Integrated Metabolomic and Transcriptomic Analysis Reveals the Pharmacological Effects and Differential Mechanisms of Isoflavone Biosynthesis in Four Species of Glycyrrhiza
Abstract
1. Introduction
2. Results and Discussion
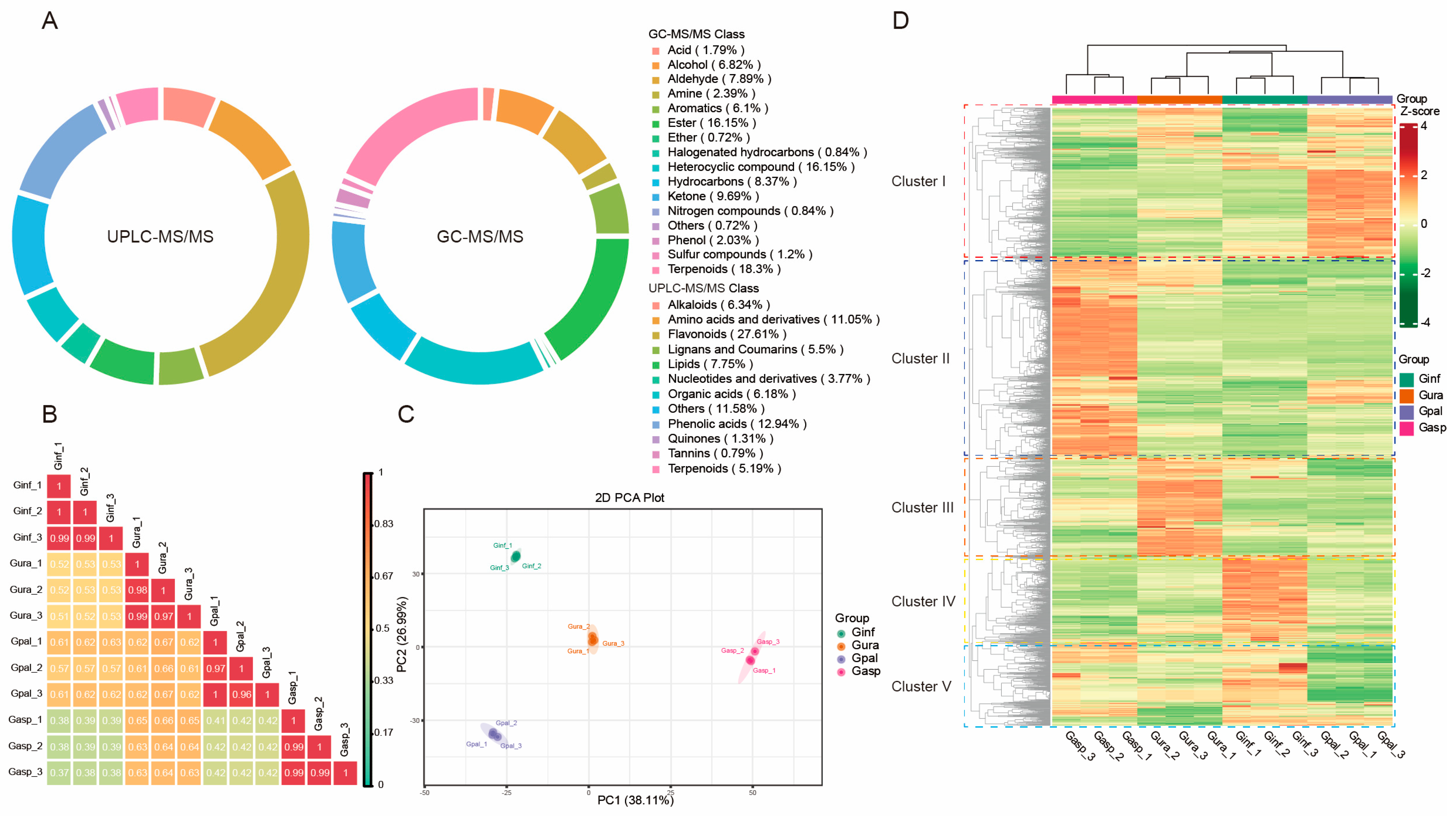
2.1. Detection of Metabolites in the Roots of Four Glycyrrhiza Species
2.2. Multivariate Statistical Analysis
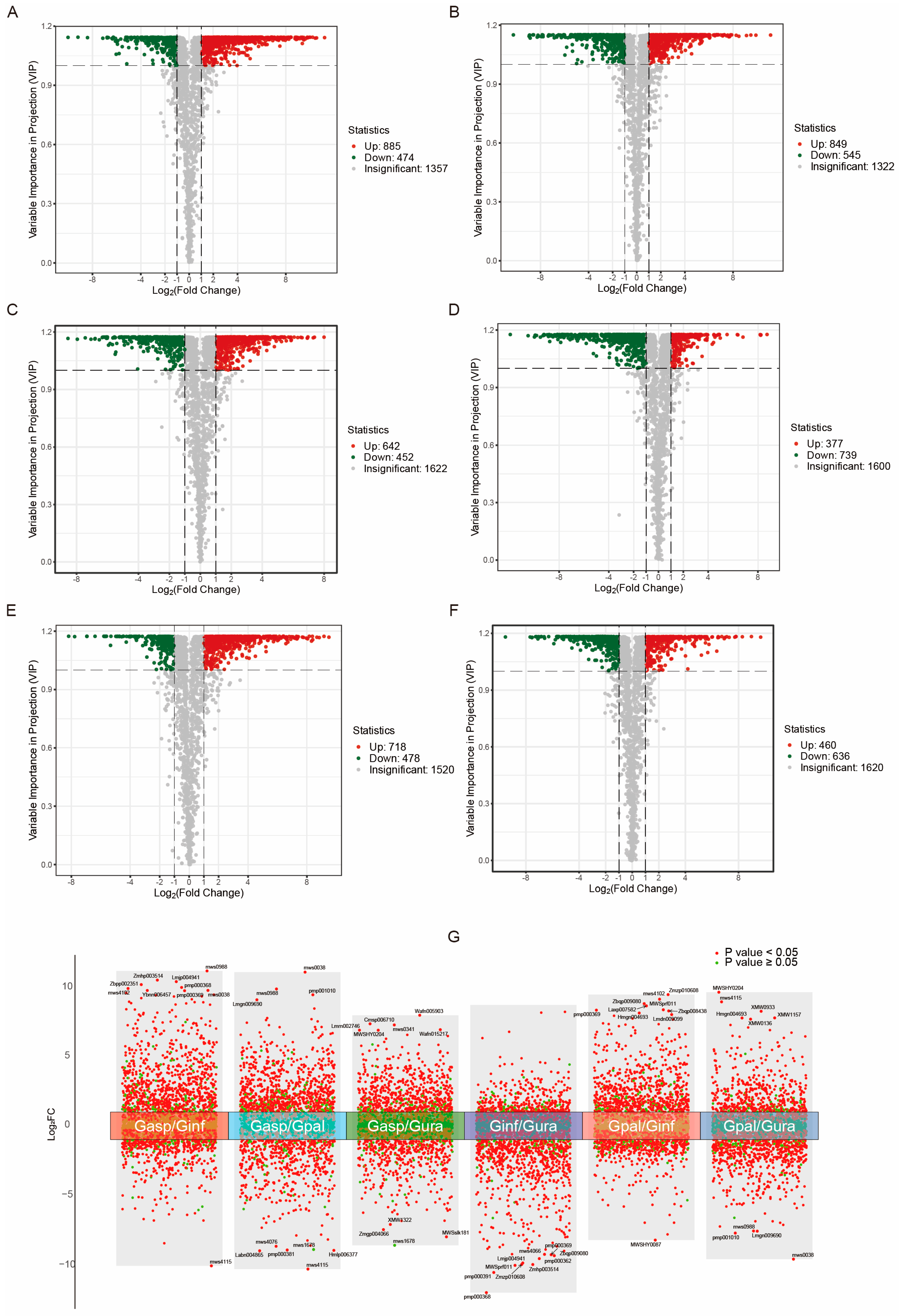
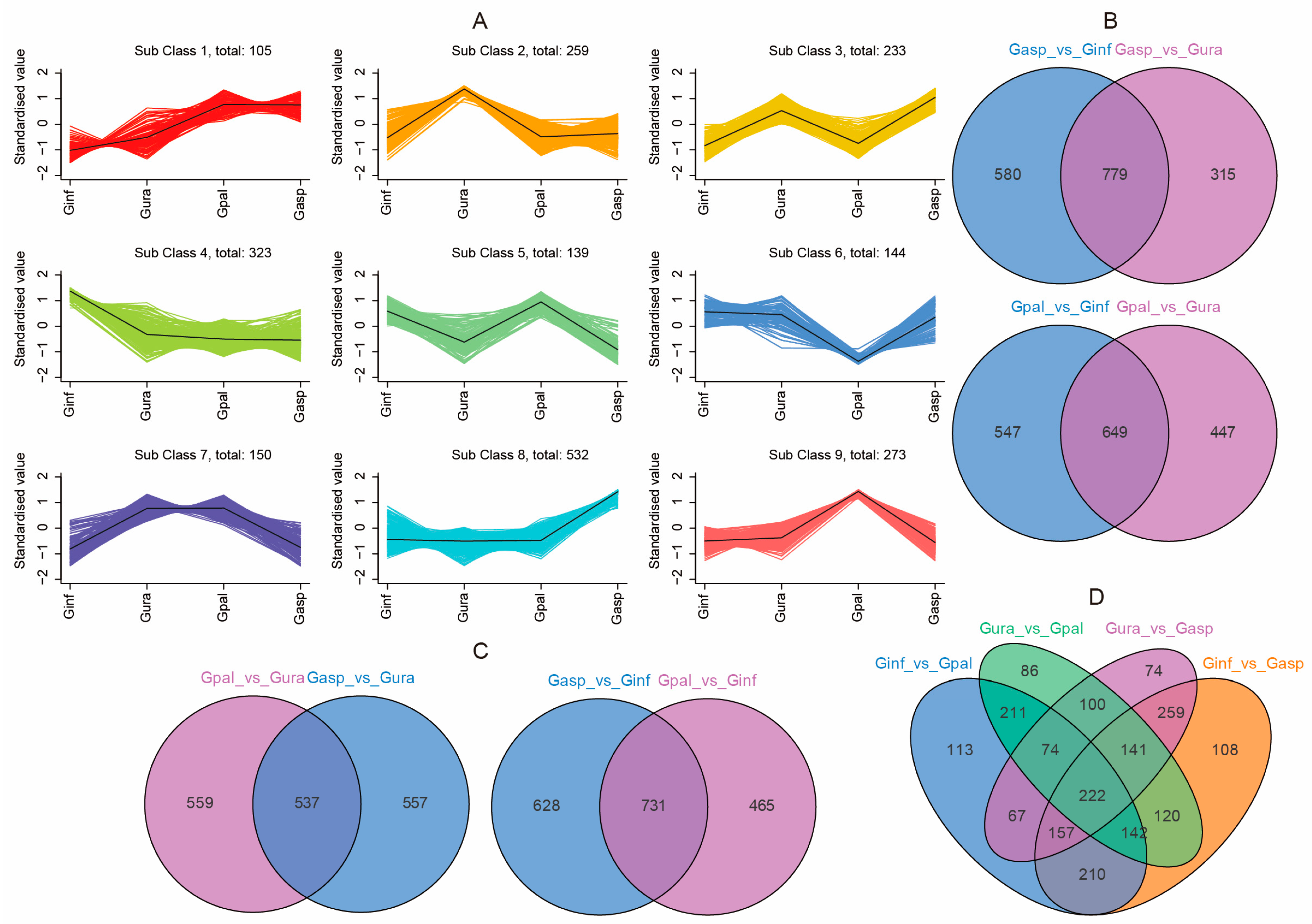
2.3. Differential Analysis of Metabolites in Different Licorice Roots
2.4. Network Pharmacology of Licorice
2.4.1. Major Medicinal Components of Licorice
2.4.2. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis
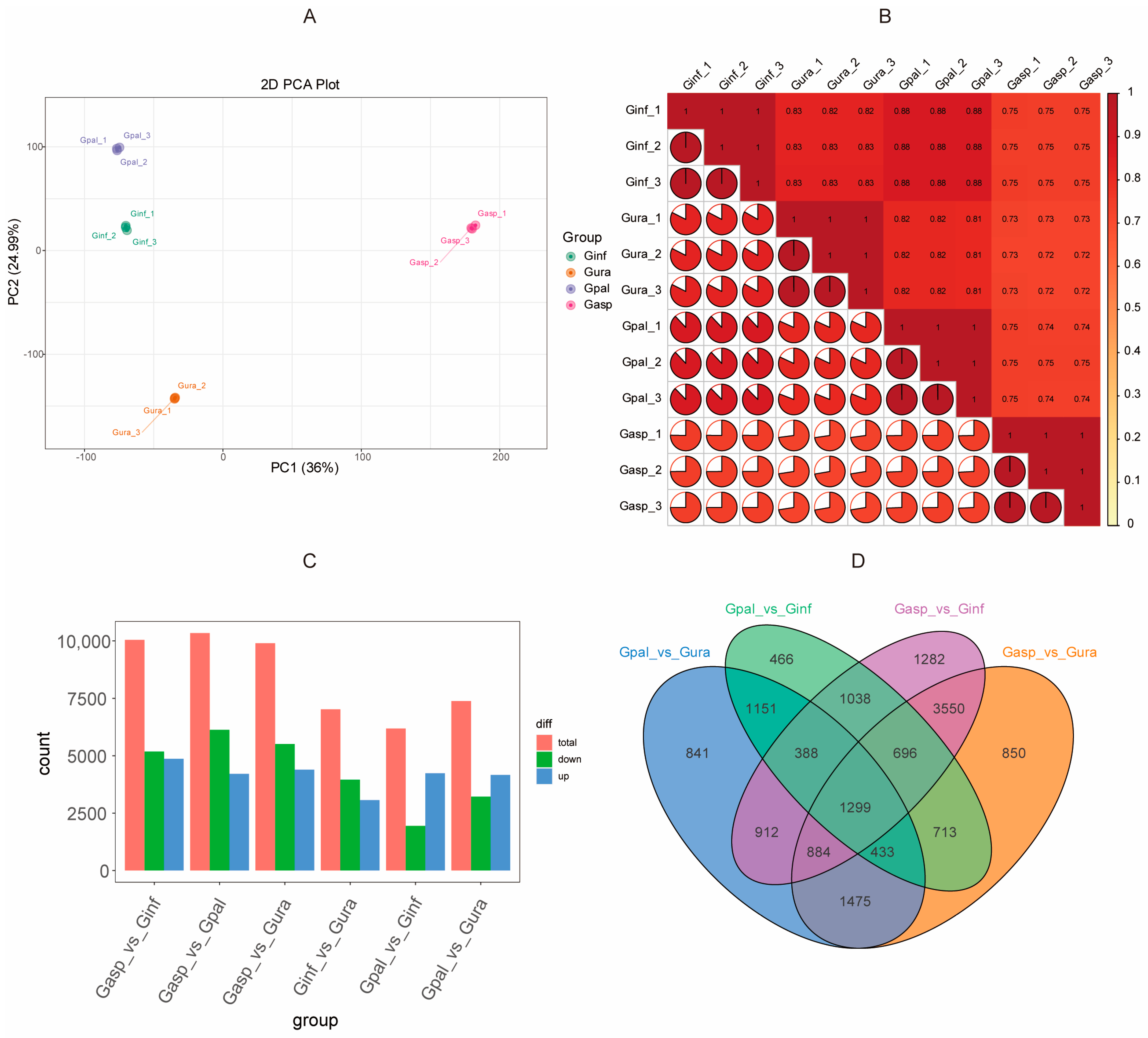
2.5. Transcriptome Analysis of Four Glycyrrhiza Species
2.6. Transcriptome Differences Among Four Glycyrrhiza Species
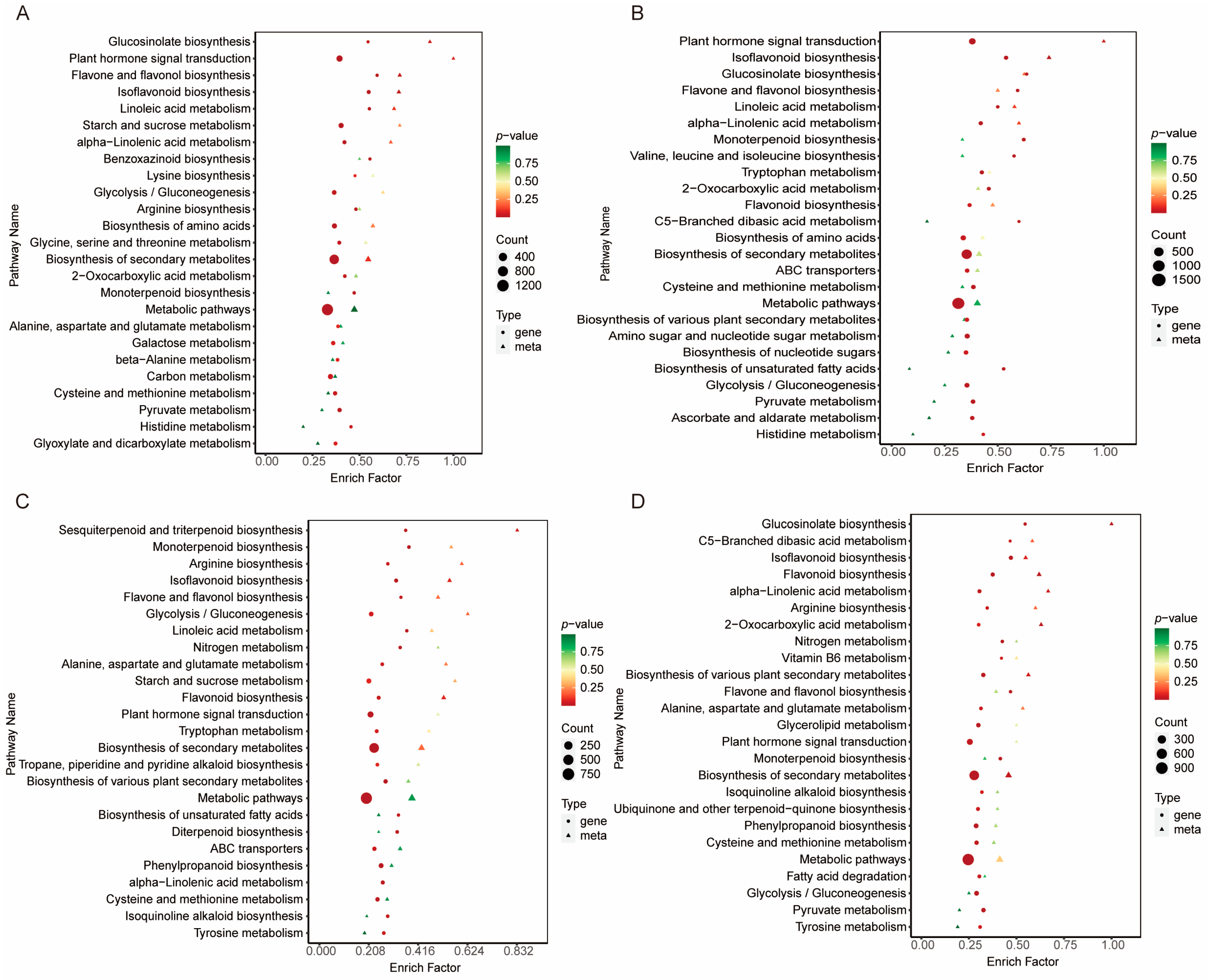
2.7. Functional and Enrichment Analysis of Differential Metabolites and Differentially Expressed Genes
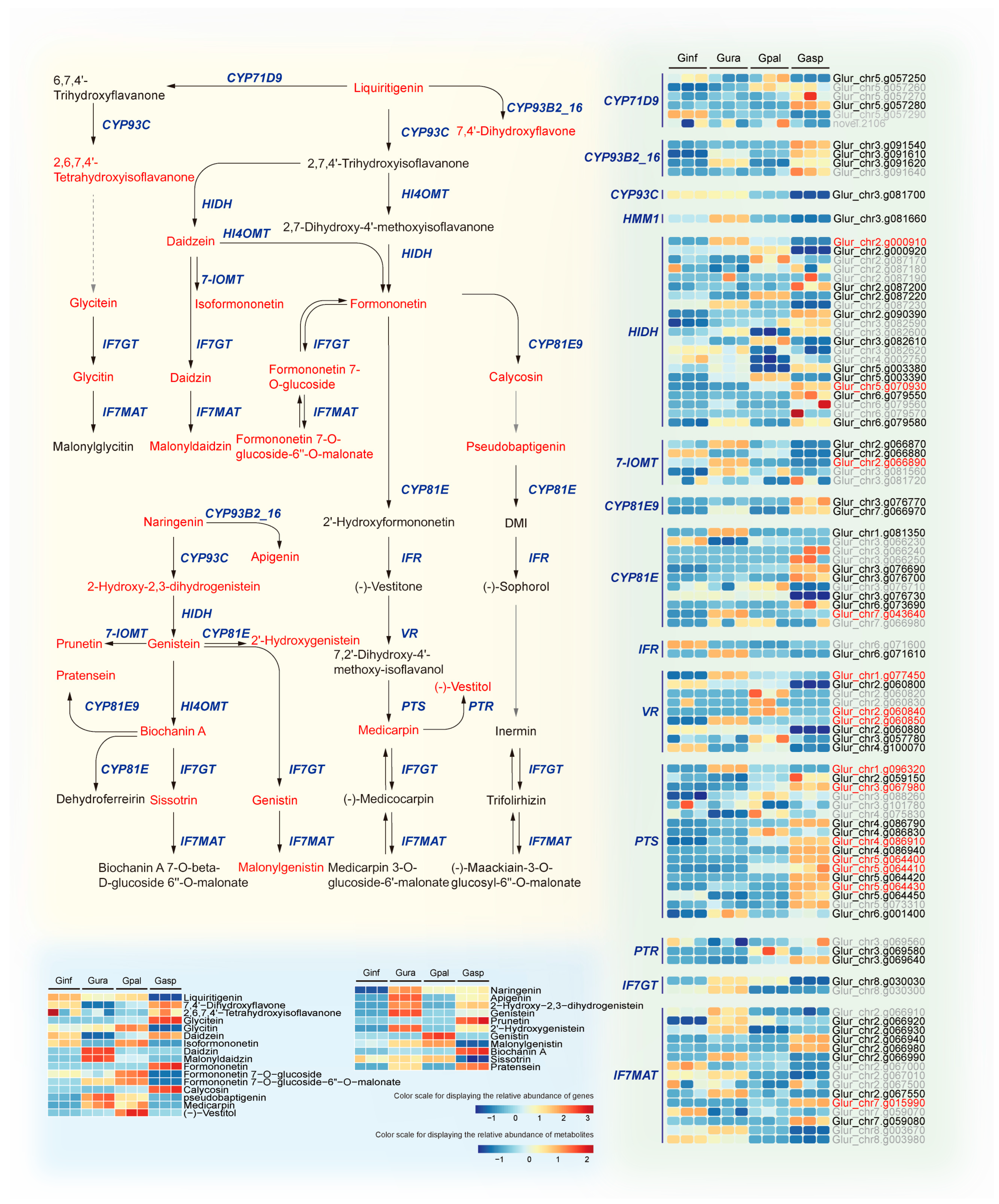
2.8. Integrative Transcriptomics and Metabolomics Analysis Reveals Isoflavonoid Biosynthesis Mechanisms in Different Glycyrrhiza Species
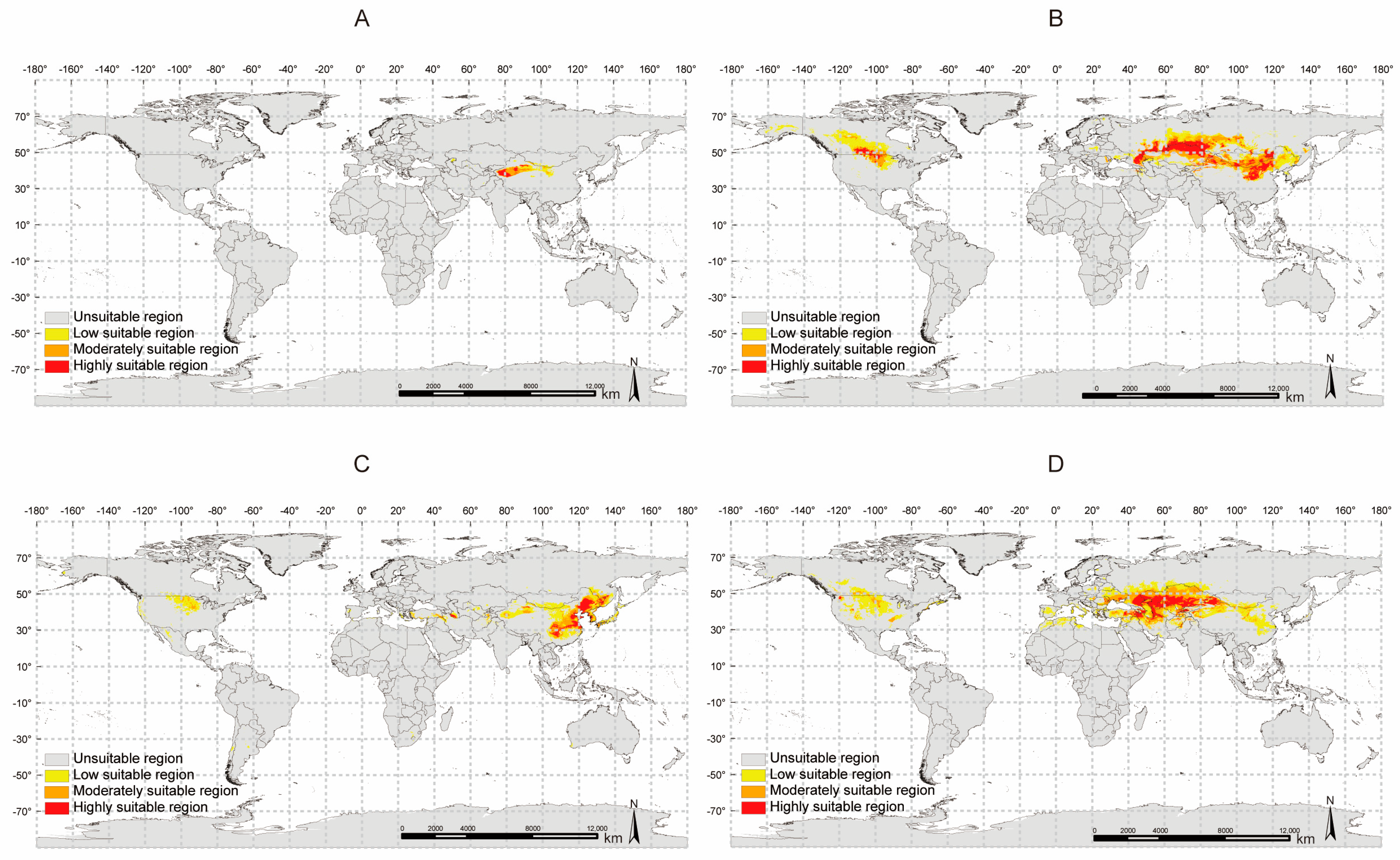
2.9. Potential Distribution Analysis of Four Glycyrrhiza Species
3. Materials and Methods
3.1. Plant Materials
3.2. Metabolomics Analysis
3.2.1. Sample Extraction
3.2.2. Metabolite Collection and Qualitative and Quantitative Analysis
3.3. Network Pharmacology
3.3.1. Prediction of the Potential Active Ingredient
3.3.2. Prediction of Diseases and Core Targets
3.3.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
3.4. Transcriptome Analysis
3.5. Potential Distribution Prediction
3.5.1. Species Distribution and Environmental Data Collection and Processing
3.5.2. Bioclimatic Suitability Zone Prediction Based on MaxEnt Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duan, L.; Han, L.-N.; Liu, B.-B.; Leostrin, A.; Harris, A.; Wang, L.; Arslan, E.; Ertuğrul, K.; Knyazev, M.; Hantemirova, E.; et al. Species Delimitation of the Liquorice Tribe (Leguminosae: Glycyrrhizeae) Based on Phylogenomic and Machine Learning Analyses. J. Syst. Evol. 2023, 61, 22–41. [Google Scholar] [CrossRef]
- Fiore, C.; Eisenhut, M.; Ragazzi, E.; Zanchin, G.; Armanini, D. A History of the Therapeutic Use of Liquorice in Europe. J. Ethnopharmacol. 2005, 99, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Buenz, E.J.; Schnepple, D.J.; Bauer, B.A.; Elkin, P.L.; Riddle, J.M.; Motley, T.J. Techniques: Bioprospecting Historical Herbal Texts by Hunting for New Leads in Old Tomes. Trends Pharmacol. Sci. 2004, 25, 494–498. [Google Scholar] [CrossRef]
- Ding, Y.; Brand, E.; Wang, W.; Zhao, Z. Licorice: Resources, Applications in Ancient and Modern Times. J. Ethnopharmacol. 2022, 298, 115594. [Google Scholar] [CrossRef]
- Pastorino, G.; Cornara, L.; Soares, S.; Rodrigues, F.; Oliveira, M.B.P.P. Liquorice (Glycyrrhiza glabra): A Phytochemical and Pharmacological Review. Phytother. Res. 2018, 32, 2323–2339. [Google Scholar] [CrossRef]
- Wang, X.; Hao, H.; Chu, L. Glycyrrhizin Inhibits LPS-Induced Inflammatory Mediator Production in Endometrial Epithelial Cells. Microb. Pathog. 2017, 109, 110–113. [Google Scholar] [CrossRef]
- Singh, V.; Pal, A.; Darokar, M.P. A Polyphenolic Flavonoid Glabridin: Oxidative Stress Response in Multidrug-Resistant Staphylococcus aureus. Free Radic. Biol. Med. 2015, 87, 48–57. [Google Scholar] [CrossRef]
- Jiang, F.; Li, Y.; Mu, J.; Hu, C.; Zhou, M.; Wang, X.; Si, L.; Ning, S.; Li, Z. Glabridin Inhibits Cancer Stem Cell-like Properties of Human Breast Cancer Cells: An Epigenetic Regulation of miR-148a/SMAd2 Signaling. Mol. Carcinog. 2016, 55, 929–940. [Google Scholar] [CrossRef]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H. Glycyrrhizin, an Active Component of Liquorice Roots, and Replication of SARS-Associated Coronavirus. Lancet 2003, 361, 2045–2046. [Google Scholar] [CrossRef]
- Oyama, K.; Kawada-Matsuo, M.; Oogai, Y.; Hayashi, T.; Nakamura, N.; Komatsuzawa, H. Antibacterial Effects of Glycyrrhetinic Acid and Its Derivatives on Staphylococcus aureus. PLoS ONE 2016, 11, e0165831. [Google Scholar] [CrossRef]
- Yin, X.; Gong, X.; Zhang, L.; Jiang, R.; Kuang, G.; Wang, B.; Chen, X.; Wan, J. Glycyrrhetinic Acid Attenuates Lipopolysaccharide-Induced Fulminant Hepatic Failure in d-Galactosamine-Sensitized Mice by up-Regulating Expression of Interleukin-1 Receptor-Associated Kinase-M. Toxicol. Appl. Pharmacol. 2017, 320, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-Y.; Ha, J.Y.; Kim, K.-M.; Jung, Y.-S.; Jung, J.-C.; Oh, S. Anti-Inflammatory Activities of Licorice Extract and Its Active Compounds, Glycyrrhizic Acid, Liquiritin and Liquiritigenin, in BV2 Cells and Mice Liver. Molecules 2015, 20, 13041–13054. [Google Scholar] [CrossRef] [PubMed]
- Hatano, T.; Kagawa, H.; Yasuhara, T.; Okuda, T. Two New Flavonoids and Other Constituents in Licorice Root: Their Relative Astringency and Radical Scavenging Effects. Chem. Pharm. Bull. 1988, 36, 2090–2097. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y. Pharmacological Activities of Coumarin Compounds in Licorice: A Review. Nat. Prod. Commun. 2020, 15, 1934578X20953954. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.-J.; Chen, J.-B.; Cao, J.-P.; Li, X.; Sun, C.-D. Citrus Flavonoids and Their Antioxidant Evaluation. Crit. Rev. Food Sci. Nutr. 2022, 62, 3833–3854. [Google Scholar] [CrossRef]
- Rakha, A.; Umar, N.; Rabail, R.; Butt, M.S.; Kieliszek, M.; Hassoun, A.; Aadil, R.M. Anti-Inflammatory and Anti-Allergic Potential of Dietary Flavonoids: A Review. Biomed. Pharmacother. 2022, 156, 113945. [Google Scholar] [CrossRef]
- Perez-Vizcaino, F.; Duarte, J. Flavonols and Cardiovascular Disease. Mol. Asp. Med. 2010, 31, 478–494. [Google Scholar] [CrossRef]
- Veitch, N.C. Isoflavonoids of the Leguminosae. Nat. Prod. Rep. 2013, 30, 988–1027. [Google Scholar] [CrossRef]
- Křížová, L.; Dadáková, K.; Kašparovská, J.; Kašparovský, T. Isoflavones. Molecules 2019, 24, 1076. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H. Soybean Isoflavones Ameliorate Ischemic Cardiomyopathy by Activating Nrf2-Mediated Antioxidant Responses. Food Funct. 2017, 8, 2935–2944. [Google Scholar] [CrossRef]
- Jahan, M.A.; Harris, B.; Lowery, M.; Coburn, K.; Infante, A.M.; Percifield, R.J.; Ammer, A.G.; Kovinich, N. The NAC Family Transcription Factor GmNAC42–1 Regulates Biosynthesis of the Anticancer and Neuroprotective Glyceollins in Soybean. BMC Genom. 2019, 20, 149. [Google Scholar] [CrossRef] [PubMed]
- Boutas, I.; Kontogeorgi, A.; Dimitrakakis, C.; Kalantaridou, S.N. Soy Isoflavones and Breast Cancer Risk: A Meta-Analysis. In Vivo 2022, 36, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Werner, N.; Werten, S.; Hoppen, J.; Palm, G.J.; Göttfert, M.; Hinrichs, W. The Induction Mechanism of the Flavonoid-Responsive Regulator FrrA. FEBS J. 2022, 289, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Sohn, S.I.; Pandian, S.; Oh, Y.J.; Kang, H.J.; Cho, W.S.; Cho, Y.S. Metabolic Engineering of Isoflavones: An Updated Overview. Front. Plant Sci. 2021, 12, 670103. [Google Scholar] [CrossRef]
- Han, J.-R.; Li, S.; Li, W.-J.; Dong, L. Mining Microbial and Metabolic Dark Matter in Extreme Environments: A Roadmap for Harnessing the Power of Multi-Omics Data. Adv. Biotechnol. 2024, 2, 26. [Google Scholar] [CrossRef]
- Lin, F.; Chhapekar, S.S.; Vieira, C.C.; Da Silva, M.P.; Rojas, A.; Lee, D.; Liu, N.; Pardo, E.M.; Lee, Y.-C.; Dong, Z.; et al. Breeding for Disease Resistance in Soybean: A Global Perspective. Theor. Appl. Genet. 2022, 135, 3773–3872. [Google Scholar] [CrossRef]
- Li, Q.; Xu, M.; Wu, F.; Guo, Z.; Yang, N.; Li, L.; Wen, W.; Xu, D. Integrated Transcriptomics and Metabolomics Provide Insights into the Biosynthesis of Militarine in the Cell Suspension Culture System of Bletilla Striata. Adv. Biotechnol. 2024, 2, 25. [Google Scholar] [CrossRef]
- Rizzato, G.; Scalabrin, E.; Radaelli, M.; Capodaglio, G.; Piccolo, O. A New Exploration of Licorice Metabolome. Food Chem. 2017, 221, 959–968. [Google Scholar] [CrossRef]
- Cheng, M.; Zhang, J.; Yang, L.; Shen, S.; Li, P.; Yao, S.; Qu, H.; Li, J.; Yao, C.; Wei, W.; et al. Recent Advances in Chemical Analysis of Licorice (Gan-Cao). Fitoterapia 2021, 149, 104803. [Google Scholar] [CrossRef]
- Schmid, C.; Dawid, C.; Peters, V.; Hofmann, T. Saponins from European Licorice Roots (Glycyrrhiza glabra). J. Nat. Prod. 2018, 81, 1734–1744. [Google Scholar] [CrossRef]
- Han, Y.J.; Kang, B.; Yang, E.-J.; Choi, M.-K.; Song, I.-S. Simultaneous Determination and Pharmacokinetic Characterization of Glycyrrhizin, Isoliquiritigenin, Liquiritigenin, and Liquiritin in Rat Plasma Following Oral Administration of Glycyrrhizae Radix Extract. Molecules 2019, 24, 1816. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Meng, T.; Wang, Y.; Tang, W. A Review of the Antiviral Activities of Glycyrrhizic Acid, Glycyrrhetinic Acid and Glycyrrhetinic Acid Monoglucuronide. Pharmaceuticals 2023, 16, 641. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, M.; Jia, Z.-C.; Liu, Y.; Wu, S.-F.; Chen, M.-X.; Hao, G.-F.; Yang, Q. Unraveling the Secrets: Evolution of Resistance Mediated by Membrane Proteins. Drug Resist. Updates 2024, 77, 101140. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Geng, Y.; Song, M.; Zhang, P.; Hou, J.; Wang, W. Genetic Structure and Diversity of Glycyrrhiza Populations Based on Transcriptome SSR Markers. Plant Mol. Biol. Rep. 2019, 37, 401–412. [Google Scholar] [CrossRef]
- Han, Y.; Pang, X.; Zhang, X.; Han, R.; Liang, Z. Resource Sustainability and Challenges: Status and Competitiveness of International Trade in Licorice Extracts under the Belt and Road Initiative. Glob. Ecol. Conserv. 2022, 34, e02014. [Google Scholar] [CrossRef]
- Huang, M.; Wang, W.; Wei, S. [Investigation on medicinal plant resources of Glycyrrhiza uralensis in China and chemical assessment of its underground part]. Zhongguo Zhong Yao Za Zhi 2010, 35, 947–952. [Google Scholar] [CrossRef]
- The Chinese Pharmacopoeia Committee (CCP). Pharmacopoeia of People Republic of China; Chemical Industry Press: Beijing, China, 2020. (In Chinese) [Google Scholar]
- Dai, C.; Wang, S.; De Souza, C.; Li, Y.-Y.; Zhou, C.; Qiu, R.; Xu, X.-Z.; Zhou, H.-L.; Wu, Y. Chemical Constituents and Chemotaxonomic Study of Glycyrrhiza pallidiflora Maxim. Biochem. Syst. Ecol. 2021, 94, 104204. [Google Scholar] [CrossRef]
- Duan, L.; Zhang, Z.-R.; Deng, S.-W.; Chen, H.-F. The Complete Chloroplast Genomes of Rare Medical Herb Glycyrrhiza inflata and Its Relative G. aspera (Fabaceae). Mitochondrial DNA Part B 2019, 4, 4083–4084. [Google Scholar] [CrossRef]
- Du, Z.; Xu, W.; Wang, Y.; Ma, Z.; Yan, P.; Huang, G.; Li, H. A Method for the Identification and Evaluation of Glycyrrhiza Germplasm Based on DNA Barcodes and Leaf Micromorphology. Flora 2025, 324, 152680. [Google Scholar] [CrossRef]
- Chen, Y.; Bai, Y.; Zhang, Z.; Zhang, Y.; Jiang, Y.; Wang, S.; Wang, Y.; Sun, Z. Transcriptomics and Metabolomics Reveal the Primary and Secondary Metabolism Changes in Glycyrrhiza uralensis with Different Forms of Nitrogen Utilization. Front. Plant Sci. 2023, 14, 1229253. [Google Scholar] [CrossRef]
- Li, Y.; Xie, Z.; Huang, Y.; Zeng, J.; Yang, C.; Yuan, L.; Wang, Y.; Li, Y. Integrated Metabolomic and Transcriptomic Analysis Provides Insights into the Flavonoid Formation in Different Glycyrrhiza Species. Ind. Crops Prod. 2024, 208, 117796. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, Y.; Yang, Z.; Song, X.; Ma, Y.; Zhao, J.; Wang, X.; Liu, H.; Fan, L. Widely Target Metabolomics Analysis of the Differences in Metabolites of Licorice under Drought Stress. Ind. Crops Prod. 2023, 202, 117071. [Google Scholar] [CrossRef]
- Shi, D.; Yang, J.; Jiang, Y.; Wen, L.; Wang, Z.; Yang, B. The Antioxidant Activity and Neuroprotective Mechanism of Isoliquiritigenin. Free Radic. Biol. Med. 2020, 152, 207–215. [Google Scholar] [CrossRef]
- Wang, K.-L.; Yu, Y.-C.; Hsia, S.-M. Perspectives on the Role of Isoliquiritigenin in Cancer. Cancers 2021, 13, 115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, X.; Zhong, B.; Liao, Q.; Wang, X.; Xie, Y.; He, X. Review on the Diverse Biological Effects of Glabridin. Drug Des. Dev. Ther. 2023, 17, 15–37. [Google Scholar] [CrossRef]
- Qin, J.; Chen, J.; Peng, F.; Sun, C.; Lei, Y.; Chen, G.; Li, G.; Yin, Y.; Lin, Z.; Wu, L.; et al. Pharmacological Activities and Pharmacokinetics of Liquiritin: A Review. J. Ethnopharmacol. 2022, 293, 115257. [Google Scholar] [CrossRef]
- Mohammed, E.; Peng, Y.; Wang, Z.; Qiang, X.; Zhao, Q. Synthesis, Antiviral, and Antibacterial Activity of the Glycyrrhizic Acid and Glycyrrhetinic Acid Derivatives. Russ. J. Bioorganic Chem. 2022, 48, 906–918. [Google Scholar] [CrossRef]
- Zhou, P.; Shi, W.; He, X.-Y.; Du, Q.-Y.; Wang, F.; Guo, J. Saikosaponin D: Review on the Antitumour Effects, Toxicity and Pharmacokinetics. Pharm. Biol. 2021, 59, 1478–1487. [Google Scholar] [CrossRef]
- Zhou, P.; Xie, W.; He, S.; Sun, Y.; Meng, X.; Sun, G.; Sun, X. Ginsenoside Rb1 as an Anti-Diabetic Agent and Its Underlying Mechanism Analysis. Cells 2019, 8, 204. [Google Scholar] [CrossRef]
- Pompei, R.; Flore, O.; Marccialis, M.A.; Pani, A.; Loddo, B. Glycyrrhizic Acid Inhibits Virus Growth and Inactivates Virus Particles. Nature 1979, 281, 689–690. [Google Scholar] [CrossRef]
- Bi, X.; Yang, L.; Lin, Y.; Deng, W.; Jiang, T.; Zhang, L.; Lu, Y.; Yi, W.; Xie, Y.; Li, M. Efficacy and Safety of Glycyrrhizic Acid in Treatment of Autoimmune Hepatitis. Am. J. Chin. Med. 2023, 51, 391–405. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sheng, Z.; Xiao, J.; Li, Y.; Huang, J.; Jia, J.; Zeng, X.; Li, L. Advances in the Roles of Glycyrrhizic Acid in Cancer Therapy. Front. Pharmacol. 2023, 14, 1265172. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wang, W.; Guo, S. GW26-E2482 Glycyrrhizic Acid and Glycyrrhetinic Acid Reduce the ROS by Regulating NADPH Oxidases. J. Am. Coll. Cardiol. 2015, 66, C46. [Google Scholar] [CrossRef][Green Version]
- Kitagawa, I. Licorice Root. A Natural Sweetener and an Important Ingredient in Chinese Medicine. Pure Appl. Chem. 2002, 74, 1189–1198. [Google Scholar] [CrossRef]
- Armanini, D.; Fiore, C.; Bielenberg, J.; Ragazzi, E. Licorice (Glycyrrhiza glabra); IRIS: New York, NY, USA, 2005; ISBN 978-0-8247-4793-0. Available online: https://www.research.unipd.it/handle/11577/1426650 (accessed on 15 October 2023).
- Asl, M.N.; Hosseinzadeh, H. Review of Pharmacological Effects of Glycyrrhiza sp. and Its Bioactive Compounds. Phytother. Res. 2008, 22, 709–724. [Google Scholar] [CrossRef]
- Karkanis, A.; Martins, N.; Petropoulos, S.A.; Ferreira, I.C.F.R. Phytochemical Composition, Health Effects, and Crop Management of Liquorice (Glycyrrhiza glabra L.): A Medicinal Plant. Food Rev. Int. 2018, 34, 182–203. [Google Scholar] [CrossRef]
- Ji, X.; Liu, N.; Huang, S.; Zhang, C. A Comprehensive Review of Licorice: The Preparation, Chemical Composition, Bioactivities and Its Applications. Am. J. Chin. Med. 2024, 52, 667–716. [Google Scholar] [CrossRef]
- Katwijk, V.M.V.; Huis In’t Veld, L.G. Metabolism of Glycyrrhetinic Acid in Human Subjects. Nature 1954, 173, 733–734. [Google Scholar] [CrossRef]
- Harding, V.; Stebbing, J. Liquorice: A Treatment for All Sorts? Lancet Oncol. 2017, 18, 1155. [Google Scholar] [CrossRef]
- Cheng, H.; Zhong, W.; Wang, L.; Zhang, Q.; Ma, X.; Wang, Y.; Wang, S.; He, C.; Wei, Q.; Fu, C. Effects of Shear Stress on Vascular Endothelial Functions in Atherosclerosis and Potential Therapeutic Approaches. Biomed. Pharmacother. 2023, 158, 114198. [Google Scholar] [CrossRef]
- Li, X.; Bechara, R.; Zhao, J.; McGeachy, M.J.; Gaffen, S.L. IL-17 Receptor–Based Signaling and Implications for Disease. Nat. Immunol. 2019, 20, 1594–1602. [Google Scholar] [CrossRef]
- Mochida, K.; Sakurai, T.; Seki, H.; Yoshida, T.; Takahagi, K.; Sawai, S.; Uchiyama, H.; Muranaka, T.; Saito, K. Draft Genome Assembly and Annotation of Glycyrrhiza uralensis, a Medicinal Legume. Plant J. 2017, 89, 181–194. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Xiong, Z.; Sun, X.; Chen, J.; Wang, C.; Chen, Y.; Zheng, D. Metabolomic Profiles and Health-Promoting Functions of Camellia Drupifera Mature-Seeds Were Revealed Relate to Their Geographical Origins Using Comparative Metabolomic Analysis and Network Pharmacology Approach. Food Chem. 2023, 426, 136619. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Xiao, Q.; Zhang, J.; Sun, X.; Guo, B.; Pei, J.; Huang, L.-F. Identification of a Secondary Q-Marker in High-Quality Ecotypes of Carthamus Tinctorius L. and Exploration of the Target Preference. Food Funct. 2023, 14, 2710–2726. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhu, Y.; Ma, W.; Shi, J.; Peng, Q.; Lin, Z.; Lv, H. Comprehensive Investigation on Non-Volatile and Volatile Metabolites in Four Types of Green Teas Obtained from the Same Tea Cultivar of Longjing 43 (Camellia sinensis var. sinensis) Using the Widely Targeted Metabolomics. Food Chem. 2022, 394, 133501. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A Database of Systems Pharmacology for Drug Discovery from Herbal Medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, F.; Yang, K.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Hu, H.; Gao, K.; Wang, W.; et al. SymMap: An Integrative Database of Traditional Chinese Medicine Enhanced by Symptom Mapping. Nucleic Acids Res. 2019, 47, D1110–D1117. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. Application of the SwissDrugDesign Online Resources in Virtual Screening. Int. J. Mol. Sci. 2019, 20, 4612. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Schriml, L.M.; Munro, J.B.; Schor, M.; Olley, D.; McCracken, C.; Felix, V.; Baron, J.A.; Jackson, R.; Bello, S.M.; Bearer, C.; et al. The Human Disease Ontology 2022 Update. Nucleic Acids Res. 2022, 50, D1255–D1261. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein–Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Luan, A.; Zhang, W.; Yang, M.; Zhong, Z.; Wu, J.; He, Y.; He, J. Unveiling the Molecular Mechanism Involving Anthocyanins in Pineapple Peel Discoloration during Fruit Maturation. Food Chem. 2023, 412, 135482. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Kong, Z.; Huan, X.; Li, L.; Zhang, P.; Wang, Q.; Guo, Y.; Zhu, W.; Qin, P. Transcriptomics and Metabolomics Analyses of the Mechanism of Flavonoid Synthesis in Seeds of Differently Colored Quinoa Strains. Genomics 2022, 114, 138–148. [Google Scholar] [CrossRef]
- Zhang, D.; Wei, X.; Liu, Z.; Wu, X.; Bao, C.; Sun, Y.; Su, N.; Cui, J. Transcriptome Analysis Reveals the Molecular Mechanism of GABA Accumulation during Quinoa (Chenopodium quinoa Willd.) Germination. J. Agric. Food Chem. 2021, 69, 12171–12186. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast One-Pass FASTQ Data Preprocessing, Quality Control, and Deduplication Using Fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Rai, A.; Hirakawa, H.; Ms, M.; Shimizu, Y.; Shirasawa, K.; Kikuchi, S.; Seki, H.; Yamazaki, M.; Toyoda, A.; Isobe, S.; et al. Chromosome-Scale Genome Assembly of Glycyrrhiza uralensis Revealed Metabolic Gene Cluster Centred Specialized Metabolites Biosynthesis. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2022, 29, dsac043. [Google Scholar] [CrossRef]
- Zhang, Y.; Park, C.; Bennett, C.; Thornton, M.; Kim, D. Rapid and Accurate Alignment of Nucleotide Conversion Sequencing Reads with HISAT-3N. Genome Res. 2021, 31, 1290–1295. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A Program for Genome-Wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-Km Spatial Resolution Climate Surfaces for Global Land Areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Sillero, N. What Does Ecological Modelling Model? A Proposed Classification of Ecological Niche Models Based on Their Underlying Methods. Ecol. Model. 2011, 222, 1343–1346. [Google Scholar] [CrossRef]

| Sample | Total Reads | Reads Mapped |
|---|---|---|
| Gasp_1 | 47,498,206 | 38,202,345 (80.43%) |
| Gasp_2 | 44,688,516 | 35,707,659 (79.90%) |
| Gasp_3 | 46,353,134 | 37,110,877 (80.06%) |
| Ginf_1 | 43,803,934 | 40,780,463 (93.10%) |
| Ginf_2 | 53,088,030 | 49,443,635 (93.14%) |
| Ginf_3 | 43,954,566 | 40,963,011 (93.19%) |
| Gpal_1 | 48,659,588 | 44,585,380 (91.63%) |
| Gpal_2 | 45,988,796 | 42,218,272 (91.80%) |
| Gpal_3 | 42,524,312 | 39,088,852 (91.92%) |
| Gura_1 | 46,388,868 | 44,009,841 (94.87%) |
| Gura_2 | 44,436,814 | 42,180,943 (94.92%) |
| Gura_3 | 45,695,936 | 43,363,295 (94.90%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Ding, Z.; Zhang, D.; Zhu, F.; Gao, B. Integrated Metabolomic and Transcriptomic Analysis Reveals the Pharmacological Effects and Differential Mechanisms of Isoflavone Biosynthesis in Four Species of Glycyrrhiza. Int. J. Mol. Sci. 2025, 26, 2539. https://doi.org/10.3390/ijms26062539
Lu Y, Ding Z, Zhang D, Zhu F, Gao B. Integrated Metabolomic and Transcriptomic Analysis Reveals the Pharmacological Effects and Differential Mechanisms of Isoflavone Biosynthesis in Four Species of Glycyrrhiza. International Journal of Molecular Sciences. 2025; 26(6):2539. https://doi.org/10.3390/ijms26062539
Chicago/Turabian StyleLu, Yuanfeng, Zhen Ding, Daoyuan Zhang, Fuyuan Zhu, and Bei Gao. 2025. "Integrated Metabolomic and Transcriptomic Analysis Reveals the Pharmacological Effects and Differential Mechanisms of Isoflavone Biosynthesis in Four Species of Glycyrrhiza" International Journal of Molecular Sciences 26, no. 6: 2539. https://doi.org/10.3390/ijms26062539
APA StyleLu, Y., Ding, Z., Zhang, D., Zhu, F., & Gao, B. (2025). Integrated Metabolomic and Transcriptomic Analysis Reveals the Pharmacological Effects and Differential Mechanisms of Isoflavone Biosynthesis in Four Species of Glycyrrhiza. International Journal of Molecular Sciences, 26(6), 2539. https://doi.org/10.3390/ijms26062539







