LXG Toxins of Bacillus Velezensis Mediate Contact-Dependent Inhibition in a T7SS-Dependent Manner to Enhance Rhizosphere Adaptability
Abstract
1. Introduction
2. Results
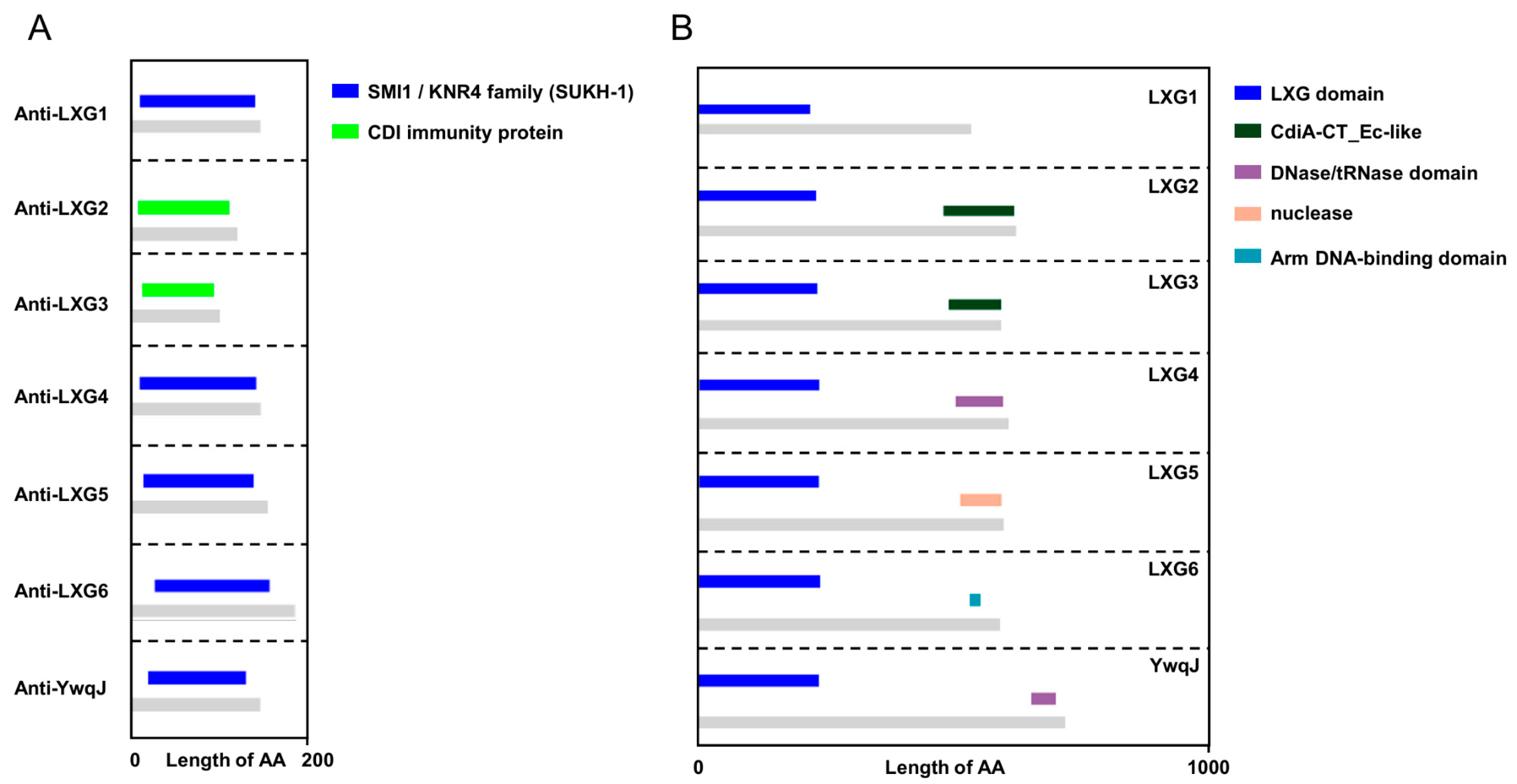
2.1. B. velezensis SQR9 Encodes LXG Domain-Containing Toxins to Inhibit Other Bacillus Strains
2.2. The Toxic Effects of the LXG Toxins Were Dependent on T7SS
2.3. lxg Genes Expressions Were Induced by Plant Root Exudates
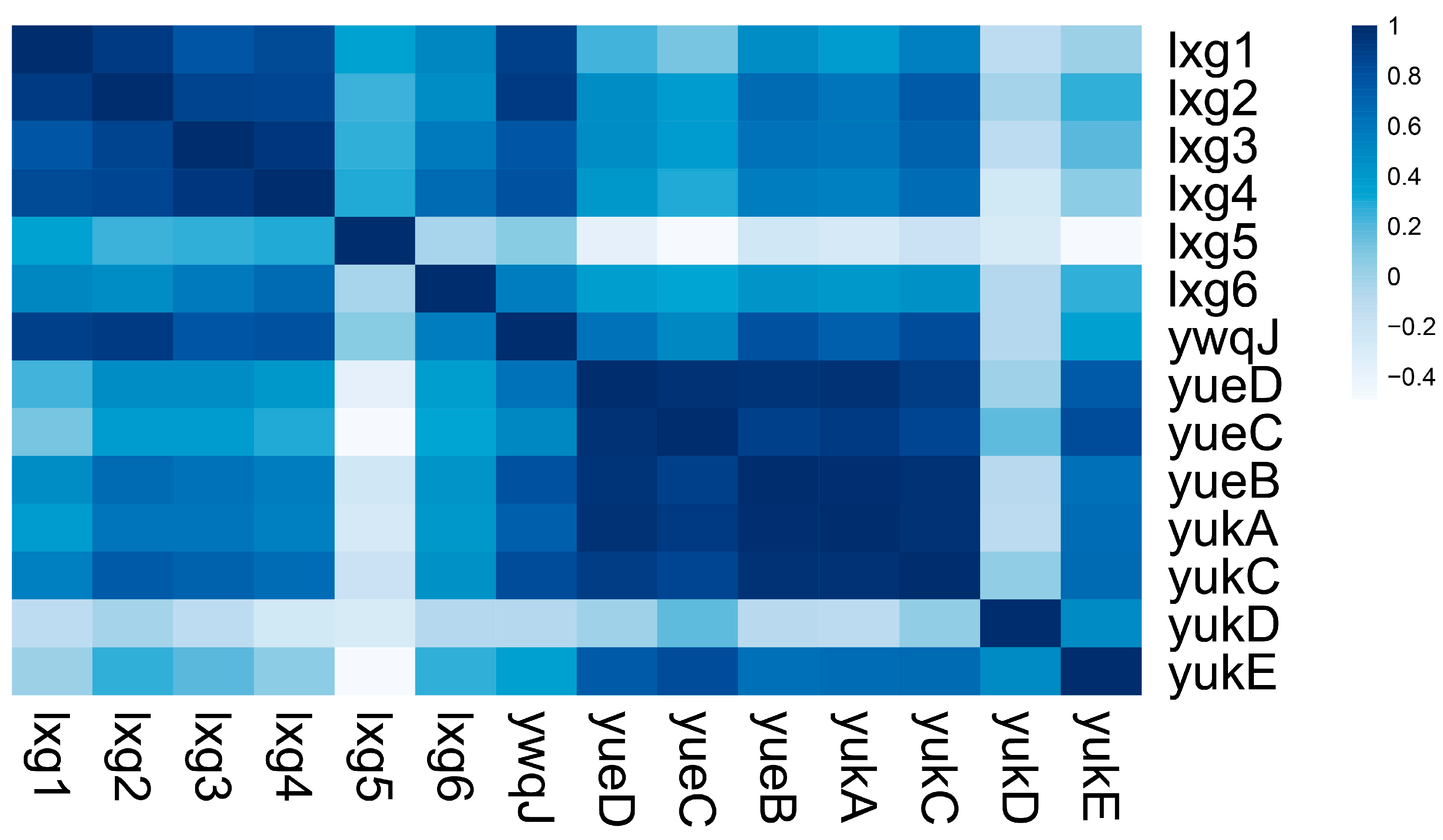
2.4. lxg Is Positively Correlated with T7SS and yukE Expression
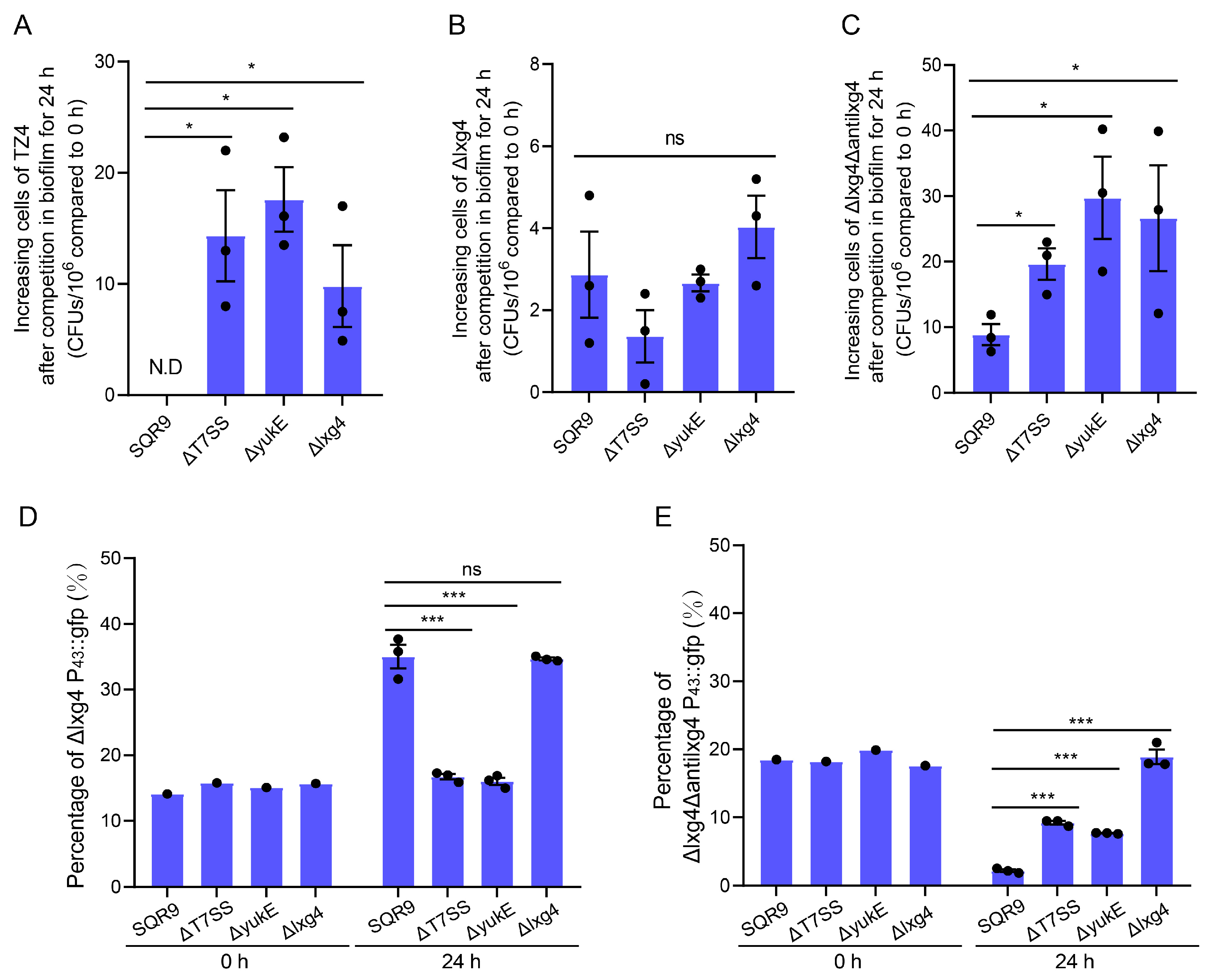
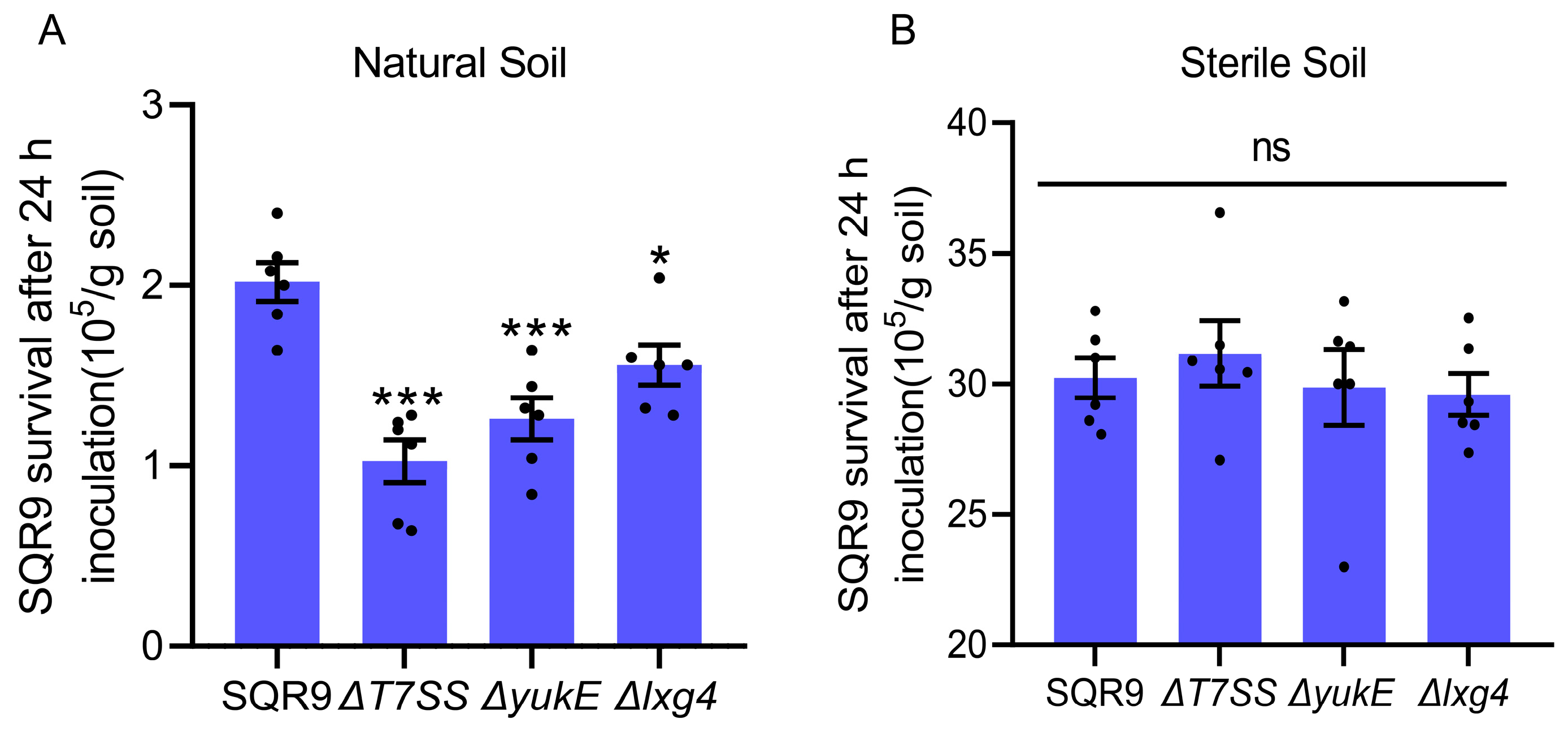
2.5. LXG Toxin Enhances B. velezensis SQR9 Ability to Compete with Rhizosphere Soil Bacteria
3. Discussion
4. Materials and Methods
4.1. Experimental Materials
4.2. Mutant Strain Construction
4.3. Growth Curve and Biofilm Formation Assay
4.4. Competition Assay
4.5. Quantitative PCR
4.6. Flow Cytometry Analysis
4.7. Rhizosphere Survival Challenge
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Qu, Q.; Zhang, Z.; Peijnenburg, W.; Liu, W.; Lu, T.; Hu, B.; Chen, J.; Chen, J.; Lin, Z.; Qian, H. Rhizosphere Microbiome Assembly and Its Impact on Plant Growth. J. Agric. Food Chem. 2020, 68, 5024–5038. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Wang, R.Y.; Song, Y.; Lu, J.; Zhou, B.J.; Song, F.; Zhang, L.J.; Huang, Q.Q.; Gong, J.; Lei, J.J.; et al. Pyoluteorin-deficient Pseudomonas protegens improves cooperation with Bacillus velezensis, biofilm formation, co-colonizing, and reshapes rhizosphere microbiome. Npj Biofilms Microbiomes 2024, 10, 145. [Google Scholar] [CrossRef]
- Little, A.E.; Robinson, C.J.; Peterson, S.B.; Raffa, K.F.; Handelsman, J. Rules of engagement: Interspecies interactions that regulate microbial communities. Annu. Rev. Microbiol. 2008, 62, 375–401. [Google Scholar] [CrossRef]
- Garcia-Bayona, L.; Comstock, L.E. Bacterial antagonism in host-associated microbial communities. Science 2018, 361, eaat2456. [Google Scholar] [CrossRef] [PubMed]
- Chepsergon, J.; Moleleki, L.N. Rhizosphere bacterial interactions and impact on plant health. Curr. Opin. Microbiol. 2023, 73, 102297. [Google Scholar] [CrossRef]
- Sugrue, I.; Ross, R.P.; Hill, C. Bacteriocin diversity, function, discovery and application as antimicrobials. Nat. Rev. Microbiol. 2024, 22, 556–571. [Google Scholar] [CrossRef]
- Zheng, S.; Sonomoto, K. Diversified transporters and pathways for bacteriocin secretion in gram-positive bacteria. Appl. Microbiol. Biotechnol. 2018, 102, 4243–4253. [Google Scholar] [CrossRef]
- Ghapanvari, P.; Taheri, M.; Jalilian, F.A.; Dehbashi, S.; Dezfuli, A.A.Z.; Arabestani, M.R. The effect of nisin on the biofilm production, antimicrobial susceptibility and biofilm formation of Staphylococcus aureus and Pseudomonas aeruginosa. Eur. J. Med. Res. 2022, 27, 173. [Google Scholar] [CrossRef]
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial biofilm: Formation, architecture, antibiotic resistance, and control strategies. Braz. J. Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef]
- Ciofu, O.; Moser, C.; Jensen, P.O.; Hoiby, N. Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 2022, 20, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.B.; Hood, R.D.; Bui, N.K.; LeRoux, M.; Vollmer, W.; Mougous, J.D. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 2011, 475, 343–347. [Google Scholar] [CrossRef]
- Souza, D.P.; Oka, G.U.; Alvarez-Martinez, C.E.; Bisson-Filho, A.W.; Dunger, G.; Hobeika, L.; Cavalcante, N.S.; Alegria, M.C.; Barbosa, L.R.; Salinas, R.K.; et al. Bacterial killing via a type IV secretion system. Nat. Commun. 2015, 6, 6453. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Bayona, L.; Guo, M.S.; Laub, M.T. Contact-dependent killing by Caulobacter crescentus via cell surface-associated, glycine zipper proteins. Elife 2017, 6, e24869. [Google Scholar] [CrossRef]
- Hofer, U. Contact-dependent killing by Myxococcus. Nat. Rev. Microbiol. 2021, 19, 744. [Google Scholar] [CrossRef]
- Garcia, E.C. Contact-dependent interbacterial toxins deliver a message. Curr. Opin. Microbiol. 2018, 42, 40–46. [Google Scholar] [CrossRef]
- Halvorsen, T.M.; Garza-Sánchez, F.; Ruhe, Z.C.; Bartelli, N.L.; Chan, N.A.; Nguyen, J.Y.; Low, D.A.; Hayes, C.S. Lipidation of Class IV CdiA Effector Proteins Promotes Target Cell Recognition during Contact-Dependent Growth Inhibition. Mbio 2021, 12, e0253021. [Google Scholar] [CrossRef]
- Willett, J.L.; Ruhe, Z.C.; Goulding, C.W.; Low, D.A.; Hayes, C.S. Contact-Dependent Growth Inhibition (CDI) and CdiB/CdiA Two-Partner Secretion Proteins. J. Mol. Biol. 2015, 427, 3754–3765. [Google Scholar] [CrossRef]
- Klein, T.A.; Ahmad, S.; Whitney, J.C. Contact-Dependent Interbacterial Antagonism Mediated by Protein Secretion Machines. Trends Microbiol. 2020, 28, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Aoki, S.K.; Pamma, R.; Hernday, A.D.; Bickham, J.E.; Braaten, B.A.; Low, D.A. Contact-dependent inhibition of growth in Escherichia coli. Science 2005, 309, 1245–1248. [Google Scholar] [CrossRef]
- Klein, T.A.; Grebenc, D.W.; Shah, P.Y.; McArthur, O.D.; Dickson, B.H.; Surette, M.G.; Kim, Y.; Whitney, J.C. Dual Targeting Factors Are Required for LXG Toxin Export by the Bacterial Type VIIb Secretion System. mBio 2022, 13, e0213722. [Google Scholar] [CrossRef] [PubMed]
- Teh, W.K.; Ding, Y.; Gubellini, F.; Filloux, A.; Poyart, C.; Givskov, M.; Dramsi, S. Characterization of TelE, a T7SS LXG Effector Exhibiting a Conserved C-Terminal Glycine Zipper Motif Required for Toxicity. Microbiol. Spectr. 2023, 11, e0148123. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.A.; Shah, P.Y.; Gkragkopoulou, P.; Grebenc, D.W.; Kim, Y.; Whitney, J.C. Structure of a tripartite protein complex that targets toxins to the type VII secretion system. Proc. Natl. Acad. Sci. USA 2024, 121, e2312455121. [Google Scholar] [CrossRef]
- Cao, Z.; Casabona, M.G.; Kneuper, H.; Chalmers, J.D.; Palmer, T. The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat. Microbiol. 2016, 2, 16183. [Google Scholar] [CrossRef]
- Garcia-Bayona, L.; Gozzi, K.; Laub, M.T. Mechanisms of Resistance to the Contact-Dependent Bacteriocin CdzC/D in Caulobacter crescentus. J. Bacteriol. 2019, 201, e00538-18. [Google Scholar] [CrossRef]
- Bayer-Santos, E.; Cenens, W.; Matsuyama, B.Y.; Oka, G.U.; Di Sessa, G.; Mininel, I.D.V.; Alves, T.L.; Farah, C.S. The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing. PLoS Pathog. 2019, 15, e1007651. [Google Scholar] [CrossRef]
- Whitney, J.C.; Chou, S.; Russell, A.B.; Biboy, J.; Gardiner, T.E.; Ferrin, M.A.; Brittnacher, M.; Vollmer, W.; Mougous, J.D. Identification, structure, and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J. Biol. Chem. 2013, 288, 26616–26624. [Google Scholar] [CrossRef]
- Russell, A.B.; Singh, P.; Brittnacher, M.; Bui, N.K.; Hood, R.D.; Carl, M.A.; Agnello, D.M.; Schwarz, S.; Goodlett, D.R.; Vollmer, W.; et al. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 2012, 11, 538–549. [Google Scholar] [CrossRef]
- Aloulou, A.; Ali, Y.B.; Bezzine, S.; Gargouri, Y.; Gelb, M.H. Phospholipases: An overview. Methods Mol. Biol. 2012, 861, 63–85. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhang, H.; Gao, Z.; Wang, D.; Liu, G.; Xu, J.; Lan, K.; Dong, Y. Structure of the type VI secretion phospholipase effector Tle1 provides insight into its hydrolysis and membrane targeting. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 2175–2185. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Zheng, Y.; Liao, N.; Wei, L.; Xu, B.; Liu, X.; Liu, J. The structural basis of the Tle4-Tli4 complex reveals the self-protection mechanism of H2-T6SS in Pseudomonas aeruginosa. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 3233–3243. [Google Scholar] [CrossRef]
- Yang, X.Y.; Li, Z.Q.; Gao, Z.Q.; Wang, W.J.; Geng, Z.; Xu, J.H.; She, Z.; Dong, Y.H. Structural and SAXS analysis of Tle5-Tli5 complex reveals a novel inhibition mechanism of H2-T6SS in Pseudomonas aeruginosa. Protein Sci. 2017, 26, 2083–2091. [Google Scholar] [CrossRef]
- Kobayashi, K. Diverse LXG toxin and antitoxin systems specifically mediate intraspecies competition in Bacillus subtilis biofilms. PLoS Genet. 2021, 17, e1009682. [Google Scholar] [CrossRef]
- Liu, Y.; Shu, X.; Chen, L.; Zhang, H.; Feng, H.; Sun, X.; Xiong, Q.; Li, G.; Xun, W.; Xu, Z.; et al. Plant commensal type VII secretion system causes iron leakage from roots to promote colonization. Nat. Microbiol. 2023, 8, 1434–1449. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, D.; Kendall, J.R.; Borriss, R.; Druzhinina, I.S.; Kubicek, C.P.; Shen, Q.; Zhang, R. Comparative Genomic Analysis of Bacillus amyloliquefaciens and Bacillus subtilis Reveals Evolutional Traits for Adaptation to Plant-Associated Habitats. Front. Microbiol. 2016, 7, 2039. [Google Scholar] [CrossRef] [PubMed]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loque, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, H.; Chen, L.; Zhang, H.; Dong, X.; Xiong, Q.; Zhang, R. Root-Secreted Spermine Binds to Bacillus amyloliquefaciens SQR9 Histidine Kinase KinD and Modulates Biofilm Formation. Mol. Plant Microbe Interact. 2020, 33, 423–432. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhang, H.; Shu, X.; Sun, X.; Feng, H.; Xu, Z.; Kovacs, A.T.; Zhang, R.; Liu, Y. Autoinducer-2 relieves soil stress-induced dormancy of Bacillus velezensis by modulating sporulation signaling. NPJ Biofilms Microbiomes 2024, 10, 117. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, H.; Fu, R.; Zhang, N.; Du, W.; Shen, Q.; Zhang, R. Induced root-secreted D-galactose functions as a chemoattractant and enhances the biofilm formation of Bacillus velezensis SQR9 in an McpA-dependent manner. Appl. Microbiol. Biotechnol. 2020, 104, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Bottery, M.J.; Passaris, I.; Dytham, C.; Wood, A.J.; van der Woude, M.W. Spatial Organization of Expanding Bacterial Colonies Is Affected by Contact-Dependent Growth Inhibition. Curr. Biol. 2019, 29, 3622–3634.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Yuan, L.S.; Tao, H.X.; Jiang, W.; Liu, C.J. pheS* as a counter-selectable marker for marker-free genetic manipulations in. J. Microbiol. Meth 2018, 151, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.; Xu, Z.; Li, X.; Li, Q.; Zhang, N.; Shen, Q.; Zhang, R. Comparative proteomics analysis of Bacillus amyloliquefaciens SQR9 revealed the key proteins involved in in situ root colonization. J. Proteome Res. 2014, 13, 5581–5591. [Google Scholar] [CrossRef]
- Kremer, J.M.; Sohrabi, R.; Paasch, B.C.; Rhodes, D.; Thireault, C.; Schulze-Lefert, P.; Tiedje, J.M.; He, S.Y. Peat-based gnotobiotic plant growth systems for Arabidopsis microbiome research. Nat. Protoc. 2021, 16, 2450–2470. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, X.; Sun, X.; Wang, K.; Duan, Y.; Liu, Y.; Zhang, R. LXG Toxins of Bacillus Velezensis Mediate Contact-Dependent Inhibition in a T7SS-Dependent Manner to Enhance Rhizosphere Adaptability. Int. J. Mol. Sci. 2025, 26, 2592. https://doi.org/10.3390/ijms26062592
Shu X, Sun X, Wang K, Duan Y, Liu Y, Zhang R. LXG Toxins of Bacillus Velezensis Mediate Contact-Dependent Inhibition in a T7SS-Dependent Manner to Enhance Rhizosphere Adaptability. International Journal of Molecular Sciences. 2025; 26(6):2592. https://doi.org/10.3390/ijms26062592
Chicago/Turabian StyleShu, Xia, Xiting Sun, Kesu Wang, Yan Duan, Yunpeng Liu, and Ruifu Zhang. 2025. "LXG Toxins of Bacillus Velezensis Mediate Contact-Dependent Inhibition in a T7SS-Dependent Manner to Enhance Rhizosphere Adaptability" International Journal of Molecular Sciences 26, no. 6: 2592. https://doi.org/10.3390/ijms26062592
APA StyleShu, X., Sun, X., Wang, K., Duan, Y., Liu, Y., & Zhang, R. (2025). LXG Toxins of Bacillus Velezensis Mediate Contact-Dependent Inhibition in a T7SS-Dependent Manner to Enhance Rhizosphere Adaptability. International Journal of Molecular Sciences, 26(6), 2592. https://doi.org/10.3390/ijms26062592





