Important Role of Bacterial Nucleoid-Associated Proteins in Discovery of Novel Secondary Metabolites
Abstract
:1. Introduction
2. The Roles of Epigenetic Modification in the Production of SMs in Microorganisms
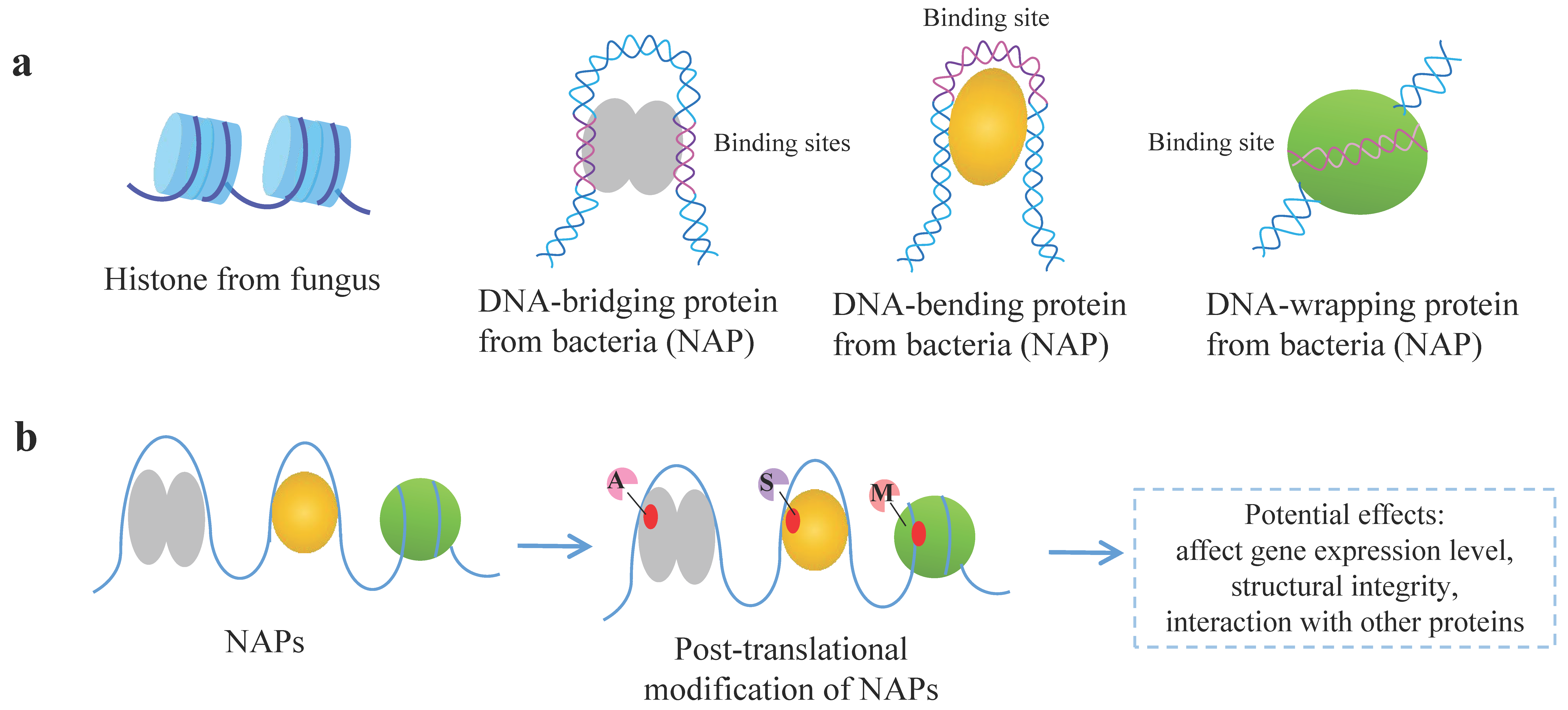
2.1. Features of Bacterial NAPs
2.2. Role of NAPs in Chromosome Dynamics During Streptomyces Life Cycles
2.3. PTMs of Bacterial NAPs
3. The Acetylation of NAP as a Primary PTM Modification
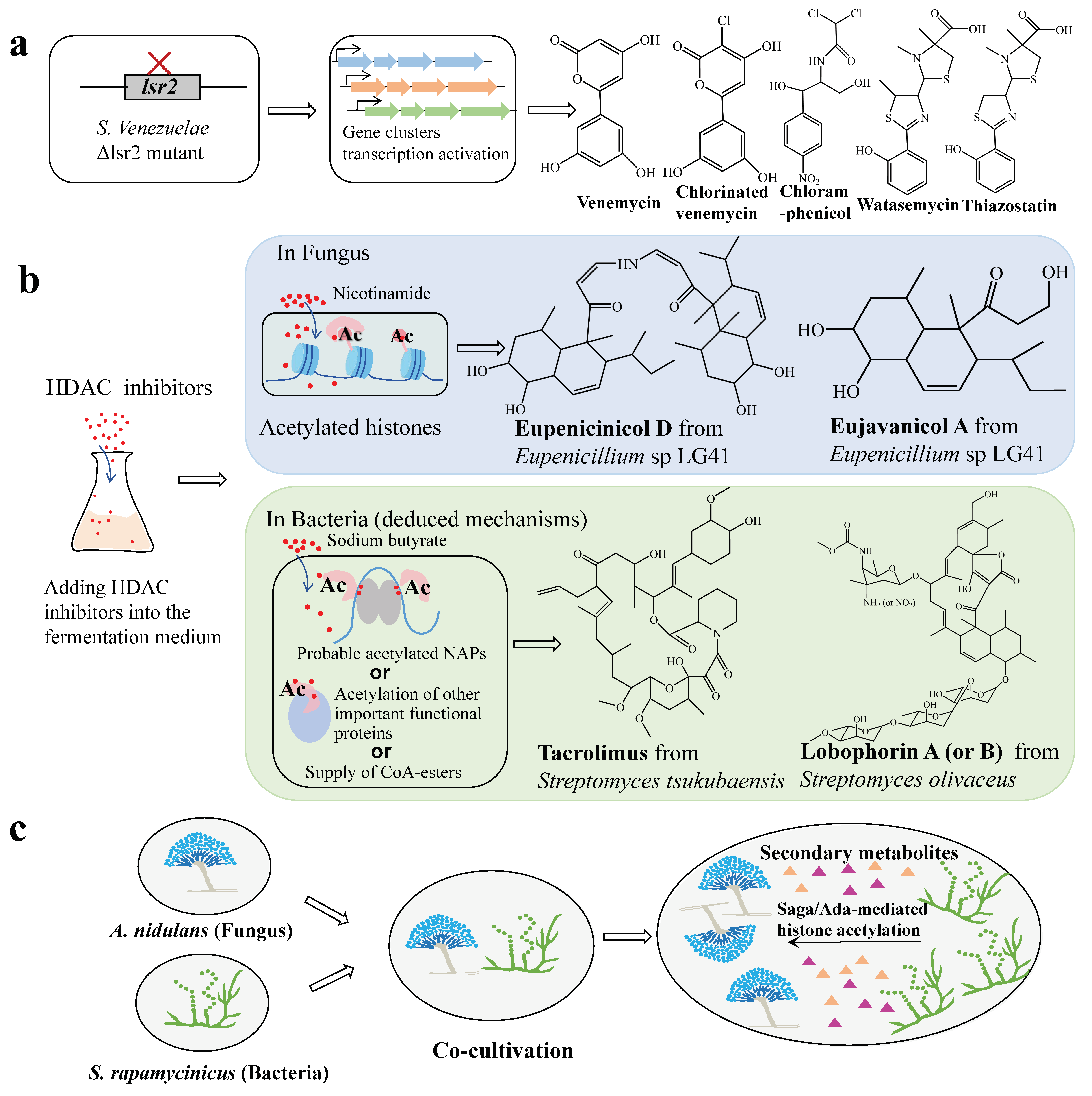
4. Effects of PTMs of Bacterial NAPs on Production of SMs
5. Activation of Silent BGCs by Adding Small Molecular Modifiers
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Li, Y.; Yu, H.; Guan, H.; Li, J.; Zhang, J.; Xiang, H.; Li, J.; Tan, H. Activation of cryptic antibiotic biosynthetic gene clusters guided by RNA-seq data from both Streptomyces ansochromogenes and ΔwblA. Antibiotics 2021, 10, 1097. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guan, H.; Li, J.; Zhang, J.; Wang, Y.; Li, J.; Tan, H. An intricate regulation of WblA controlling production of silent tylosin analogues and abolishment of expressible nikkomycin. Sci. China Life Sci. 2023, 66, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tan, H. Microbial quorum sensing signaling molecules and their roles in the biosynthesis of natural products. Sci. China Life Sci. 2023, 66, 2429–2432. [Google Scholar] [CrossRef]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef]
- Li, Y.; Tan, H. Biosynthesis and molecular regulation of secondary metabolites in microorganisms. Sci. China Life Sci. 2017, 60, 935–938. [Google Scholar] [CrossRef]
- Williams, R.B.; Henrikson, J.C.; Hoover, A.R.; Lee, A.E.; Cichewicz, R.H. Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem. 2008, 6, 1895–1897. [Google Scholar] [CrossRef]
- Yang, K.L.; Tian, J.; Keller, N.P. Post-translational modifications drive secondary metabolite biosynthesis in Aspergillus: A review. Environ. Microbiol. 2022, 24, 2857–2881. [Google Scholar] [CrossRef]
- Makhwitine, J.P.; Kumalo, H.M.; Ndlovu, S.I.; Mkhwanazi, N.P. Epigenetic induction of secondary metabolites production in endophytic fungi Penicillium chrysogenum and GC-MS analysis of crude metabolites with anti-HIV-1 activity. Microorganisms 2023, 11, 1404. [Google Scholar] [CrossRef]
- Wang, X.; Sena, J.G.; Hoover, A.; King, J.; Ellis, T.K.; Powell, D.R.; Cichewicz, R.H. Chemical epigenetics alters the secondary metabolite composition of guttate excreted by an atlantic-forest-soil-derived Penicillium citreonigrum. J. Nat. Prod. 2010, 73, 942–948. [Google Scholar] [CrossRef]
- Asai, T.; Chung, Y.M.; Sakurai, H.; Ozeki, T.; Chang, F.R.; Yamashita, K.; Oshima, Y. Tenuipyrone, a novel skeletal polyketide from the entomopathogenic fungus, Isaria tenuipes, cultivated in the presence of epigenetic modifiers. Org. Lett. 2012, 14, 513–515. [Google Scholar] [CrossRef]
- Muhammad, J.S.; Khan, N.A.; Maciver, S.K.; Alharbi, A.M.; Alfahemi, H.; Siddiqui, R. Epigenetic-mediated antimicrobial resistance: Host versus pathogen epigenetic alterations. Antibiotics 2022, 11, 809. [Google Scholar] [CrossRef] [PubMed]
- Dillon, S.C.; Dorman, C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010, 8, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, H.; Cui, T.; Zhou, X.; Jia, Y.; Zhang, H.; He, Z. NapM, a new nucleoid-associated protein, broadly regulates gene expression and affects mycobacterial resistance to anti-tuberculosis drugs. Mol. Microbiol. 2016, 101, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Dadinova, L.A.; Petoukhov, M.V.; Gordienko, A.M.; Manuvera, V.A.; Lazarev, V.N.; Rakitina, T.V.; Mozhaev, A.A.; Peters, G.S.; Shtykova, E.V. Nucleoid-associated proteins HU and IHF: Oligomerization in solution and hydrodynamic properties. Biochemistry 2023, 88, 640–654. [Google Scholar]
- Bradshaw, E.; Saalbach, G.; McArthur, M. Proteomic survey of the Streptomyces coelicolor nucleoid. J. Proteom. 2013, 83, 37–46. [Google Scholar] [CrossRef]
- Dilweg, I.W.; Dame, R.T. Post-translational modification of nucleoid-associated proteins: An extra layer of functional modulation in bacteria? Biochem. Soc. Trans. 2018, 46, 1381–1392. [Google Scholar] [CrossRef]
- van der Valk, R.A.; Vreede, J.; Qin, L.; Moolenaar, G.F.; Hofmann, A.; Goosen, N.; Dame, R.T. Mechanism of environmentally driven conformational changes that modulate H-NS DNA-bridging activity. eLife 2017, 6, e27369. [Google Scholar] [CrossRef]
- Riccardi, E.; van Mastbergen, E.C.; Navarre, W.W.; Vreede, J. Predicting the mechanism and rate of H-NS binding to AT-rich DNA. PLoS Comput. Biol. 2019, 15, e1006845. [Google Scholar] [CrossRef]
- Gordon, B.R.G.; Li, Y.; Wang, L.; Sintsova, A.; van Bakel, H.; Tian, S.H.; Navarre, W.W.; Xia, B.; Liu, J. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 5154–5159. [Google Scholar] [CrossRef]
- Swinger, K.K.; Rice, P.A. IHF and HU: Flexible architects of bent DNA. Curr. Opin. Struc. Biol. 2004, 14, 28–35. [Google Scholar] [CrossRef]
- Peterson, S.N.; Reich, N.O. LRP: A nucleoid-associated protein with gene regulatory functions. In Bacterial Chromatin; Dame, R.T., Dorman, C.J., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 353–364. [Google Scholar]
- Kostrewa, D.; Granzin, J.; Koch, C.; Choe, H.W.; Raghunathan, S.; Wolf, W.; Labahn, J.; Kahmann, R.; Saenger, W. Three-dimensional structure of the E. Coli DNA-binding protein Fis. Nature 1991, 349, 178–180. [Google Scholar] [PubMed]
- Brinkman, A.B.; Ettema, T.J.G.; de Vos, W.M.; van der Oost, J. The Lrp family of transcriptional regulators. Mol. Microbiol. 2003, 48, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Willemse, J.; Erkelens, A.M.; Carrion, V.J.; Dame, R.T.; Van Wezel, G.P. System-wide analysis of the GATC-binding nucleoid-associated protein Gbn and its impact on Streptomyces development. mSystems 2022, 7, e0006122. [Google Scholar] [CrossRef] [PubMed]
- Sunita; Singhvi, N.; Gupta, V.; Singh, Y.; Shukla, P. Computational approaches for the structure-based identification of novel inhibitors targeting nucleoid-associated proteins in Mycobacterium Tuberculosis. Mol. Biotechnol. 2024, 66, 814–823. [Google Scholar] [CrossRef]
- Zhang, X.; Andres, S.N.; Elliot, M.A. Interplay between nucleoid-associated proteins and transcription factors in controlling specialized metabolism in Streptomyces. mBio 2021, 12, e0107721. [Google Scholar] [CrossRef]
- Hindra; Elliot, M.A. A complex regulatory network governs the production of an antibiotic with unusual cell-density-dependence. bioRxiv 2023, 13, 571536. [Google Scholar]
- Gehrke, E.J.; Zhang, X.; Pimentel-Elardo, S.M.; Johnson, A.R.; Rees, C.A.; Jones, S.E.; Hindra; Gehrke, S.S.; Turvey, S.; Boursalie, S.; et al. Silencing cryptic specialized metabolism in Streptomyces by the nucleoid-associated Protein Lsr2. eLife 2019, 8, e47691. [Google Scholar] [CrossRef]
- Deng, L.; Wang, R.; Wang, G.; Liu, M.; Liao, Z.; Liao, G.; Chen, M. Roseosporol A, the first isolation of a novel sesquiterpenoid from Streptomyces roseosporus. Nat. Prod. Res. 2019, 33, 2038–2043. [Google Scholar] [CrossRef]
- Deng, L.; Wang, R.; Wang, G.; Liu, M.; Liao, G.; Liao, Z.; Chen, M. Targeted isolation of sulfur-containing metabolites from Lsr2-deletion mutant strain of Streptomyces roseosporus. RSC Adv. 2017, 7, 37771–37777. [Google Scholar] [CrossRef]
- Strzałka, A.; Mikołajczyk, J.; Kowalska, K.; Skurczyński, M.; Holmes, N.; Jakimowicz, D. The role of two major nucleoid associated proteins in Streptomyces, HupA and HupS, in stress survival and gene expression regulation. bioRxiv 2024, 23, 275. [Google Scholar] [CrossRef]
- Yang, Y.; Song, E.; Willemse, J.; Park, S.H.; Kim, W.S.; Kim, E.J.; Lee, B.R.; Kim, J.N.; van Wezel, G.P.; Kim, B.G. A novel function of Streptomyces integration host factor (sIHF) in the control of antibiotic production and sporulation in Streptomyces coelicolor. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2012, 101, 479–492. [Google Scholar] [CrossRef]
- Facey, P.D.; Hitchings, M.D.; Saavedra-Garcia, P.; Fernandez-Martinez, L.; Dyson, P.J.; Del Sol, R. Streptomyces coelicolor Dps-like proteins: Differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol. Microbiol. 2009, 73, 1186–1202. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, M.; Facey, P.; Francis, L.; Bayliss, S.; Del Sol, R.; Dyson, P. A novel bifunctional histone protein in Streptomyces: A candidate for structural coupling between DNA conformation and transcription during development and stress? Nucleic Acids Res. 2013, 41, 4813–4824. [Google Scholar] [CrossRef] [PubMed]
- Szafran, M.J.; Jakimowicz, D.; Elliot, M.A. Compaction and control-the role of chromosome-organizing proteins in Streptomyces. FEMS Microbiol. Rev. 2020, 44, 725–739. [Google Scholar] [CrossRef]
- Szafran, M.J.; Gongerowska, M.; Małecki, T.; Elliot, M.; Jakimowicz, D. Transcriptional response of Streptomyces coelicolor to rapid chromosome relaxation or long-term supercoiling imbalance. Front. Microbiol. 2019, 10, 1605. [Google Scholar] [CrossRef]
- Salerno, P.; Larsson, J.; Bucca, G.; Laing, E.; Smith, C.P.; Flärdh, K. One of the two genes encoding nucleoid-associated HU proteins in Streptomyces coelicolor is developmentally regulated and specifically involved in spore maturation. J. Bacteriol. 2009, 191, 6489–6500. [Google Scholar] [CrossRef]
- Swiercz, J.P.; Nanji, T.; Gloyd, M.; Guarné, A.; Elliot, M.A. A novel nucleoid-associated protein specific to the actinobacteria. Nucleic Acids Res. 2013, 41, 4171–4184. [Google Scholar] [CrossRef]
- Nanji, T.; Gehrke, E.J.; Shen, Y.; Gloyd, M.; Zhang, X.; Firby, C.D.; Huynh, A.; Razi, A.; Ortega, J.; Elliot, M.A.; et al. Streptomyces IHF uses multiple interfaces to bind DNA. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 129405. [Google Scholar] [CrossRef]
- Szafran, M.J.; Małecki, T.; Strzałka, A.; Pawlikiewicz, K.; Duława, J.; Zarek, A.; Kois-Ostrowska, A.; Findlay, K.C.; Le, T.; Jakimowicz, D. Spatial rearrangement of the Streptomyces venezuelae linear chromosome during sporogenic development. Nat. Commun. 2021, 12, 5222. [Google Scholar] [CrossRef]
- Carabetta, V.J. Addressing the possibility of a histone-like code in bacteria. J. Proteome Res. 2021, 20, 27–37. [Google Scholar] [CrossRef]
- Macek, B.; Forchhammer, K.; Hardouin, J.; Weber-Ban, E.; Grangeasse, C.; Mijakovic, I. Protein post-translational modifications in bacteria. Nat. Rev. Microbiol. 2019, 17, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Arold, S.T.; Leonard, P.G.; Parkinson, G.N.; Ladbury, J.E. H-NS forms a superhelical protein scaffold for DNA condensation. Proc. Natl. Acad. Sci. USA 2010, 107, 15728–15732. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, D.H.; Chen, L. Antimicrobial resistance and mechanisms of epigenetic regulation. Front. Cell Infect. Microbiol. 2023, 13, 1199646. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Xie, L.; Li, X.; Cheng, Z.; Xie, J. Unexpected extensive lysine acetylation in the trump-card antibiotic producer Streptomyces roseosporus revealed by proteome-wide profiling. J. Proteom. 2014, 106, 260–269. [Google Scholar] [CrossRef]
- Manteca, A.; Ye, J.; Sánchez, J.; Jensen, O.N. Phosphoproteonne analysis of development reveals extensive protein phosphorylation accompanying bacterial differentiation. J. Proteome Res. 2011, 10, 5481–5492. [Google Scholar] [CrossRef]
- Sun, C.; Xu, W.; Zhao, Q.; Luo, S.; Chen, X.; Li, Y.; Mao, X. Crotonylation of key metabolic enzymes regulates carbon catabolite repression in Streptomyces roseosporus. Commun. Biol. 2020, 3, 192. [Google Scholar] [CrossRef]
- Zhang, H.; Li, P.; Ren, S.; Cheng, Z.; Zhao, G.; Zhao, W. CobB2-mediated lysine desuccinylation regulates protein biosynthesis and carbon metabolism in Streptomyces coelicolor. Mol. Cell. Proteom. 2019, 18, 2003–2017. [Google Scholar] [CrossRef]
- Bok, J.W.; Chiang, Y.M.; Szewczyk, E.; Reyes-Domingez, Y.; Davidson, A.D.; Sanchez, J.F.; Lo, H.C.; Watanabe, K.; Strauss, J.; Oakley, B.R.; et al. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 2009, 5, 462–464. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Nakayama, T.; Takami, Y. Participation of histones and histone-modifying enzymes in cell functions through alterations in chromatin structure. J. Biochem. 2001, 129, 491–499. [Google Scholar] [CrossRef]
- Shwab, E.K.; Bok, J.W.; Tribus, M.; Galehr, J.; Graessle, S.; Keller, N.P. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 2007, 6, 1656–1664. [Google Scholar] [CrossRef] [PubMed]
- Carr, R.A.; Tucker, T.; Newman, P.M.; Jadalla, L.; Jaludi, K.; Reid, B.E.; Alpheaus, D.N.; Korrapati, A.; Pivonka, A.E.; Carabetta, V.J. Nε-lysine acetylation of the histone-like protein HBsu influences antibiotic survival and persistence in Bacillus subtilis. Front. Microbiol. 2024, 15, 1356733. [Google Scholar] [CrossRef] [PubMed]
- Green, K.D.; Biswas, T.; Pang, A.; Willby, M.J.; Reed, M.S.; Stuchlik, O.; Pohl, J.; Posey, J.E.; Tsodikov, O.V.; Garneau-Tsodikova, S. Acetylation by Eis and deacetylation by Rv1151c of Mycobacterium tuberculosis HupB: Biochemical and structural insight. Biochemistry 2018, 57, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Dulawa-Kobeluszczyk, J.; Strzalka, A.; Tracz, M.; Bartynska, M.; Pawlikiewicz, K.; Lebkowski, T.; Wróbel, S.; Szymczak, J.; Zarek, A.; Malecki, T.; et al. The activity of CobB1 protein deacetylase contributes to nucleoid compaction in Streptomyces venezuelae spores by increasing HupS affinity for DNA. Nucleic Acids Res. 2024, 52, 7112–7128. [Google Scholar] [CrossRef]
- Sun, C.; Li, Y.; Mao, X. Regulation of protein post-translational modifications on metabolism of Actinomycetes. Biomolecules 2020, 10, 1122. [Google Scholar] [CrossRef]
- Li, G.; Kusari, S.; Golz, C.; Laatsch, H.; Strohmann, C.; Spiteller, M. Epigenetic modulation of endophytic eupenicillium sp LG41 by a histone deacetylase inhibitor for production of decalin-containing compounds. J. Nat. Prod. 2017, 80, 983–988. [Google Scholar] [CrossRef]
- Wang, C.; Huang, D.; Liang, S. Identification and metabolomic analysis of chemical elicitors for tacrolimus accumulation in Streptomyces tsukubaensis. Appl. Microbiol. Biotechnol. 2018, 102, 7541–7553. [Google Scholar] [CrossRef]
- Zheng, J.; Li, Y.; Liu, N.; Zhang, J.; Liu, S.J.; Tan, H. Multi-omics data reveal the effect of sodium butyrate on gene expression and protein modification in Streptomyces. Genom. Proteom. Bioinform. 2023, 21, 1149–1162. [Google Scholar] [CrossRef]
- Nützmann, H.W.; Reyes-Dominguez, Y.; Scherlach, K.; Schroeckh, V.; Horn, F.; Gacek, A.; Schümann, J.; Hertweck, C.; Strauss, J.; Brakhage, A.A. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc. Natl. Acad. Sci. USA 2011, 108, 14282–14287. [Google Scholar] [CrossRef]
- Mathur, S.; Hoskins, C. Drug development: Lessons from nature. Biomed. Rep. 2017, 66, 612–614. [Google Scholar] [CrossRef]
- Bind, S.; Bind, S.; Sharma, A.K.; Chaturvedi, P. Epigenetic modification: A key tool for secondary metabolite production in microorganisms. Front. Microbiol. 2022, 13, 1122. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, W.; Shao, C.; Chi, Z.; Wang, C. DNA methyltransferase inhibitor induced fungal biosynthetic products: Diethylene glycol phthalate ester oligomers from the marine-derived fungus. Mar. Biotechnol. 2016, 18, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Zhuang, Z.; Zhang, F.; Song, F.; Zhong, H.; Ran, F.; Yu, S.; Xu, G.; Lan, F.; Wang, S. Inhibition of aflatoxin metabolism and growth of Aspergillus flavus in liquid culture by a DNA methylation inhibitor. Food Addit. Contam. A 2015, 32, 554–563. [Google Scholar] [CrossRef] [PubMed]
- De Ruijter, A.; Van Gennip, A.H.; Caron, H.N.; Kemp, S.; Van Kuilenburg, A. Histone deacetylases (HDACs): Characterization of the classical HDAC family. Biochem. J. 2003, 370, 737–749. [Google Scholar] [CrossRef]
- Xue, M.; Hou, X.; Fu, J.; Zhang, J.; Wang, J.; Zhao, Z.; Xu, D.; Lai, D.; Zhou, L. Recent advances in search of bioactive secondary metabolites from fungi triggered by chemical epigenetic modifiers. J. Fungi. 2023, 9, 172. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, N.; Hwang, S.; Kim, W.; Lee, Y.; Cho, S.; Palsson, B.O.; Cho, B.K. Discovery of novel secondary metabolites encoded in actinomycete genomes through coculture. J. Ind. Microbiol. Biotechnol. 2021, 48, kuaa001. [Google Scholar] [CrossRef]
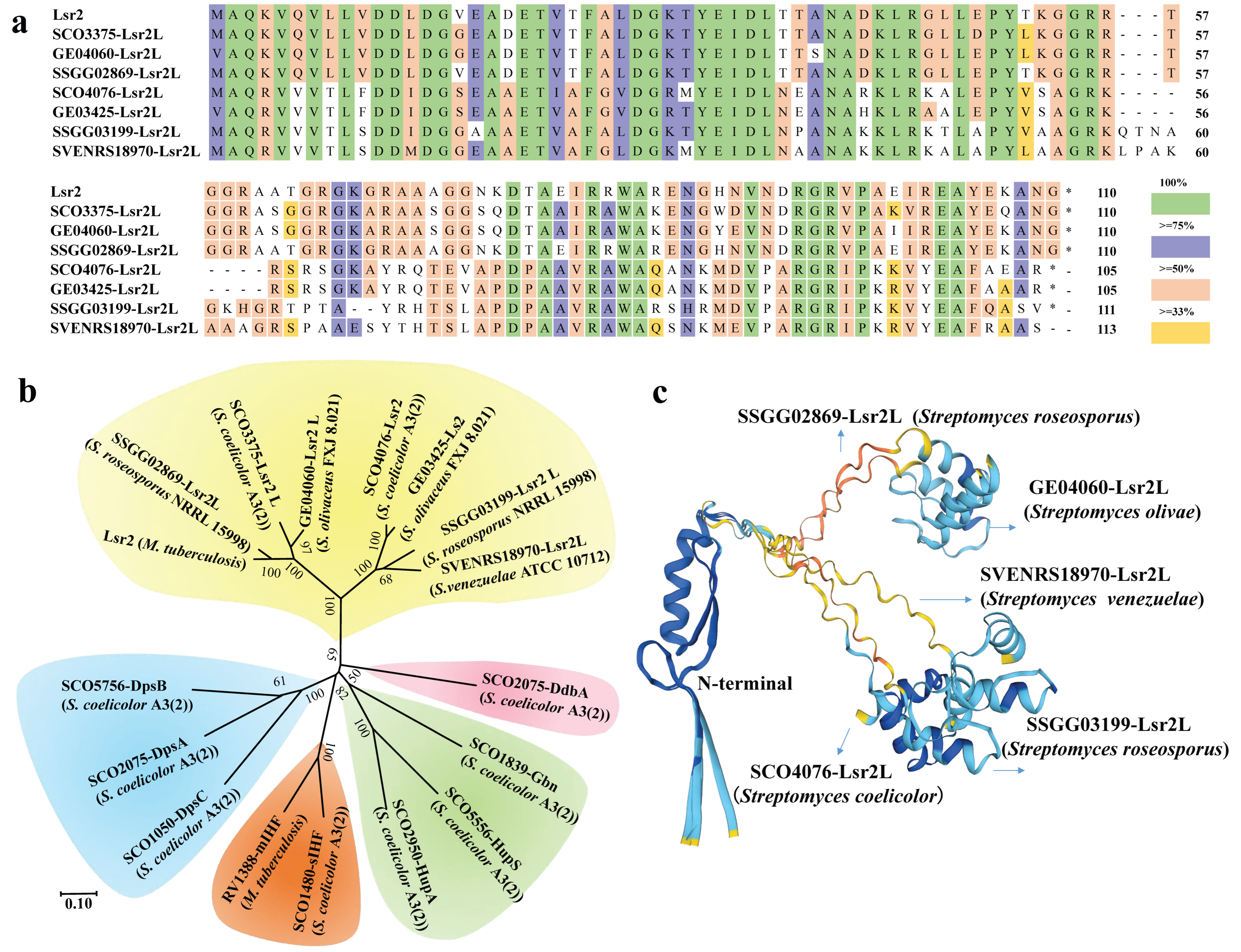
| NAP Types | Proteins | Strain Resources (Example) | Effects on SMs Production | References |
|---|---|---|---|---|
| Lsr2 | SVEN-RS18970, SSGG02869, SSGG03199, GE04060, GE03425, SCO3375, SCO4076, | Streptomyces venezuelae, Streptomyces sp. WAC07094, Streptomyces roseosporus, Streptomyces olivaceus, Streptomyces coelicolor | Chloramphenicol/Venemycin/CI-venemycin/Thiazostatin/Watasemycin, Saquayamycin, Roseosporol A/pyrismycins A-F, Lobophorins | [26,27,28,29,30] |
| HupA/ HupS | SCO2950, SCO5556 | Streptomyces coelicolor | Carotenoid compounds, undecyloprodigiosin, ectoine, | [31] |
| sIHF | SCO1480 | Streptomyces coelicolor | Actinorhodin, undecylprodigiosin | [32] |
| Gbn | SCO1839 | Streptomyces coelicolor | Actinorhodin, sporulation | [24] |
| Dps-like proteins | SCO0596, SCO5756, SCO1050 | Streptomyces coelicolor | Condensation of spore nucleoids | [33] |
| DdbA | SCO2075 | Streptomyces coelicolor | Modulate RNA polymerase activity combined with ppGpp, organize DNA conformation in response to stress | [34] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, X.; Zhang, J.; Zheng, J.; Liao, G.; Ding, Y.; Li, Y. Important Role of Bacterial Nucleoid-Associated Proteins in Discovery of Novel Secondary Metabolites. Int. J. Mol. Sci. 2025, 26, 2393. https://doi.org/10.3390/ijms26062393
Xia X, Zhang J, Zheng J, Liao G, Ding Y, Li Y. Important Role of Bacterial Nucleoid-Associated Proteins in Discovery of Novel Secondary Metabolites. International Journal of Molecular Sciences. 2025; 26(6):2393. https://doi.org/10.3390/ijms26062393
Chicago/Turabian StyleXia, Xiulei, Jihui Zhang, Jiazhen Zheng, Guojian Liao, Yanqin Ding, and Yue Li. 2025. "Important Role of Bacterial Nucleoid-Associated Proteins in Discovery of Novel Secondary Metabolites" International Journal of Molecular Sciences 26, no. 6: 2393. https://doi.org/10.3390/ijms26062393
APA StyleXia, X., Zhang, J., Zheng, J., Liao, G., Ding, Y., & Li, Y. (2025). Important Role of Bacterial Nucleoid-Associated Proteins in Discovery of Novel Secondary Metabolites. International Journal of Molecular Sciences, 26(6), 2393. https://doi.org/10.3390/ijms26062393






