Development of a Pre-Modification Strategy to Overcome Restriction–Modification Barriers and Enhance Genetic Engineering in Lactococcus lactis for Nisin Biosynthesis
Abstract
1. Introduction
2. Results
2.1. Identification of RM Systems in L. lactis C20
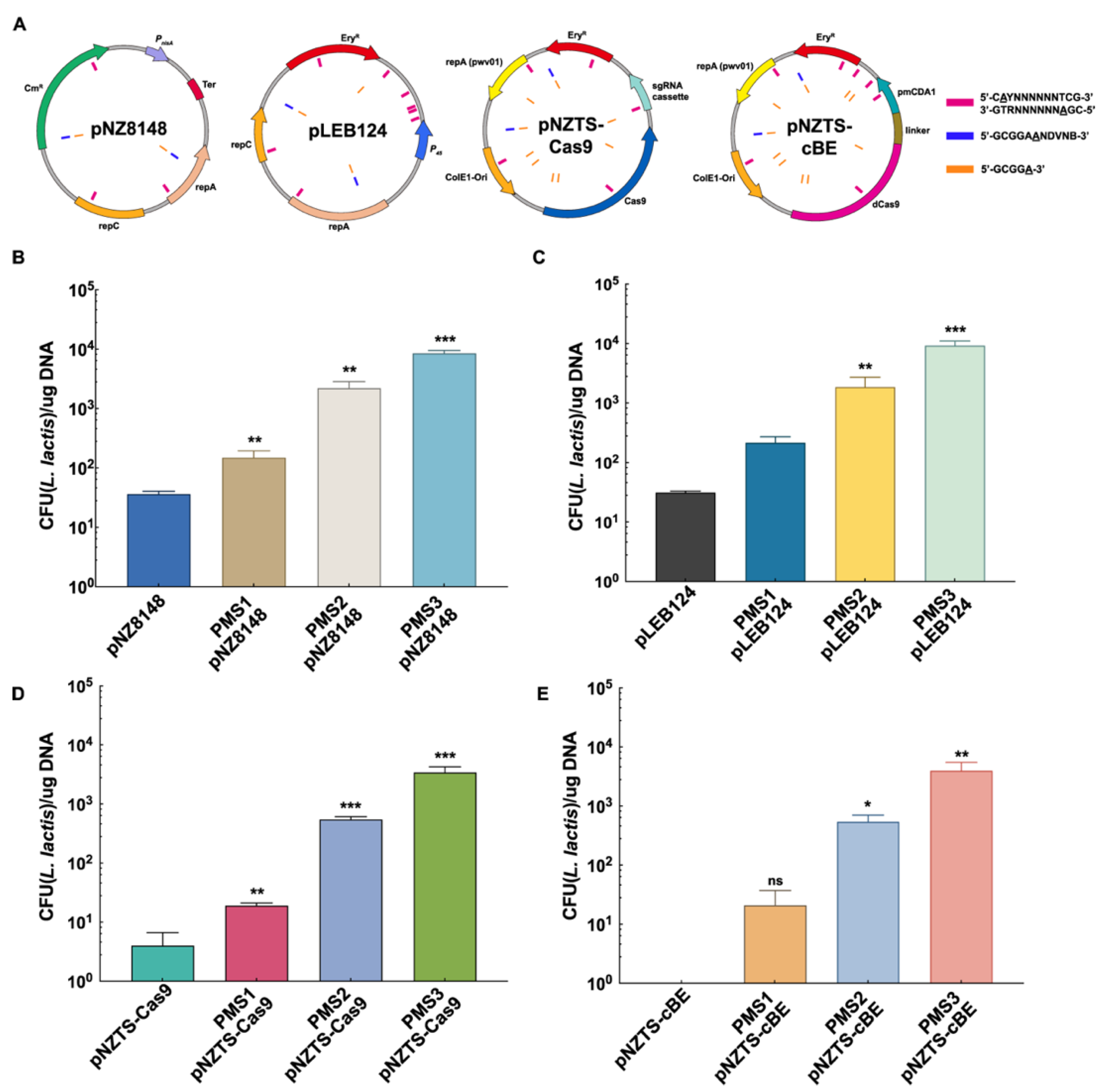
2.2. Development of PMS Tools and Evasion of RM Systems for High Transformation Efficiency
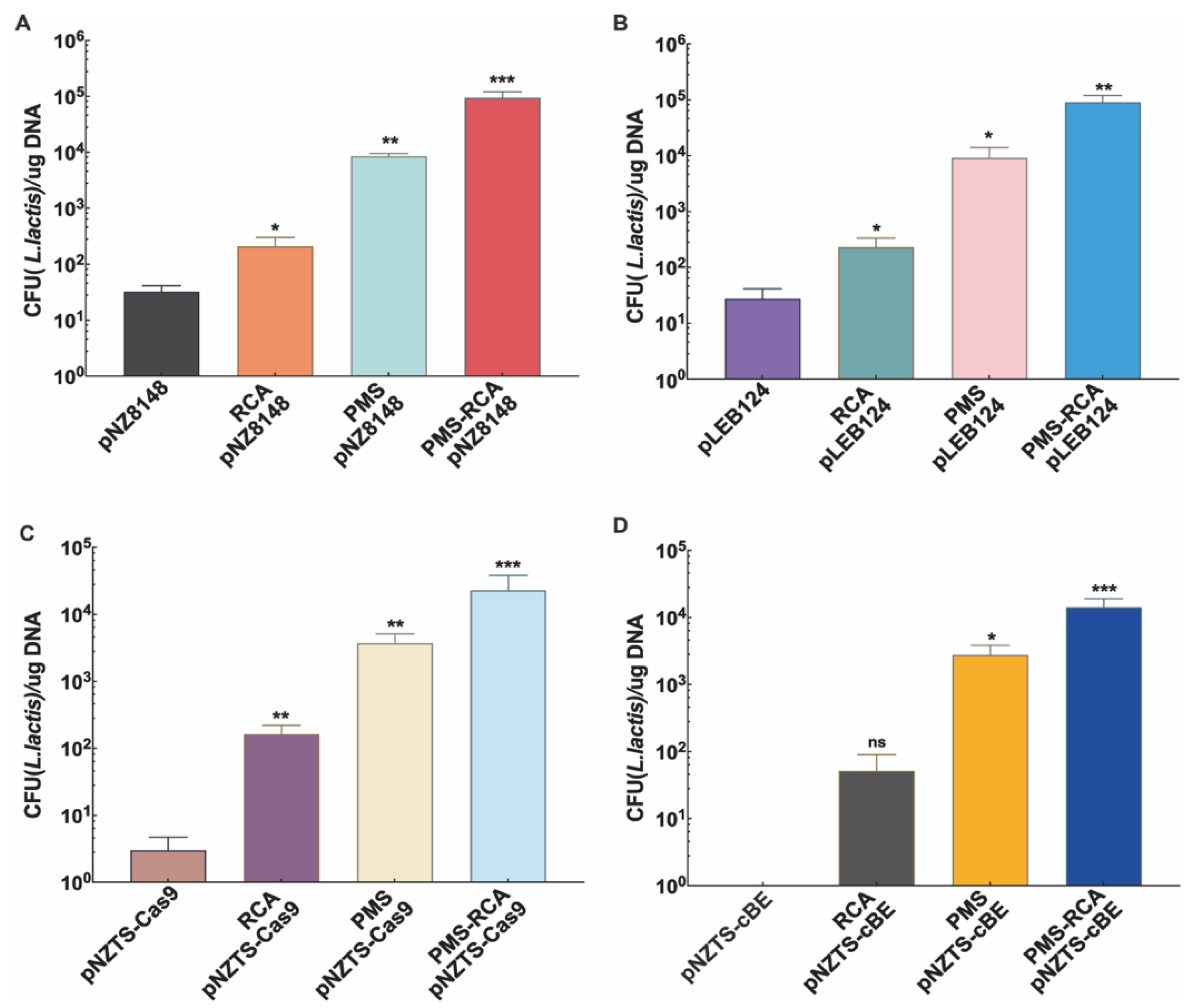
2.3. Further Enhancing Transformation Efficiency Through PMS-RCA
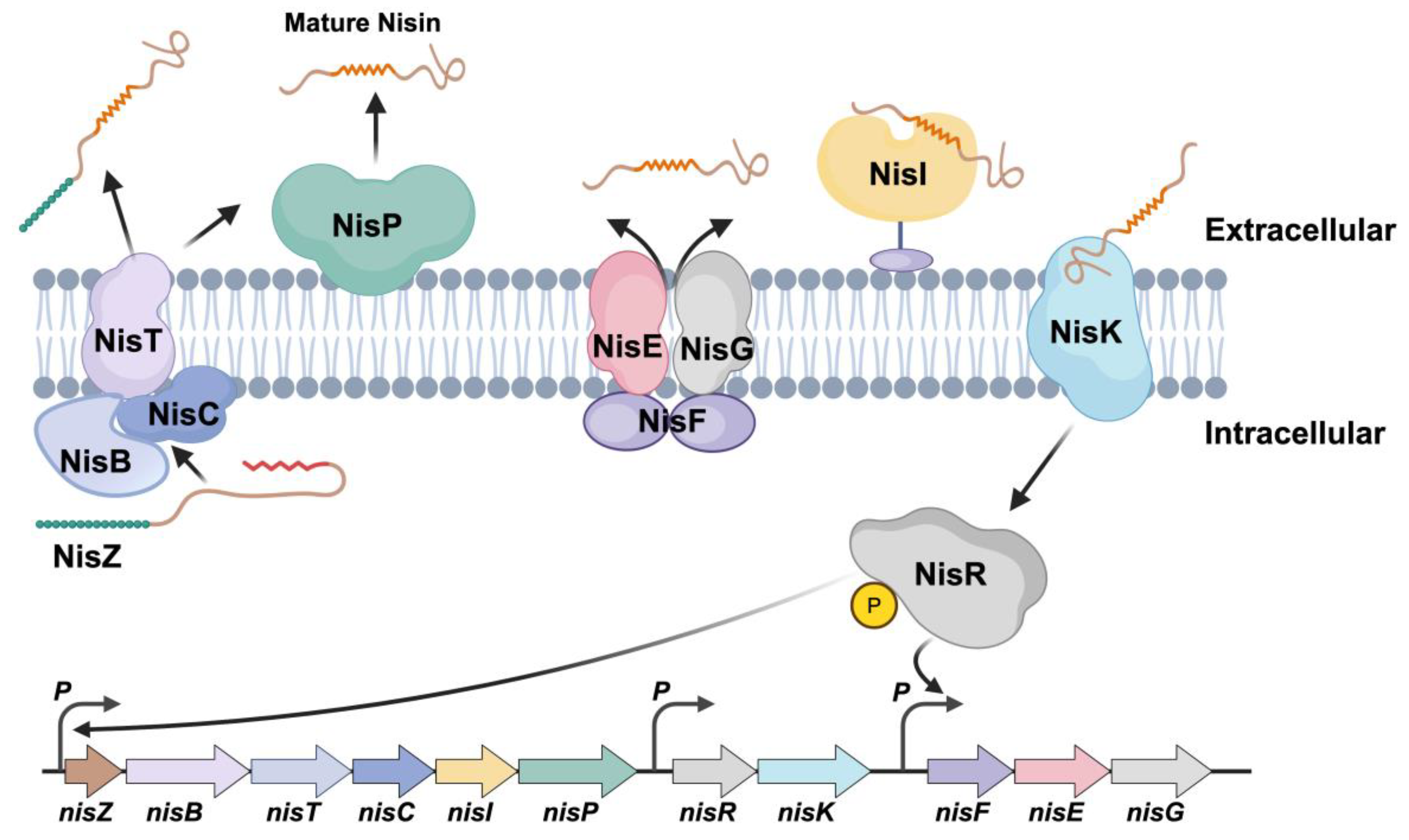
2.4. Enhancing Nisin Titers by Introducing Large Nisin Biosynthetic Gene Cluster
2.5. Expression Profiles of Genes Within Nisin Biosynthetic Gene Clusters in Engineered L. lactis Strains
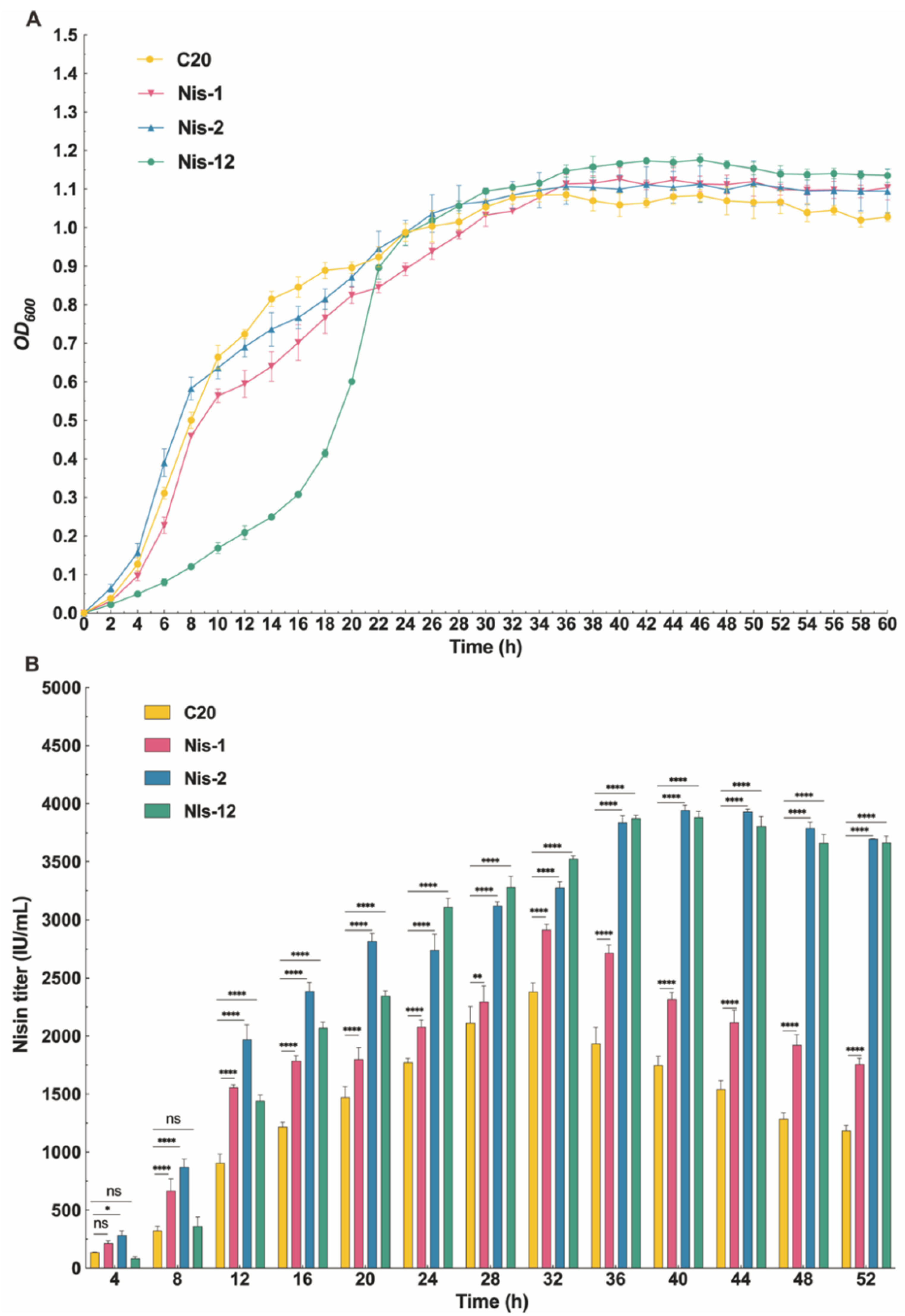
2.6. Nisin Titers of L. lactis Nis-2 in a 5 L Bioreactor
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, Culture Media, and Conditions
4.2. SMRT Sequencing and Methylome Analysis
4.3. Classification of RM System
4.4. Construction of PMS System
4.5. Overexpression of Nisin Biosynthesis Gene Cluster and Nisin Titer Assessment
4.6. Rolling Circle Amplification (RCA)-Combined PMS
4.7. L. lactis Transformations and Determination of Transformation Efficiency
4.8. Quantitative Real-Time PCR
4.9. Flask Fermentation and Fed-Batch Fermentation
4.10. Quantification and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tian, X.; Chen, H.; Liu, H.; Chen, J. Recent Advances in Lactic Acid Production by Lactic Acid Bacteria. Appl. Biochem. Biotechnol. 2021, 193, 4151–4171. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Chen, M.; Jia, J.; Bao, J. Increasing Acid Tolerance of an Engineered Lactic Acid Bacterium Pediococcus Acidilactici for L-Lactic Acid Production. Fermentation 2022, 8, 96. [Google Scholar] [CrossRef]
- Tran, Q.N.M.; Mimoto, H.; Koyama, M.; Nakasaki, K. Lactic Acid Bacteria Modulate Organic Acid Production during Early Stages of Food Waste Composting. Sci. Total Environ. 2019, 687, 341–347. [Google Scholar] [CrossRef]
- Kanafusa, S.; Uhlig, E.; Uemura, K.; Galindo, F.G.; Hakansson, A. The Effect of Nanosecond Pulsed Electric Field on the Production of Metabolites from Lactic Acid Bacteria in Fermented Watermelon Juice. Innov. Food Sci. Emerg. Technol. 2021, 72, 102749. [Google Scholar] [CrossRef]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical Applications of Nisin. J. Appl. Microbiol. 2016, 120, 1449–1465. [Google Scholar] [CrossRef]
- Li, B.; Yu, J.P.J.; Brunzelle, J.S.; Moll, G.N.; van der Donk, W.A.; Nair, S.K. Structure and Mechanism of the Lantibiotic Cyclase Involved in Nisin Biosynthesis. Science 2006, 311, 1464–1467. [Google Scholar] [CrossRef]
- Topisirovic, L.; Kojic, M.; Fira, D.; Golic, N.; Strahinic, I.; Lozo, J. Potential of Lactic Acid Bacteria Isolated from Specific Natural Niches in Food Production and Preservation. Int. J. Food Microbiol. 2006, 112, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Welker, D.L.; Coburn, B.M.; McClatchy, J.H.; Broadbent, J.R. Multiple Pulse Electroporation of Lactic Acid Bacteria Lactococcus Lactis and Lactobacillus Casei. J. Microbiol. Methods 2019, 166, 105741. [Google Scholar] [CrossRef]
- Papagianni, M.; Avramidis, N.; Filioussis, G. High Efficiency Electrotransformation of Lactococcus Lactis Spp. Lactis Cells Pretreated with Lithium Acetate and Dithiothreitol. BMC Biotechnol. 2007, 7, 15. [Google Scholar] [CrossRef]
- Mugwanda, K.; Hamese, S.; Zyl, W.F.V.; Prinsloo, E.; Plessis, M.D.; Dicks, L.M.; Raj, D.B.T.G. Recent Advances in Genetic Tools for Engineering Probiotic Lactic Acid Bacteria. Biosci. Rep. 2023, 43, BSR20211299. [Google Scholar] [CrossRef]
- Meng, Q.; Yuan, Y.; Li, Y.; Wu, S.; Shi, K.; Liu, S. Optimization of Electrotransformation Parameters and Engineered Promoters for Lactobacillus Plantarum from Wine. ACS Synth. Biol. 2021, 10, 1728–1738. [Google Scholar] [CrossRef] [PubMed]
- Kotnik, T.; Frey, W.; Sack, M.; Haberl Meglič, S.; Peterka, M.; Miklavčič, D. Electroporation-Based Applications in Biotechnology. Trends Biotechnol. 2015, 33, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Cui, Y.; Qu, X. Optimization of Electrotransformation (ETF) Conditions in Lactic Acid Bacteria (LAB). J. Microbiol. Methods 2020, 174, 105944. [Google Scholar] [CrossRef]
- Pleška, M.; Qian, L.; Okura, R.; Bergmiller, T.; Wakamoto, Y.; Kussell, E.; Guet, C.C. Bacterial Autoimmunity Due to a Restriction-Modification System. Curr. Biol. 2016, 26, 404–409. [Google Scholar] [CrossRef]
- Tock, M.; Dryden, D. The Biology of Restriction and Anti-Restriction. Curr. Opin. Microbiol. 2005, 8, 466–472. [Google Scholar] [CrossRef]
- Floyd, M.; Tang, J.; Kane, M.; Emerson, D. Captured Diversity in a Culture Collection: Case Study of the Geographic and Habitat Distributions of Environmental Isolates Held at the American Type Culture Collection. Appl. Environ. Microbiol. 2005, 71, 2813–2823. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.; Vincze, T.; Posfai, J.; Macelis, D. REBASE-a Database for DNA Restriction and Modification: Enzymes, Genes and Genomes. Nucleic Acids Res. 2015, 43, D298–D299. [Google Scholar] [CrossRef]
- Johnston, C.D.; Cotton, S.L.; Rittling, S.R.; Starr, J.R.; Borisy, G.G.; Dewhirst, F.E.; Lemon, K.P. Systematic Evasion of the Restriction-Modification Barrier in Bacteria. Proc. Natl. Acad. Sci. USA 2019, 116, 11454–11459. [Google Scholar] [CrossRef]
- Johnston, C.D.; Skeete, C.A.; Fomenkov, A.; Roberts, R.J.; Rittling, S.R. Restriction-Modification Mediated Barriers to Exogenous DNA Uptake and Incorporation Employed by Prevotella Intermedia. PLoS ONE 2017, 12, e0185234. [Google Scholar] [CrossRef]
- Oliveira, P.H.; Touchon, M.; Rocha, E.P.C. The Interplay of Restriction-Modification Systems with Mobile Genetic Elements and Their Prokaryotic Hosts. Nucleic Acids Res. 2014, 42, 10618–10631. [Google Scholar] [CrossRef]
- Vasu, K.; Nagaraja, V. Diverse Functions of Restriction-Modification Systems in Addition to Cellular Defense. Microbiol. Mol. Biol. Rev. 2013, 77, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Okada, R.; Matsumoto, M.; Zhang, Y.; Isaka, M.; Tatsuno, I.; Hasegawa, T. Emergence of Type I Restriction Modification System-Negative Emm 1 Type Streptococcus Pyogenes Clinical Isolates in Japan. Apmis 2014, 122, 914–921. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.A.; Ji, L.; Schmitz, R.J.; Westpheling, J.; Guss, A.M. Rational Development of Transformation in Clostridium thermocellum ATCC 27405 via Complete Methylome Analysis and Evasion of Native Restriction–Modification Systems. J. Ind. Microbiol. Biotechnol. 2019, 46, 1435–1443. [Google Scholar] [CrossRef]
- Ando, T.; Xu, Q.; Torres, M.; Kusugami, K.; Israel, D.A.; Blaser, M.J. Restriction-Modification System Differences in Helicobacter Pylori Are a Barrier to Interstrain Plasmid Transfer. Mol. Microbiol. 2000, 37, 1052–1065. [Google Scholar] [CrossRef]
- Liang, Y.; Jiao, S.; Wang, M.; Yu, H.; Shen, Z. A CRISPR/Cas9-Based Genome Editing System for Rhodococcus Ruber TH. Metab. Eng. 2020, 57, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Zautner, A.E.; Goldschmidt, A.-M.; Thürmer, A.; Schuldes, J.; Bader, O.; Lugert, R.; Groß, U.; Stingl, K.; Salinas, G.; Lingner, T. SMRT Sequencing of the Campylobacter Coli BfR-CA-9557 Genome Sequence Reveals Unique Methylation Motifs. BMC Genom. 2015, 16, 1088. [Google Scholar] [CrossRef]
- Chen, Z.; Shen, M.; Mao, C.; Wang, C.; Yuan, P.; Wang, T.; Sun, D. A Type I Restriction Modification System Influences Genomic Evolution Driven by Horizontal Gene Transfer in Paenibacillus Polymyxa. Front. Microbiol. 2021, 12, 709571. [Google Scholar] [CrossRef]
- Monk, I.R.; Shah, I.M.; Xu, M.; Tan, M.-W.; Foster, T.J. Transforming the Untransformable: Application of Direct Transformation to Manipulate Genetically Staphylococcus Aureus and Staphylococcus Epidermidis. mBio 2012, 3, e00277-11. [Google Scholar] [CrossRef]
- Monk, I.R.; Tree, J.J.; Howden, B.P.; Stinear, T.P.; Foster, T.J. Complete Bypass of Restriction Systems for Major Staphylococcus Aureus Lineages. mBio 2015, 6, e00308-15. [Google Scholar] [CrossRef]
- Morozova, N.; Sabantsev, A.; Bogdanova, E.; Fedorova, Y.; Maikova, A.; Vedyaykin, A.; Rodic, A.; Djordjevic, M.; Khodorkovskii, M.; Severinov, K. Temporal Dynamics of Methyltransferase and Restriction Endonuclease Accumulation in Individual Cells after Introducing a Restriction-Modification System. Nucleic Acids Res. 2016, 44, 790–800. [Google Scholar] [CrossRef]
- Fryer, T.; Wolff, D.S.; Overath, M.D.; Schäfer, E.; Laustsen, A.H.; Jenkins, T.P.; Andersen, C. Simplified post-assembly plasmid library amplification for increased transformation yields in E. coli and S. cerevisiae. bioRxiv 2023. biorxiv:2023.11.02.565362. [Google Scholar] [CrossRef]
- Salyers, A.; Bonheyo, G.; Shoemaker, N. Starting a New Genetic System: Lessons from Bacteroides. Methods 2000, 20, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Rowsell, E. Quick Identification of Type I Restriction Enzyme Isoschizomers Using Newly Developed pTypeI and Reference Plasmids. Nucleic Acids Res. 2008, 36, e81. [Google Scholar] [CrossRef]
- Bai, H.; Deng, A.; Liu, S.; Cui, D.; Qiu, Q.; Wang, L.; Yang, Z.; Wu, J.; Shang, X.; Zhang, Y.; et al. A Novel Tool for Microbial Genome Editing Using the Restriction-Modification System. ACS Synth. Biol. 2018, 7, 98–106. [Google Scholar] [CrossRef]
- Nye, T.M.; Jacob, K.M.; Holley, E.K.; Nevarez, J.M.; Dawid, S.; Simmons, L.A.; Watson, M.E. DNA Methylation from a Type I Restriction Modification System Influences Gene Expression and Virulence in Streptococcus Pyogenes. PLoS Pathog. 2019, 15, e1007841. [Google Scholar] [CrossRef]
- Kita, K.; Suisha, M.; Kotani, H.; Yanase, H.; Kato, N. Cloning and Sequence Analysis of the Stsl Restriction-Modification Gene: Presence of Homology to Fokl Restriction-Modification Enzymes. Nucleic Acids Res. 1992, 20, 4167–4172. [Google Scholar] [CrossRef]
- Finn, M.B.; Ramsey, K.M.; Tolliver, H.J.; Dove, S.L.; Wessels, M.R. Improved Transformation Efficiency of Group A Streptococcus by Inactivation of a Type I Restriction Modification System. PLoS ONE 2021, 16, e0248201. [Google Scholar] [CrossRef]
- Yang, P.; Yang, J.; Lin, T.; Liu, Q.; Yin, Y.; Chen, D.; Yang, S. Efficient Genome Editing in Most Staphylococcus aureus by Using the Restriction-Modification System Silent CRISPR-Cas9 Toolkit. ACS Synth. Biol. 2023, 12, 3340–3351. [Google Scholar] [CrossRef]
- Monk, I.R. Genetic Manipulation of Staphylococci—Breaking through the Barrier. Front. Cell. Infect. Microbiol. 2012, 2, 49. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, W.; Deng, A.; Sun, Z.; Zhang, Y.; Liang, Y.; Che, Y.; Wen, T. A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of Bacteria. PLoS Genet. 2012, 8, e1002987. [Google Scholar] [CrossRef]
- Yao, C.; Tang, H.; Wu, W.; Tang, J.; Guo, W.; Luo, D.; Yang, D. Double Rolling Circle Amplification Generates Physically Cross-Linked DNA Network for Stem Cell Fishing. J. Am. Chem. Soc. 2020, 142, 3422–3429. [Google Scholar] [CrossRef] [PubMed]
- Soares, R.R.G.; Madaboosi, N.; Nilsson, M. Rolling Circle Amplification in Integrated Microsystems: An Uncut Gem toward Massively Multiplexed Pathogen Diagnostics and Genotyping. Acc. Chem. Res. 2021, 54, 3979–3990. [Google Scholar] [CrossRef]
- Fu, Y.; Mu, D.; Qiao, W.; Zhu, D.; Wang, X.; Liu, F.; Xu, H.; Saris, P.; Kuipers, O.P.; Qiao, M. Co-Expression of Nisin Z and Leucocin C as a Basis for Effective Protection Against Listeria Monocytogenes in Pasteurized Milk. Front. Microbiol. 2018, 9, 547. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Qiao, Y.; Gao, G.; Liao, Z.; Wu, Z.; Saris, P.E.J.; Xu, H.; Qiao, M. A Novel Co-Cultivation Strategy to Generate Low-Crystallinity Bacterial Cellulose and Increase Nisin Yields. Int. J. Biol. Macromol. 2022, 202, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Liu, F.; Wan, X.; Qiao, Y.; Li, R.; Wu, Z.; Saris, P.E.J.; Xu, H.; Qiao, M. Genomic Features and Construction of Streamlined Genome Chassis of Nisin Z Producer Lactococcus Lactis N8. Microorganisms 2021, 10, 47. [Google Scholar] [CrossRef]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef]
- Poorinmohammad, N.; Hamedi, J.; Masoudi-Nejad, A. Genome-Scale Exploration of Transcriptional Regulation in the Nisin Z Producer Lactococcus Lactis Subsp. Lactis IO-1. Sci. Rep. 2020, 10, 3787. [Google Scholar] [CrossRef]
- Cases, I.; Lorenzo, V.d.; Ouzounis, C.A. Transcription Regulation and Environmental Adaptation in Bacteria. Trends Microbiol. 2003, 11, 248–253. [Google Scholar] [CrossRef]
- Yeung, K.Y.; Medvedovic, M.; Bumgarner, R.E. From Co-Expression to Co-Regulation: How Many Microarray Experiments Do We Need? Genome Biol. 2004, 5, R48. [Google Scholar] [CrossRef]
- De Smet, R.; Marchal, K. Advantages and Limitations of Current Network Inference Methods. Nat. Rev. Microbiol. 2010, 8, 717–729. [Google Scholar] [CrossRef]
- Dussault, D.; Vu, K.D.; Lacroix, M. Enhancement of Nisin Production by Lactococcus Lactis Subsp. Lactis. Probiotics Antimicrob. Proteins 2016, 8, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Fernandez de Ullivarri, M.; Ross, R.P.; Hill, C. After a Century of Nisin Research-Where Are We Now? FEMS Microbiol. Rev. 2023, 47, fuad023. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Yu, L.; Tian, F.; Zhai, Q.; Fan, L.; Chen, W. Antibiotic-Induced Gut Dysbiosis and Barrier Disruption and the Potential Protective Strategies. Crit. Rev. Food Sci. Nutr. 2022, 62, 1427–1452. [Google Scholar] [CrossRef] [PubMed]
- Flusberg, B.A.; Webster, D.R.; Lee, J.H.; Travers, K.J.; Olivares, E.C.; Clark, T.A.; Korlach, J.; Turner, S.W. Direct Detection of DNA Methylation during Single-Molecule, Real-Time Sequencing. Nat. Methods 2010, 7, 461–465. [Google Scholar] [CrossRef]
- Murray, I.A.; Clark, T.A.; Morgan, R.D.; Boitano, M.; Anton, B.P.; Luong, K.; Fomenkov, A.; Turner, S.W.; Korlach, J.; Roberts, R.J. The Methylomes of Six Bacteria. Nucleic Acids Res. 2012, 40, 11450–11462. [Google Scholar] [CrossRef]
- Machanick, P.; Bailey, T.L. MEME-ChIP: Motif Analysis of Large DNA Datasets. Bioinformatics 2011, 27, 1696–1697. [Google Scholar] [CrossRef]
- Li, Y.; Lin, Z.; Huang, C.; Zhang, Y.; Wang, Z.; Tang, Y.; Chen, T.; Zhao, X. Metabolic Engineering of Escherichia Coli Using CRISPR–Cas9 Meditated Genome Editing. Metab. Eng. 2015, 31, 13–21. [Google Scholar] [CrossRef]
- Wang, H.H.; Isaacs, F.J.; Carr, P.A.; Sun, Z.Z.; Xu, G.; Forest, C.R.; Church, G.M. Programming Cells by Multiplex Genome Engineering and Accelerated Evolution. Nature 2009, 460, 894–898. [Google Scholar] [CrossRef]
- Qiao, W.; Qiao, Y.; Liu, F.; Zhang, Y.; Li, R.; Wu, Z.; Xu, H.; Saris, P.E.J.; Qiao, M. Engineering Lactococcus Lactis as a Multi-Stress Tolerant Biosynthetic Chassis by Deleting the Prophage-Related Fragment. Microb. Cell Fact. 2020, 19, 225. [Google Scholar] [CrossRef]
- Ahrné, S.; Molin, G.; Axelsson, L. Transformation ofLactobacillus Reuteri with Electroporation: Studies on the Erythromycin Resistance Plasmid pLUL631. Curr. Microbiol. 1992, 24, 199–205. [Google Scholar] [CrossRef]
- Holo, H.; Nes, I.F. High-Frequency Transformation, by Electroporation, of Lactococcus Lactis Subsp. Cremoris Grown with Glycine in Osmotically Stabilized Media. Appl. Environ. Microbiol. 1989, 55, 3119–3123. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. 16S rRNA Gene Sequencing for Bacterial Identification in the Diagnostic Laboratory: Pluses, Perils, and Pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef] [PubMed]
- Tian, K.; Hong, X.; Guo, M.; Li, Y.; Wu, H.; Caiyin, Q.; Qiao, J. Development of Base Editors for Simultaneously Editing Multiple Loci in Lactococcus Lactis. ACS Synth. Biol. 2022, 11, 3644–3656. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Zhang, Y.; Chen, R.; Liu, K.; Wu, H.; Qiao, J.; Caiyin, Q. Development of a Pre-Modification Strategy to Overcome Restriction–Modification Barriers and Enhance Genetic Engineering in Lactococcus lactis for Nisin Biosynthesis. Int. J. Mol. Sci. 2025, 26, 2200. https://doi.org/10.3390/ijms26052200
Chen C, Zhang Y, Chen R, Liu K, Wu H, Qiao J, Caiyin Q. Development of a Pre-Modification Strategy to Overcome Restriction–Modification Barriers and Enhance Genetic Engineering in Lactococcus lactis for Nisin Biosynthesis. International Journal of Molecular Sciences. 2025; 26(5):2200. https://doi.org/10.3390/ijms26052200
Chicago/Turabian StyleChen, Chen, Yue Zhang, Ruiqi Chen, Ke Liu, Hao Wu, Jianjun Qiao, and Qinggele Caiyin. 2025. "Development of a Pre-Modification Strategy to Overcome Restriction–Modification Barriers and Enhance Genetic Engineering in Lactococcus lactis for Nisin Biosynthesis" International Journal of Molecular Sciences 26, no. 5: 2200. https://doi.org/10.3390/ijms26052200
APA StyleChen, C., Zhang, Y., Chen, R., Liu, K., Wu, H., Qiao, J., & Caiyin, Q. (2025). Development of a Pre-Modification Strategy to Overcome Restriction–Modification Barriers and Enhance Genetic Engineering in Lactococcus lactis for Nisin Biosynthesis. International Journal of Molecular Sciences, 26(5), 2200. https://doi.org/10.3390/ijms26052200





