New Hepatocellular Carcinoma (HCC) Primary Cell Cultures as Models for Exploring Personalized Anti-TGF-β Therapies Based on Tumor Characteristics
Abstract
1. Introduction
2. Results
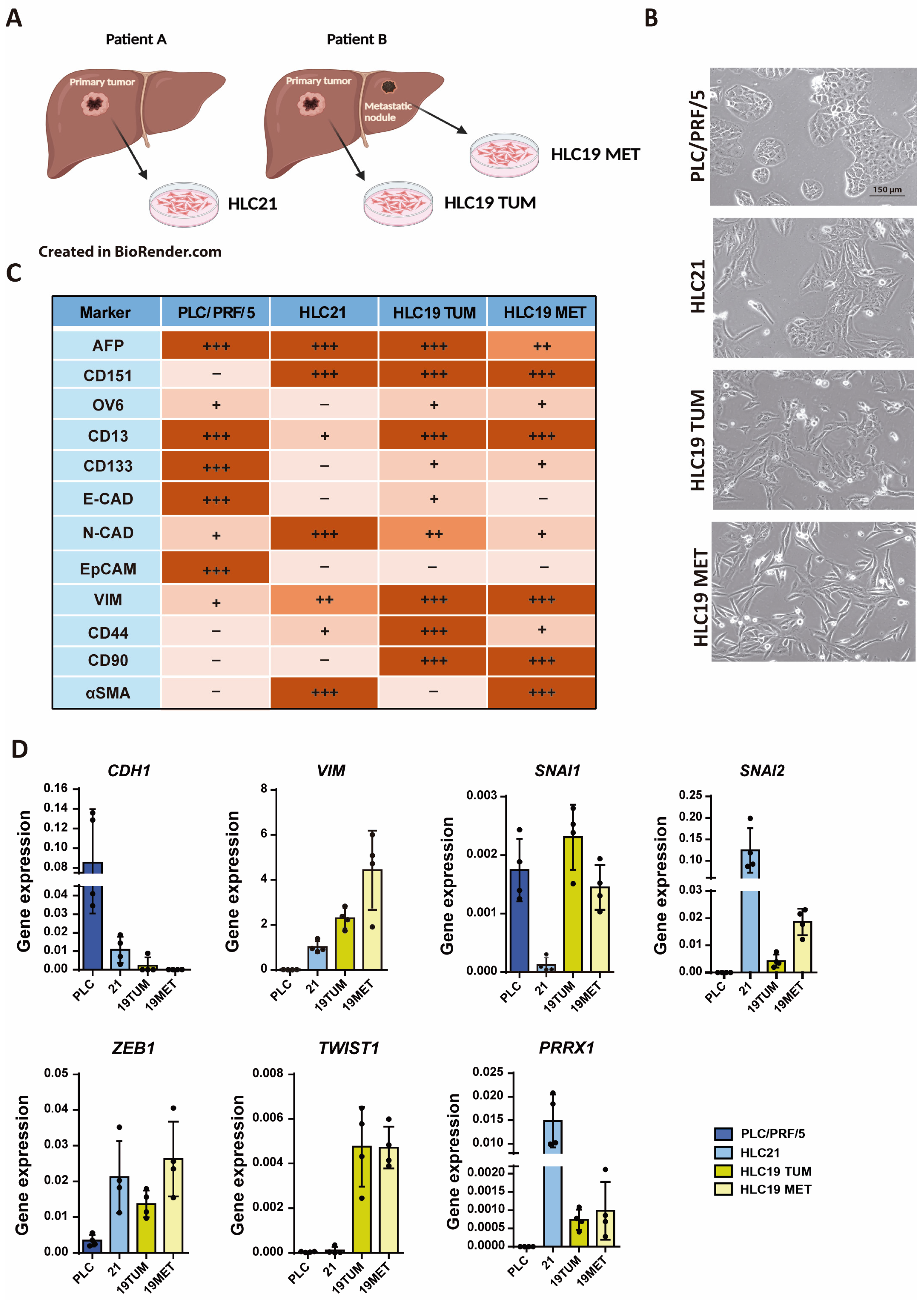
2.1. Characterization of New Liver Tumor Cells Isolated from HCC Patients
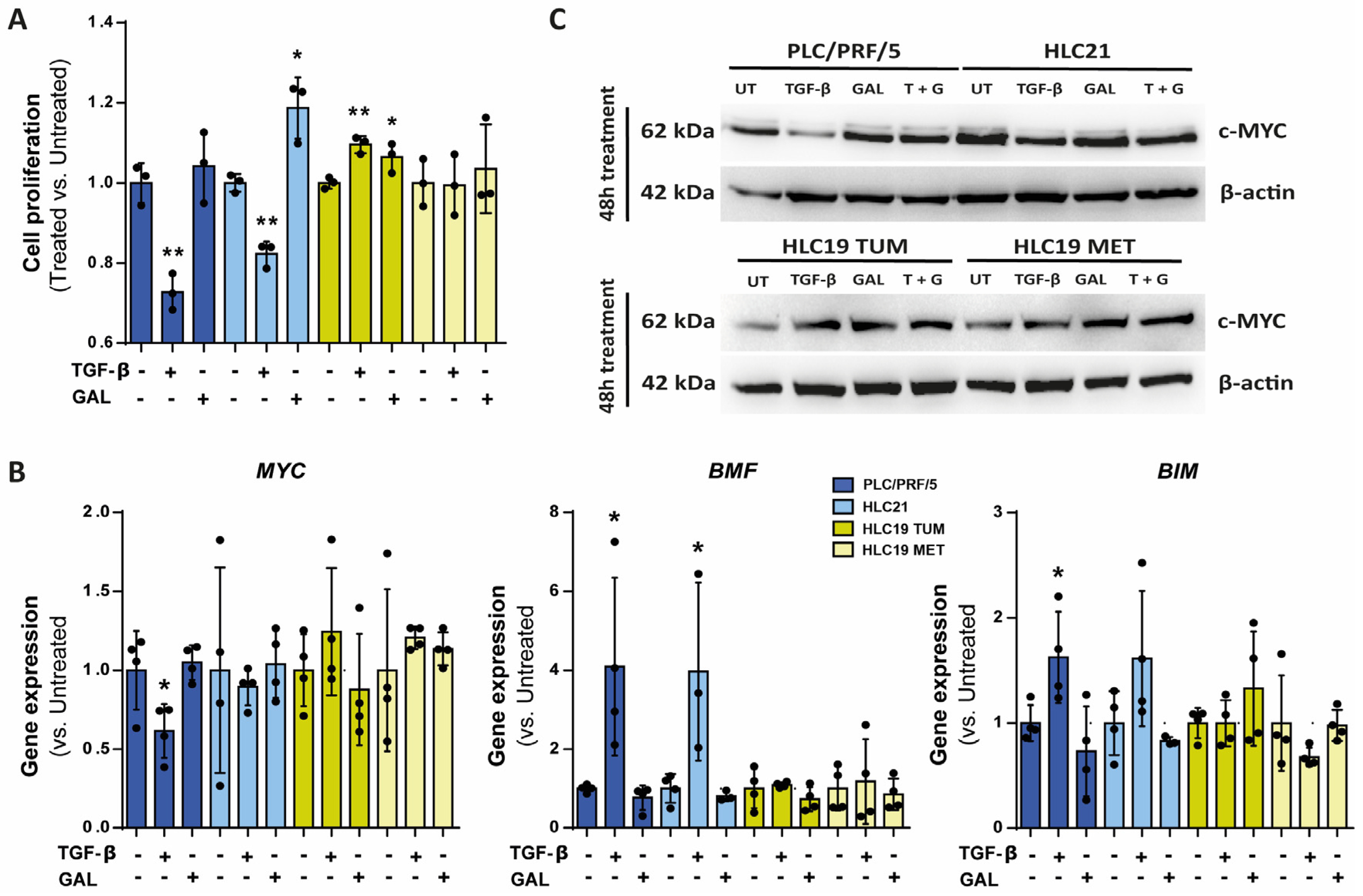
2.2. Response to TGF-β in Terms of SMAD Phosphorylation, Growth Inhibition and Apoptosis
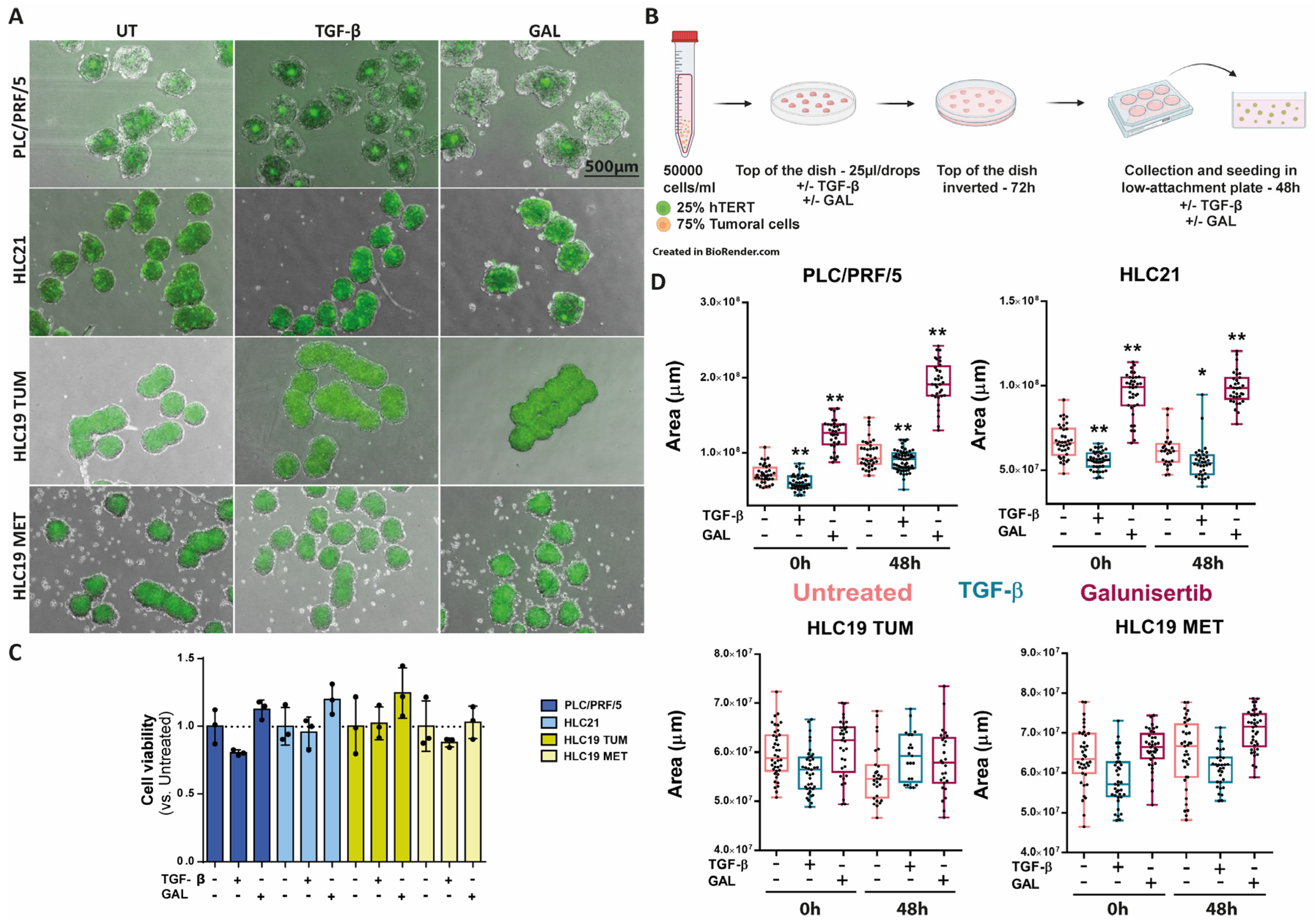
2.3. Response to TGF-β in Terms of Cell Migration
2.4. Response to TGF-β in 3D Cultures
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Reagents, and Ethics Statement
4.2. Isolation and Characterization of the Primary HCC Cell Lines
4.3. Flow Cytometry
4.4. Cell Viability Analysis
4.4.1. Crystal Violet Staining
4.4.2. MTS Assay
4.5. Transwell Migration Assay
4.6. Immunofluorescence and Image Acquisition
4.7. Formation of 3D Spheroid Cultures
4.8. Western Blot Analysis
4.9. Gene Expression Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Massague, J.; Sheppard, D. TGF-beta signaling in health and disease. Cell 2023, 186, 4007–4037. [Google Scholar] [CrossRef] [PubMed]
- Massague, J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Sanchez, E.; Gonzalez-Sanchez, E.; Vaquero, J.; Férnandez-Barrena, M.G.; Lasarte, J.J.; Avila, M.A.; Sarobe, P.; Reig, M.; Calvo, M.; Fabregat, I. The TGF-beta Pathway: A Pharmacological Target in Hepatocellular Carcinoma? Cancers 2021, 13, 3248. [Google Scholar] [CrossRef]
- Carr, B.I.; Carr, B.I.; Hayashi, I.; Branum, E.L.; Moses, H.L. Inhibition of DNA synthesis in rat hepatocytes by platelet-derived type beta transforming growth factor. Cancer Res. 1986, 46, 2330–2334. [Google Scholar]
- Sánchez, A.; Alvarez, A.M.; Benito, M.; Fabregat, I. Apoptosis induced by transforming growth factor-beta in fetal hepatocyte primary cultures: Involvement of reactive oxygen intermediates. J. Biol. Chem. 1996, 271, 7416–7422. [Google Scholar] [CrossRef]
- Caja, L.; Sancho, P.; Bertran, E.; Fabregat, I. Dissecting the effect of targeting the epidermal growth factor receptor on TGF-beta-induced-apoptosis in human hepatocellular carcinoma cells. J. Hepatol. 2011, 55, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Caja, L.; Sancho, P.; Bertran, E.; Iglesias-Serret, D.; Gil, J.; Fabregat, I. Overactivation of the MEK/ERK pathway in liver tumor cells confers resistance to TGF-beta-induced cell death through impairing up-regulation of the NADPH oxidase NOX4. Cancer Res. 2009, 69, 7595–7602. [Google Scholar] [CrossRef]
- Malfettone, A.; Soukupova, J.; Bertran, E.; Crosas-Molist, E.; Lastra, R.; Fernando, J.; Koudelkova, P.; Rani, B.; Fabra, Á.; Serrano, T.; et al. Transforming growth factor-beta-induced plasticity causes a migratory stemness phenotype in hepatocellular carcinoma. Cancer Lett. 2017, 392, 39–50. [Google Scholar] [CrossRef]
- Yoshida, K.; Matsuzaki, K.; Murata, M.; Yamaguchi, T.; Suwa, K.; Okazaki, K. Clinico-Pathological Importance of TGF-beta/Phospho-Smad Signaling during Human Hepatic Fibrocarcinogenesis. Cancers 2018, 10, 183. [Google Scholar] [CrossRef]
- Caja, L.; Dituri, F.; Mancarella, S.; Caballero-Diaz, D.; Moustakas, A.; Giannelli, G.; Fabregat, I. TGF-beta and the Tissue Microenvironment: Relevance in Fibrosis and Cancer. Int. J. Mol. Sci. 2018, 19, 1294. [Google Scholar] [CrossRef]
- Thangaraj, J.L.; Coffey, M.; Lopez, E.; Kaufman, D.S. Disruption of TGF-beta signaling pathway is required to mediate effective killing of hepatocellular carcinoma by human iPSC-derived NK cells. Cell Stem Cell 2024, 31, 1327–1343.e5. [Google Scholar] [CrossRef] [PubMed]
- Piqué-Gili, M.; Andreu-Oller, C.; Mesropian, A.; Esteban-Fabró, R.; Bárcena-Varela, M.; de Galarreta, M.R.; Montironi, C.; Martinez-Quetglas, I.; Cappuyns, S.; Peix, J.; et al. Oncogenic role of PMEPA1 and its association with immune exhaustion and TGF-beta activation in HCC. JHEP Rep. 2024, 6, 101212. [Google Scholar] [CrossRef]
- Giannelli, G.; Mikulits, W.; Dooley, S.; Fabregat, I.; Moustakas, A.; ten Dijke, P.; Portincasa, P.; Winter, P.; Janssen, R.; Leporatti, S.; et al. The rationale for targeting TGF-beta in chronic liver diseases. Eur. J. Clin. Investig. 2016, 46, 349–361. [Google Scholar] [CrossRef]
- Kelley, R.K.; Gane, E.; Assenat, E.; Siebler, J.; Galle, P.R.; Merle, P.; Hourmand, I.O.; Cleverly, A.; Zhao, Y.; Gueorguieva, I.; et al. A Phase 2 Study of Galunisertib (TGF-beta1 Receptor Type I Inhibitor) and Sorafenib in Patients With Advanced Hepatocellular Carcinoma. Clin. Transl. Gastroenterol. 2019, 10, e00056. [Google Scholar] [CrossRef]
- Faivre, S.; Santoro, A.; Kelley, R.K.; Gane, E.D.; Costentin, C.E.; Gueorguieva, I.; Smith, C.; Cleverly, A.; Lahn, M.M.; Raymond, E.; et al. Novel transforming growth factor beta receptor I kinase inhibitor galunisertib (LY2157299) in advanced hepatocellular carcinoma. Liver Int. 2019, 39, 1468–1477. [Google Scholar] [CrossRef]
- Dituri, F.; Scialpi, R.; Schmidt, T.A.; Frusciante, M.; Mancarella, S.; Lupo, L.G.; Villa, E.; Giannelli, G. Proteoglycan-4 is correlated with longer survival in HCC patients and enhances sorafenib and regorafenib effectiveness via CD44 In Vitro. Cell Death Dis. 2020, 11, 984. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Nguyen, N.T.; Eun, J.R.; Zhang, Y.; Jung, Y.J.; Tschudy-Seney, B.; Trotsyuk, A.; Lam, A.; Ramsamooj, R.; Zhang, Y.; et al. Identification of cancer stem cell subpopulations of CD34(+) PLC/PRF/5 that result in three types of human liver carcinomas. Stem Cells Dev. 2015, 24, 1008–1021. [Google Scholar] [CrossRef] [PubMed]
- Ke, A.W.; Shi, G.M.; Zhou, J.; Huang, X.Y.; Shi, Y.H.; Ding, Z.B.; Wang, X.Y.; Devbhandari, R.P.; Fan, J. CD151 amplifies signaling by integrin alpha6beta1 to PI3K and induces the epithelial-mesenchymal transition in HCC cells. Gastroenterology 2011, 140, 1629–1641.e15. [Google Scholar] [CrossRef]
- Wang, S.; Zhu, M.; Wang, Q.; Hou, Y.; Li, L.; Weng, H.; Zhao, Y.; Chen, D.; Ding, H.; Guo, J.; et al. Alpha-fetoprotein inhibits autophagy to promote malignant behaviour in hepatocellular carcinoma cells by activating PI3K/AKT/mTOR signalling. Cell Death Dis. 2018, 9, 1027. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- Şïmşek, G.Ö. Immunotherapy in the Treatment of Cancer: Today and Tomorrow. Curr. Mol. Biol. Rep. 2024, 10, 54–64. [Google Scholar] [CrossRef]
- van den Bulk, J.; de Miranda, N.; Ten Dijke, P. Therapeutic targeting of TGF-beta in cancer: Hacking a master switch of immune suppression. Clin. Sci. 2021, 135, 35–52. [Google Scholar] [CrossRef]
- Piorońska, W.; Nwosu, Z.C.; Han, M.; Büttner, M.; Ebert, M.P.; Dooley, S.; Meyer, C. Dysregulated paired related homeobox 1 impacts on hepatocellular carcinoma phenotypes. BMC Cancer 2021, 21, 1006. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Shen, J.; Liu, H.; Wen, Z.; Yang, J.; Yang, P.; Liu, K.; Chang, Y.; Duan, J.; Lu, K. Downregulation of PRRX1 via the p53-dependent signaling pathway predicts poor prognosis in hepatocellular carcinoma. Oncol. Rep. 2017, 38, 1083–1090. [Google Scholar] [CrossRef]
- Meng, J.; Chen, S.; Han, J.X.; Qian, B.; Wang, X.R.; Zhong, W.L.; Qin, Y.; Zhang, H.; Gao, W.F.; Lei, Y.Y.; et al. Twist1 Regulates Vimentin through Cul2 Circular RNA to Promote EMT in Hepatocellular Carcinoma. Cancer Res. 2018, 78, 4150–4162. [Google Scholar] [CrossRef]
- Hirata, H.; Sugimachi, K.; Takahashi, Y.; Ueda, M.; Sakimura, S.; Uchi, R.; Kurashige, J.; Takano, Y.; Nanbara, S.; Komatsu, H.; et al. Downregulation of PRRX1 Confers Cancer Stem Cell-Like Properties and Predicts Poor Prognosis in Hepatocellular Carcinoma. Ann. Surg. Oncol. 2015, 22 (Suppl. S3), S1402–S1409. [Google Scholar] [CrossRef]
- Park, S.A.; Kim, M.J.; Park, S.Y.; Kim, J.S.; Lim, W.; Nam, J.S.; Yhong Sheen, Y. TIMP-1 mediates TGF-beta-dependent crosstalk between hepatic stellate and cancer cells via FAK signaling. Sci. Rep. 2015, 5, 16492. [Google Scholar] [CrossRef] [PubMed]
- Morén, A.; Bellomo, C.; Tsubakihara, Y.; Kardassis, D.; Mikulits, W.; Heldin, C.H.; Moustakas, A. LXRalpha limits TGFbeta-dependent hepatocellular carcinoma associated fibroblast differentiation. Oncogenesis 2019, 8, 36. [Google Scholar] [CrossRef]
- Akkiz, H. Emerging Role of Cancer-Associated Fibroblasts in Progression and Treatment of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 3941. [Google Scholar] [CrossRef]
- Angulo, P.; Keach, J.C.; Batts, K.P.; Lindor, K.D. Independent predictors of liver fibrosis in patients with nonalcoholic steatohepatitis. Hepatology 1999, 30, 1356–1362. [Google Scholar] [CrossRef]
- Rani, B.; Malfettone, A.; Dituri, F.; Soukupova, J.; Lupo, L.; Mancarella, S.; Fabregat, I.; Giannelli, G. Galunisertib suppresses the staminal phenotype in hepatocellular carcinoma by modulating CD44 expression. Cell Death Dis. 2018, 9, 373. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Sharma, H.; Chowdhury, S.; Chowdhury, R.; Mukherjee, S. Transforming growth factor- beta mediated regulation of epigenome is required for epithelial to mesenchymal transition associated features in liver cancer cells. Heliyon 2023, 9, e14665. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, Á.M.; Palao, N.; Bragado, P.; Gutierrez-Uzquiza, A.; Herrera, B.; Sánchez, A.; Porras, A. New and Old Key Players in Liver Cancer. Int. J. Mol. Sci. 2023, 24, 17152. [Google Scholar] [CrossRef] [PubMed]
- Schnabl, B.; Choi, Y.H.; Olsen, J.C.; Hagedorn, C.H.; Brenner, D.A. Immortal activated human hepatic stellate cells generated by ectopic telomerase expression. Lab. Investig. 2002, 82, 323–333. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scialpi, R.; Espinosa-Sotelo, R.; Bertran, E.; Dituri, F.; Giannelli, G.; Fabregat, I. New Hepatocellular Carcinoma (HCC) Primary Cell Cultures as Models for Exploring Personalized Anti-TGF-β Therapies Based on Tumor Characteristics. Int. J. Mol. Sci. 2025, 26, 2430. https://doi.org/10.3390/ijms26062430
Scialpi R, Espinosa-Sotelo R, Bertran E, Dituri F, Giannelli G, Fabregat I. New Hepatocellular Carcinoma (HCC) Primary Cell Cultures as Models for Exploring Personalized Anti-TGF-β Therapies Based on Tumor Characteristics. International Journal of Molecular Sciences. 2025; 26(6):2430. https://doi.org/10.3390/ijms26062430
Chicago/Turabian StyleScialpi, Rosanna, Rut Espinosa-Sotelo, Esther Bertran, Francesco Dituri, Gianluigi Giannelli, and Isabel Fabregat. 2025. "New Hepatocellular Carcinoma (HCC) Primary Cell Cultures as Models for Exploring Personalized Anti-TGF-β Therapies Based on Tumor Characteristics" International Journal of Molecular Sciences 26, no. 6: 2430. https://doi.org/10.3390/ijms26062430
APA StyleScialpi, R., Espinosa-Sotelo, R., Bertran, E., Dituri, F., Giannelli, G., & Fabregat, I. (2025). New Hepatocellular Carcinoma (HCC) Primary Cell Cultures as Models for Exploring Personalized Anti-TGF-β Therapies Based on Tumor Characteristics. International Journal of Molecular Sciences, 26(6), 2430. https://doi.org/10.3390/ijms26062430







