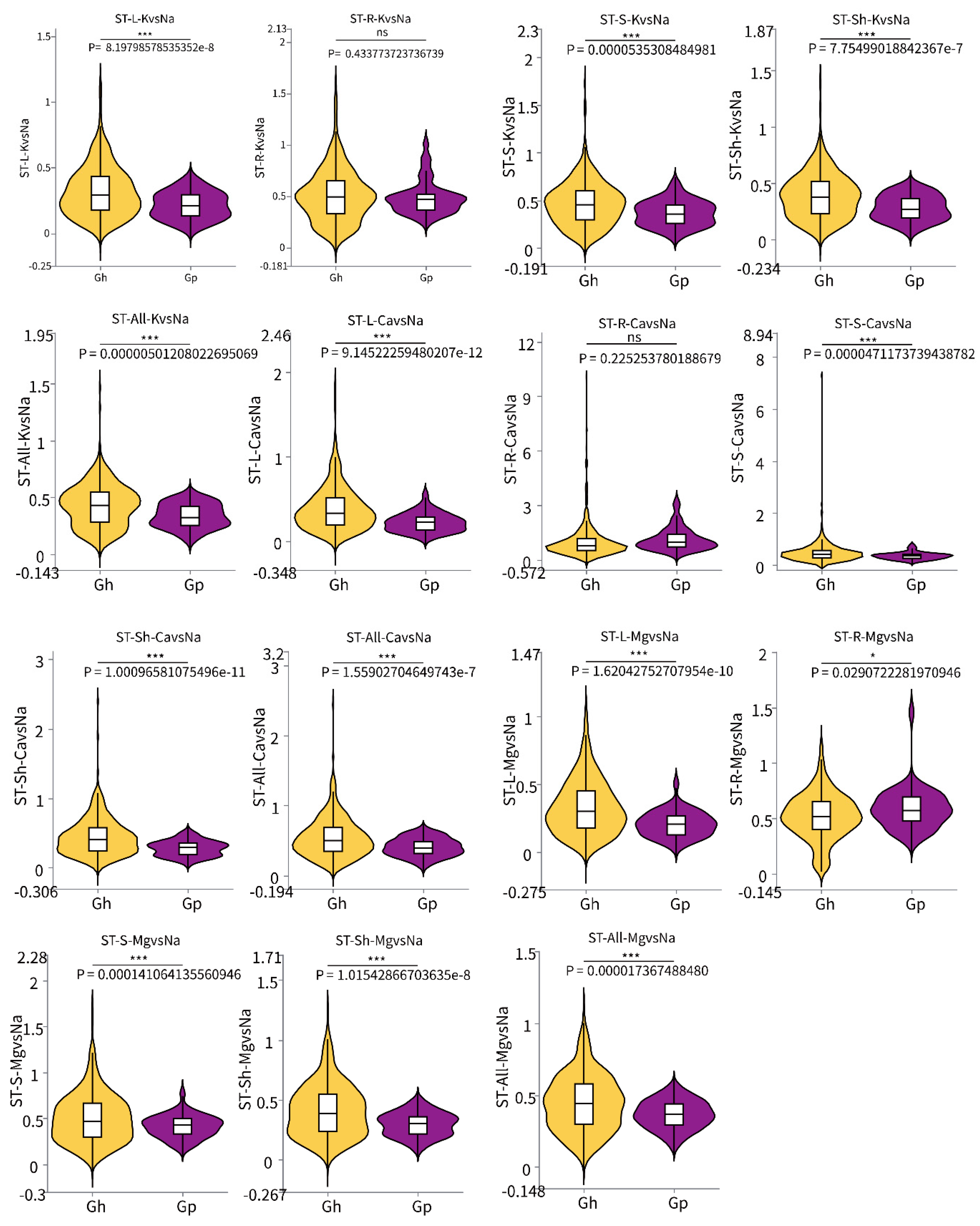
2.4. Analysis of Significant Differences in Salt Tolerance Coefficient Between G. purpurascens Cotton and Cultivated Upland Cotton
2.5. Principal Component Analysis of 430 Offspring Cotton Traits Under Salt Stress at Seedling Stage
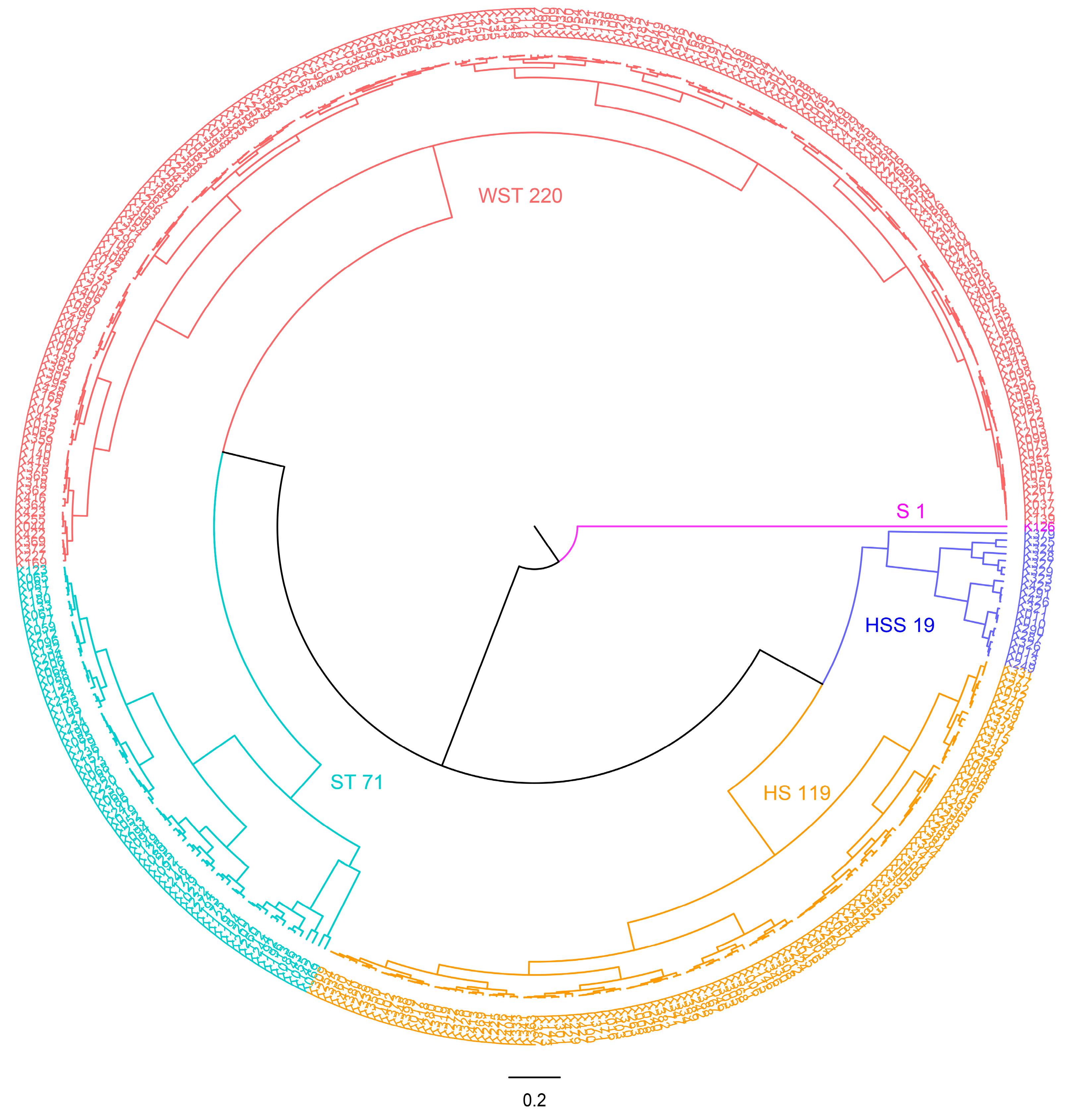
2.6. Evaluation of Salt Tolerance at Seedling Stage of 430 Upland Cotton Populations
2.7. Salt Tolerance Identification Results of 430 Upland Cotton Germplasms
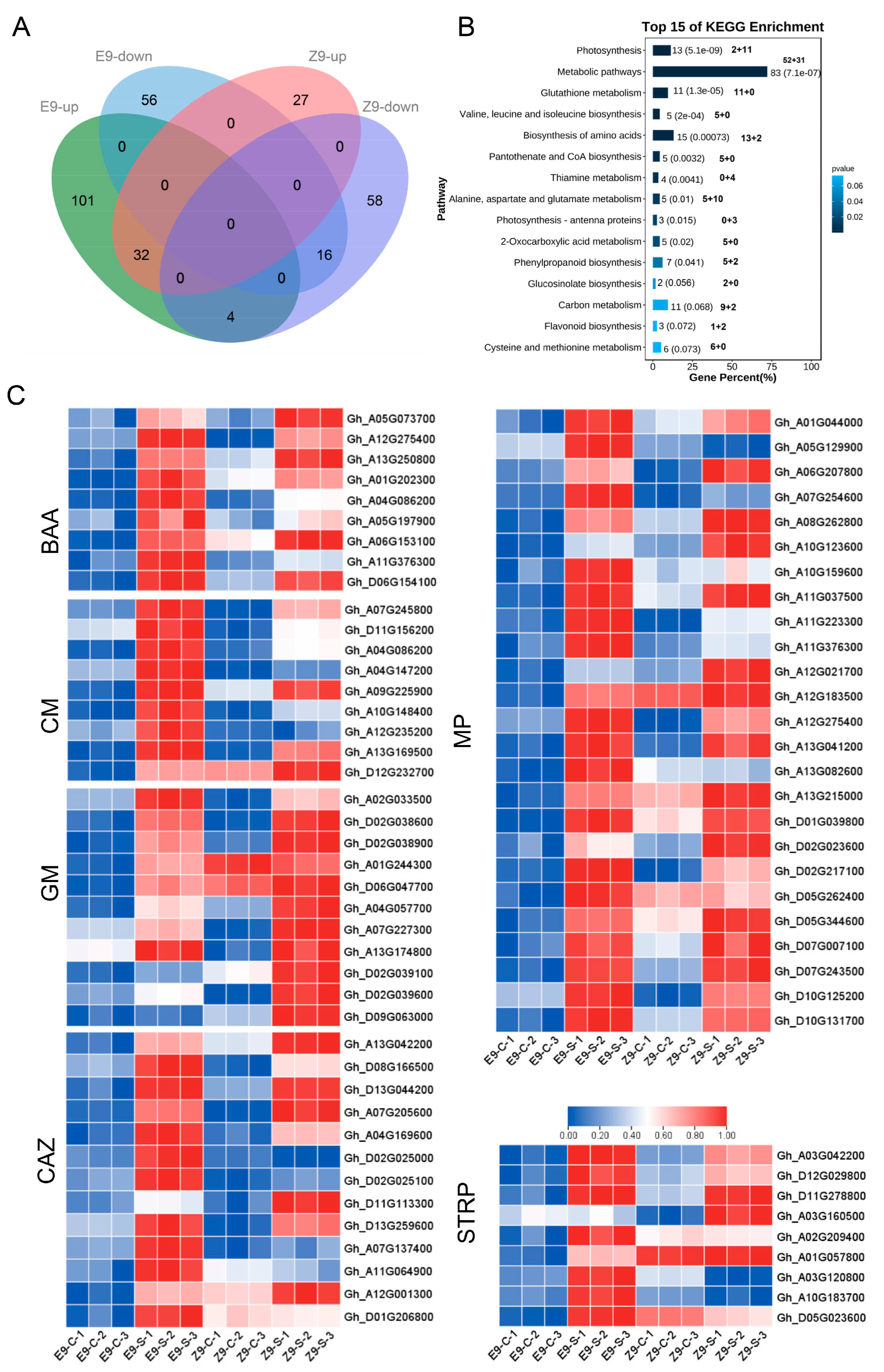
2.8. Quantitative Proteomic Responses to Salt Stress
2.9. Enrichment of DAPs by KEGG Pathway and Identification of Salt Tolerance-Related Proteins Under Salt Stress
3. Discussion
4. Materials and Methods
4.1. Experimental Material
4.2. Experimental Design
4.3. Measurement Indicators and Methods
4.3.1. Preparation of Sample Solution
4.3.2. Measurement Method
- (1)
- Determination Method of Sample Solution
- (2)
- Determination of Two-Step Dilution Sample Solution
4.3.3. Standard Curve
4.4. Phenotypic Data Calculation
4.5. TMT Quantitative Proteomics
4.5.1. Growth Conditions and Treatment
4.5.2. Protein Extraction and Digestion for Proteomic Sequencing Analysis
4.5.3. TMT Labeling
4.5.4. Peptide Identification by Nano UPLC-MS/MS
4.5.5. Protein Identification
4.6. Data Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Farooq, T.; Zain-ul-Hudda, Q.F.; Fatima, R.; Muneer, R.; Ali, A.; Shehzad, S.; Imran, A.; Sohail, S.; Iftikhar, A.; Mazhar, M. Overexpression of gene ΔGH_A07G1537 associated with fiber quality in upland cotton (Gossypium hirsutum L.) Through pollen tube transformation method. Agrobiol. Rec. 2024, 17, 129–137. [Google Scholar]
- Gupta, B.; Huang, B. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef] [PubMed]
- Yichie, Y.; Brien, C.; Berger, B.; Roberts, T.H.; Atwell, B.J. Salinity tolerance in Australian wild Oryza species varies widely and matches that observed in O-sativa. Rice 2018, 11, 66. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Duan, Z. Signal Regulation Mechanism of Plant Salt Stress. Genom. Appl. Biol. 2019, 38, 3706–3713. [Google Scholar]
- Gerona, M.E.B.; Deocampo, M.P.; Egdane, J.A.; Ismail, A.M.; Dionisio-Sese, M.L. Physiological Responses of Contrasting Rice Genotypes to Salt Stress at Reproductive Stage. Rice Sci. 2019, 26, 207–219. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Salt stress signaling and mechanisms of plant salt tolerance. Genet. Eng. 2006, 27, 141–177. [Google Scholar] [CrossRef]
- Hosseini, M.K.; Powell, A.A.; Bingham, I.J. Comparison of the seed germination and early seedling growth of soybean in saline conditions. Seed Sci. Res. 2002, 12, 165–172. [Google Scholar] [CrossRef]
- Etukuri, S.P. A Comparative Gene Expression and Regulation Analyses Among Four Gossypium Species. Ph.D. Thesis, Alabama Agricultural and Mechanical University, Huntsville, AL, USA, 2021. [Google Scholar]
- Zafar, M.; Ijaz, A.; Anwar, Z.; Shakeel, A.; Qadir, F.; Kamal, H.; Razzaq, A.; Jiang, X. Ancient to modern origins: The evolutionary journey of Gossypium genus and its implications for cotton breeding. Agrobiol. Rec. 2024, 17, 13–22. [Google Scholar]
- Xu, Y.; Wei, Y.; Li, Z.; Cai, X.; Wang, Y.; Wang, X.; Zhang, Z.; Wang, K.; Liu, F.; Zhou, Z. Integrated Evaluation and the Physiological and Biochemical Responses of Semi-Wild Cotton under Complex Salt-Alkali Stress. Cotton Sci. 2018, 30, 231–241. [Google Scholar]
- Nazir, M.F.; He, S.; Ahmed, H.; Sarfraz, Z.; Jia, Y.; Li, H.; Sun, G.; Iqbal, M.S.; Pan, Z.; Du, X. Genomic insight into the divergence and adaptive potential of a forgotten landrace G. hirsutum L. purpurascens. J. Genet. Genom. 2021, 48, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Zhu, G.; Zhang, T.; Guo, W. High-density 80 K SNP array is a powerful tool for genotyping G. hirsutum accessions and genome analysis. BMC Genom. 2017, 18, 654. [Google Scholar] [CrossRef]
- Ding, G.; Liu, J.; Shi, L.; Xu, F. Plant inomics: A new field in plant nutrition. J. Plant Nutr. Fertil. 2010, 16, 479–484. [Google Scholar]
- Guo, X. Current status and development of special cotton breeding in China. Cotton Sci. 1995, 7, 65–69. [Google Scholar]
- Xu, Z.; Lu, X.; Wei, Y.; Meng, C.; Zhang, M.; Zhang, Y.; Wang, M.; Wang, J.; Zhang, C.; Li, Y. Identification and evaluation of salt tolerance of space-induced wild soybean SP1 population at the seedling stage. Acta Prataculturae Sin. 2023, 32, 168–178. [Google Scholar]
- Bai, D.; Xue, Y.; Huang, L.; Huai, D.; Tian, Y.; Wang, P.; Zhang, X.; Zhang, H.; Li, N.; Jiang, H.; et al. Identification of cold tolerance and screening of evaluation indices for different peanut varieties at the germination stage. Acta Agron. Sin. 2022, 48, 2066–2079. [Google Scholar]
- Gu, X.; Wu, F.; Liu, H.; Nie, J.; Gao, H.; Ma, Q. Identification and evaluation of salt tolerance of 30 rice materials. J. Trop. Biol. 2020, 11, 314–323. [Google Scholar] [CrossRef]
- Sun, X.; Jiang, Q.; Hu, Z.; Li, H.; Pang, B.; Zhang, F.; Zhang, S.; Zhang, H. Identification and evaluation of salt tolerance of wheat germplasm resources at the seedling stage. Acta Agron. Sin. 2023, 49, 1132–1139. [Google Scholar]
- Hu, L.; Wang, S.; Wang, L.; Cheng, X.; Chen, H. Identification of salt tolerance and screening of salt-tolerant germplasm of mung bean at the seedling stage. Acta Agron. Sin. 2022, 48, 367–379. [Google Scholar] [CrossRef]
- Li, P.; Yan, J.; Zhang, H.; Zhang, Y.; Tao, S.; Zhang, Q.; Aldiyar, X.A.; Huang, Z. Screening and evaluation of salt tolerance of 146 Brassica napus germplasms at the germination stage. Acta Agric. Boreali-Occident. Sin. 2021, 30, 848–859. [Google Scholar]
- Jamshidi Goharrizi, K.; Baghizadeh, A.; Kalantar, M.; Fatehi, F. Assessment of changes in some biochemical traits and proteomic profile of UCB-1 pistachio rootstock leaf under salinity stress. J. Plant Growth Regul. 2020, 39, 608–630. [Google Scholar] [CrossRef]
- Nazari, M.; Moosavi, S.S.; Maleki, M.; Jamshidi Goharrizi, K. Chloroplastic acyl carrier protein synthase I and chloroplastic 20 kDa chaperonin proteins are involved in wheat (Triticum aestivum) in response to moisture stress. J. Plant Interact. 2020, 15, 180–187. [Google Scholar] [CrossRef]
- Jamshidi Goharrizi, K.; Fatehi, F.; Nazari, M.; Salehi, F.; Maleki, M. Assessment of changes in the content of sulforaphane and expression levels of CYP79F1 and myrosinase genes and proteomic profile of Lepidium draba plant under water-deficit stress induced by polyethylene glycol. Acta Physiol. Plant. 2020, 42, 101. [Google Scholar] [CrossRef]
- Long, R.; Gao, Y.; Sun, H.; Zhang, T.; Li, X.; Li, M.; Sun, Y.; Kang, J.; Wang, Z.; Ding, W. Quantitative proteomic analysis using iTRAQ to identify salt-responsive proteins during the germination stage of two Medicago species. Sci. Rep. 2018, 8, 9553. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Mu, Y.; Hu, X.; Ma, B.; Wang, Z.; Zhu, L.; Xu, J.; Huang, C.; Pan, Y. Comparative proteomic analysis reveals that the Heterosis of two maize hybrids is related to enhancement of stress response and photosynthesis respectively. BMC Plant Biol. 2021, 21, 34. [Google Scholar] [CrossRef] [PubMed]
- Aranjuelo, I.; Molero, G.; Erice, G.; Avice, J.C.; Nogues, S. Plant physiology and proteomics reveals the leaf response to drought in alfalfa (Medicago sativa L.). J. Exp. Bot. 2011, 62, 111–123. [Google Scholar] [CrossRef]
- Rahman, M.A.; Yong-Goo, K.; Iftekhar, A.; Liu, G.-s.; Hyoshin, L.; Joo, L.J.; Byung-Hyun, L. Proteome analysis of alfalfa roots in response to water deficit stress. J. Integr. Agric. 2016, 15, 1275–1285. [Google Scholar] [CrossRef]
- Xiong, J.; Sun, Y.; Yang, Q.; Tian, H.; Zhang, H.; Liu, Y.; Chen, M. Proteomic analysis of early salt stress responsive proteins in alfalfa roots and shoots. Proteome Sci. 2017, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wei, Z.; Qiao, Z.; Wu, Z.; Cheng, L.; Wang, Y. Proteomics analysis of alfalfa response to heat stress. PLoS ONE 2013, 8, e82725. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shi, S. Physiological and proteomic responses of contrasting alfalfa (Medicago sativa L.) varieties to PEG-induced osmotic stress. Front. Plant Sci. 2018, 9, 242. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, Z.; Song, X.; Zhang, F.; Liu, Y. TMT-based proteomic analysis of liquorice root in response to drought stress. BMC Genom. 2022, 23, 524. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Liu, L.; Zhang, Y.; Sun, H.; Zhang, K.; Bai, Z.; Dong, H.; Liu, Y.; Li, C. Tandem mass tag-based (TMT) quantitative proteomics analysis reveals the response of fine roots to drought stress in cotton (Gossypium hirsutum L.). BMC Plant Biol. 2020, 20, 328. [Google Scholar] [CrossRef]
- Pagel, O.; Kollipara, L.; Sickmann, A. Tandem mass tags for comparative and discovery proteomics. In Quantitative Methods in Proteomics; Springer: Berlin/Heidelberg, Germany, 2021; pp. 117–131. [Google Scholar]
- Peng, Z.; He, S.; Sun, J.; Xu, F.; Jia, Y.; Pan, Z.; Wang, L.; Du, X. Efficient identification method of salt tolerance of upland cotton at the seedling stage. Acta Agron. Sin. 2014, 40, 476–486. [Google Scholar] [CrossRef]
- Baxter, I.R.; Ziegler, G.; Lahner, B.; Mickelbart, M.V.; Foley, R.; Danku, J.; Armstrong, P.; Salt, D.E.; Hoekenga, O.A. Single-Kernel Ionomic Profiles Are Highly Heritable Indicators of Genetic and Environmental Influences on Elemental Accumulation in Maize Grain (Zea mays). PLoS ONE 2014, 9, e87628. [Google Scholar] [CrossRef]
- Li, C.; Dong, Z.; Guan, Y.; Liu, J.; Li, H.; Mei, Y. Genetic contribution and decision coefficient analysis of agronomic characters and lint yield traits of upland cotton in southern Xinjiang. Acta Agron. Sin. 2024, 50, 1486–1502. [Google Scholar]
- Teo, H.M.; Aziz, A.; Wahizatul, A.A.; Bhubalan, K.; Nordahliawate, S.M.S.; Syazlie, M.C.I.; Ng, L.C. Setting a Plausible Route for Saline Soil-Based Crop Cultivations by Application of Beneficial Halophyte-Associated Bacteria: A Review. Microorganisms 2022, 10, 657. [Google Scholar] [CrossRef]
- Han, X.; Wu, Z.; Liu, F.; Wang, Y.; Wei, X.; Tian, P.; Ling, F. Transcriptomic Analysis and Salt-Tolerance Gene Mining during Rice Germination. Genes 2023, 14, 1556. [Google Scholar] [CrossRef]
- Peng, Z.; Rehman, A.; Li, X.; Jiang, X.; Tian, C.; Wang, X.; Li, H.; Wang, Z.; He, S.; Du, X. Comprehensive Evaluation and Transcriptome Analysis Reveal the Salt Tolerance Mechanism in Semi-Wild Cotton (Gossypium purpurascens). Int. J. Mol. Sci. 2023, 24, 12853. [Google Scholar] [CrossRef]
- Liu, G.; Lu, L.; Liu, J. Identification of salt tolerance of cotton variety resources. Crop Var. Resour. 1993, 2, 21–22. [Google Scholar] [CrossRef]
- Sun, X.; Liu, Y.; Chen, Q. Research progress on salt tolerance of cotton. Cotton Sci. 1998, 10, 7–13. [Google Scholar]
- Wang, J.; Wang, D.; Fan, W.; Song, G.; Wang, S.; Ye, W. The characters of salt-tolerance at different growth stages in cotton. Acta Ecol. Sin. 2011, 31, 3720–3727. [Google Scholar]
- Xiong, S.; Wang, Y.; Chen, Y.; Gao, M.; Zhao, Y.; Wu, L. Effects of Drought Stress and Rehydration on Physiological and Biochemical Properties of Four Oak Species in China. Plants 2022, 11, 679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Hussain, S.; Wang, Y.; Liu, Y.; Li, Q.; Chen, Y.; Wei, H.; Gao, P.; Dai, Q. Comprehensive Evaluation of Salt Tolerance in Rice (Oryza sativa L.) Germplasm at the Germination Stage. Agronomy 2021, 11, 1569. [Google Scholar] [CrossRef]
- Zhou, Q.; Lu, Q.; Zhao, Z.; Wu, C.; Fu, X.; Zhao, Y.; Han, Y.; Lin, H.; Chen, W.; Mou, L.; et al. Identification of drought tolerance of 244 spring wheat varieties at the seedling stage. Sci. Agric. Sin. 2024, 57, 1646–1663. [Google Scholar]
- Sun, J.-b.; Sun, G.-y.; Liu, X.-d.; Hu, Y.-b.; Zhao, Y.-s. Effects of salt stress on mulberry seedlings growth, leaf water status, and ion distribution in various organs. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2009, 20, 543–548. [Google Scholar]
- Zhang, J.; Li, J.; Song, Y.; Xing, S.; Chi, J.; Ma, B. Advances in Research on the Mechanism of Plant Salinity Tolerance and Breeding of Salt-tolerant Plants. World For. Res. 2003, 16, 16–22. [Google Scholar]
- Wu, H.; Su, W.; Shi, M.; Xue, X.; Ren, H.; Wang, Y.; Zhao, A.; Li, D. Diversity Analysis and Comprehensive Evaluation of Jujube Fruit Traits. J. Plant Genet. Resour. 2022, 23, 1613–1625. [Google Scholar]
- Zhang, X.; Chen, C.; Zhang, J.; Zeng, Y.; Bao, M.; Zhang, S.; Shang, J.; Sha, X.; Wu, J.; Zhang, G.; et al. Analysis and Comprehensive Evaluation of Agronomic and Yield Traits of 55Alfalfa Varieties. Acta Agrestia Sin. 2023, 31, 3453–3461. [Google Scholar]
- Ma, X.; Liu, Y.; Shang, W.; Gao, N.; Zhang, T.; Piao, F.; Wang, Y.; Zhao, W. Comprehensive evaluation of salt tolerance of 44 watermelon germplasm resources at the seedling stage. China Cucurbits Veg. 2024, 37, 45–53. [Google Scholar] [CrossRef]
- Ma, S.; Tian, R.; Hu, H.; Lü, J.; Tian, L.; Luo, C.; Zhang, Y.; Li, P. Comprehensive evaluation and screening of salt tolerance of japonica rice germplasm resources at the seedling stage. J. Plant Genet. Resour. 2020, 21, 1089–1101. [Google Scholar] [CrossRef]
- Wang, W.; Wang, W.; Zou, J.; Yu, L.; Wang, Z.; Wang, F.; Lu, L.; Niu, L. Research progress on comprehensive evaluation methods of salt tolerance in wheat. Mod. Agric. Res. 2022, 28, 111–113. [Google Scholar] [CrossRef]
- Haque, M.S.; Hasanuzzaman, M.; Rahman, M.T.; Islam, N.; Begum, S.N.; Yasmin, S. Hydroponic and in vitro screening of wheat varieties for salt-tolerance. Plant Sci. Today 2022, 9, 844–854. [Google Scholar] [CrossRef]
- Dong, Y.; Hu, G.; Yu, J.; Thu, S.W.; Grover, C.E.; Zhu, S.; Wendel, J.F. Salt-tolerance diversity in diploid and polyploid cotton (Gossypium) species. Plant J. 2020, 101, 1135–1151. [Google Scholar] [CrossRef]
- Joshi, N.; Reddy, S.P.P.; Kumar, N.; Bharadwaj, C.; Tapan, K.; Patil, B.; Jain, P.K.; Nimmy, M.; Roorkiwal, M.; Verma, P. Siphoning novel sources of seedling salinity tolerance from the diverse chickpea landraces. Crop Pasture Sci. 2023, 74, 1080–1093. [Google Scholar] [CrossRef]
- Iniative, A.G. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar]
- Shiu, S.-H.; Bleecker, A.B. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003, 132, 530–543. [Google Scholar] [CrossRef] [PubMed]
- Piovesana, S.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Montone, C.M.; Zenezini Chiozzi, R.; Laganà, A. Recent trends and analytical challenges in plant bioactive peptide separation, identification and validation. Anal. Bioanal. Chem. 2018, 410, 3425–3444. [Google Scholar] [CrossRef] [PubMed]
- Ziemann, S.; van der Linde, K.; Lahrmann, U.; Acar, B.; Kaschani, F.; Colby, T.; Kaiser, M.; Ding, Y.; Schmelz, E.; Huffaker, A. An apoplastic peptide activates salicylic acid signalling in maize. Nat. Plants 2018, 4, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Tavormina, P.; De Coninck, B.; Nikonorova, N.; De Smet, I.; Cammue, B.P. The plant peptidome: An expanding repertoire of structural features and biological functions. Plant Cell 2015, 27, 2095–2118. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Li, K.; Yan, R.; Xu, F.; Ni, L.; Jiang, M. The UDP-glucuronic acid decarboxylase OsUXS3 regulates Na+ ion toxicity tolerance under salt stress by interacting with OsCATs in rice. Plant Physiol. Biochem. 2023, 196, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Muslu, S.; Kasapoğlu, A.G.; Güneş, E.; Aygören, A.S.; Yiğider, E.; İlhan, E.; Aydın, M. Genome-Wide Analysis of Glutathione S-Transferase Gene Family in P. vulgaris Under Drought and Salinity Stress. Plant Mol. Biol. Report. 2024, 42, 57–76. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, F.; Yan, Q.; Duan, Z.; Wang, S.; Ao, B.; Han, Y.; Zhang, J. Genome-Wide Analysis of the Rab Gene Family in Melilotus albus Reveals Their Role in Salt Tolerance. Int. J. Mol. Sci. 2023, 24, 126. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, T.; Li, J.; Chen, S.; Grin, I.R.; Zharkov, D.O.; Yu, B.; Li, H. Genome-wide analysis of WD40 protein family and functional characterization of BvWD40-82 in sugar beet. Front. Plant Sci. 2023, 14, 1185440. [Google Scholar] [CrossRef]
- Guan, H.; Zhang, Y.; Li, J.; Zhu, Z.; Chang, J.; Bakari, A.; Chen, S.; Zheng, K.; Cao, S. Analysis of the UDP-Glucosyltransferase (UGT) Gene Family and Its Functional Involvement in Drought and Salt Stress Tolerance in Phoebe bournei. Plants 2024, 13, 722. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yang, J.; Wang, B.; Liu, W.; Hua, Z.; Jiang, H. Influence of salinity on the diversity and composition of carbohydrate metabolism, nitrogen and sulfur cycling genes in lake surface sediments. Front. Microbiol. 2022, 13, 1019010. [Google Scholar] [CrossRef]
- Aiana; Chauhan, H.; Singh, K. Glycoside hydrolases reveals their differential role in response to drought and salt stress in potato (Solanum tuberosum). Funct. Plant Biol. 2024, 51, FP24114. [Google Scholar] [CrossRef]
- Wang, Z.; Li, N.; Xu, Y.; Wang, W.; Liu, Y. Functional activity of endophytic bacteria G9H01 with high salt tolerance and anti-Magnaporthe oryzae that isolated from saline-alkali-tolerant rice. Sci. Total Environ. 2024, 926, 171822. [Google Scholar] [CrossRef] [PubMed]
- Noh, G.; Kim, J.-H.; Cho, S.W.; Kim, Y.-H.; Jung, J.-Y.; Hong, W.-J.; Jung, K.-H.; Park, G.; Son, H.-J.; Jo, I.H.; et al. Overexpression of the ginseng GH18 gene confers salinity tolerance in Arabidopsis. Plant Biotechnol. Rep. 2022, 16, 683–696. [Google Scholar] [CrossRef]
- Zhang, K.; Sun, Y.; Li, M.; Long, R. CrUGT87A1, a UDP-sugar glycosyltransferases (UGTs) gene from Carex rigescens, increases salt tolerance by accumulating flavonoids for antioxidation in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 159, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Song, J.; Sheng, S.; Wang, D.; Wang, T.; Wang, N.; Chen, A.; Wang, L.; Peng, Y.; Ma, Y.; et al. Genome-wide characterization of Solanum tuberosum UGT gene family and functional analysis of StUGT178 in salt tolerance. BMC Genom. 2024, 25, 1206. [Google Scholar] [CrossRef]
- Jadamba, C.; Kang, K.; Paek, N.-C.; Lee, S.I.; Yoo, S.-C. Overexpression of rice expansin7 (Osexpa7) confers enhanced tolerance to salt stress in rice. Int. J. Mol. Sci. 2020, 21, 454. [Google Scholar] [CrossRef]
- Dong, B.; Wang, Q.; Zhou, D.; Wang, Y.; Miao, Y.; Zhong, S.; Fang, Q.; Yang, L.; Xiao, Z.; Zhao, H. Abiotic stress treatment reveals expansin like A gene OfEXLA1 improving salt and drought tolerance of Osmanthus fragrans by responding to abscisic acid. Hortic. Plant J. 2024, 10, 573–585. [Google Scholar] [CrossRef]
- Yin, D.; Zhang, X.; Wang, Y.; Cui, D. Principal component analysis and comprehensive evaluation of main quality traits in peanut. J. Plant Genet. Resour. 2011, 12, 507–512+518. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, D.; Luo, Z.; Li, S.; Tan, X.; Tan, Z.; Ma, L.; Dai, Y. Application of principal component analysis in walnut selection. Chin. Agric. Sci. Bull. 2011, 27, 45–48. [Google Scholar]
- Chen, S.C.; Zhu, Y.L. Subpattern-based principle component analysis. Pattern Recognit. 2004, 37, 1081–1083. [Google Scholar] [CrossRef]


| Grade | Salt-Tolerant Grade | Phenotypic Characteristics | Number of Materials | Frequency (%) |
|---|---|---|---|---|
| A1 | Salt-resistant | No wilting occurred after salt stress treatment. On the 10th day after salt stress, the growth was still normal, and there were basically no symptoms of salt damage. | 2 | 0.47 |
| A2 | Salt-tolerant | There was no obvious wilting of leaves 6 h after salt stress. A few leaves appeared wilted or drooping at 96 h after salt stress. On the 10th day after salt stress, a few leaves showed yellowing. | 93 | 21.63 |
| A3 | Weakly salt-tolerant | There was no obvious wilting of leaves 6 h after salt stress. After salt stress, about half of the leaf area appeared wilted or drooped at 96 h. On the 10th day after salt stress, about half of the leaves turned yellow. | 199 | 46.28 |
| A4 | Salt sensitivity | Leaf wilting was obvious at 96 h after salt stress. On the 10th day after salt stress, most of the leaf area dried up and lost water, and the growth was seriously hindered. | 92 | 21.39 |
| A5 | Highly sensitive | Leaf wilting was obvious at 96 h after salt stress. On the 10th day after salt stress, the plants were severely damaged, and the leaves and stems were basically completely wilted. | 44 | 10.23 |
| Grade | Salt Resistance | CSTI | Number of Materials | Frequency (%) | Proportion of Gp Cotton (%) |
|---|---|---|---|---|---|
| L1 | Salt-resistant (SR) | 5.633 | 1 | 0.23 | 0 |
| L2 | Salt-tolerant (ST) | 3.652–5.108 | 71 | 16.51 | 0 |
| L3 | Weakly salt-tolerant (WST) | 2.503–3.629 | 220 (28) | 51.16 | 12.73 |
| L4 | Salt sensitivity (SS) | 1.563–2.478 | 119 (15) | 27.67 | 12.61 |
| L5 | Highly sensitive (HSS) | 0.411–1.495 | 19 (2) | 4.42 | 10.53 |
| Phenotypic Grading | A1 | A2 | A3 | A4 | A5 | Total |
|---|---|---|---|---|---|---|
| L1 | 1 | 1 | ||||
| L2 | 30 | 37 | 4 | 71 | ||
| L3 | 1 | 47 | 114 | 38 | 20 | 220 |
| L4 | 1 | 16 | 43 | 39 | 20 | 119 |
| L5 | 4 | 11 | 4 | 19 | ||
| Total | 2 | 93 | 199 | 92 | 44 | 430 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Rehman, A.; Wang, Z.; Li, H.; Ma, J.; Du, X.; Peng, Z.; He, S. Evaluation of Salt-Tolerant Germplasms and Identification of Salt Tolerance-Related Proteins in Upland Cotton at the Seedling Stage. Int. J. Mol. Sci. 2025, 26, 1982. https://doi.org/10.3390/ijms26051982
Li X, Rehman A, Wang Z, Li H, Ma J, Du X, Peng Z, He S. Evaluation of Salt-Tolerant Germplasms and Identification of Salt Tolerance-Related Proteins in Upland Cotton at the Seedling Stage. International Journal of Molecular Sciences. 2025; 26(5):1982. https://doi.org/10.3390/ijms26051982
Chicago/Turabian StyleLi, Xiawen, Abdul Rehman, Zhenzhen Wang, Hongge Li, Jun Ma, Xiongming Du, Zhen Peng, and Shoupu He. 2025. "Evaluation of Salt-Tolerant Germplasms and Identification of Salt Tolerance-Related Proteins in Upland Cotton at the Seedling Stage" International Journal of Molecular Sciences 26, no. 5: 1982. https://doi.org/10.3390/ijms26051982
APA StyleLi, X., Rehman, A., Wang, Z., Li, H., Ma, J., Du, X., Peng, Z., & He, S. (2025). Evaluation of Salt-Tolerant Germplasms and Identification of Salt Tolerance-Related Proteins in Upland Cotton at the Seedling Stage. International Journal of Molecular Sciences, 26(5), 1982. https://doi.org/10.3390/ijms26051982





