Exploration of Small Non-Coding RNAs as Molecular Markers of Ram Sperm Fertility †
Abstract
:1. Introduction
2. Results
2.1. Data Quality Summary
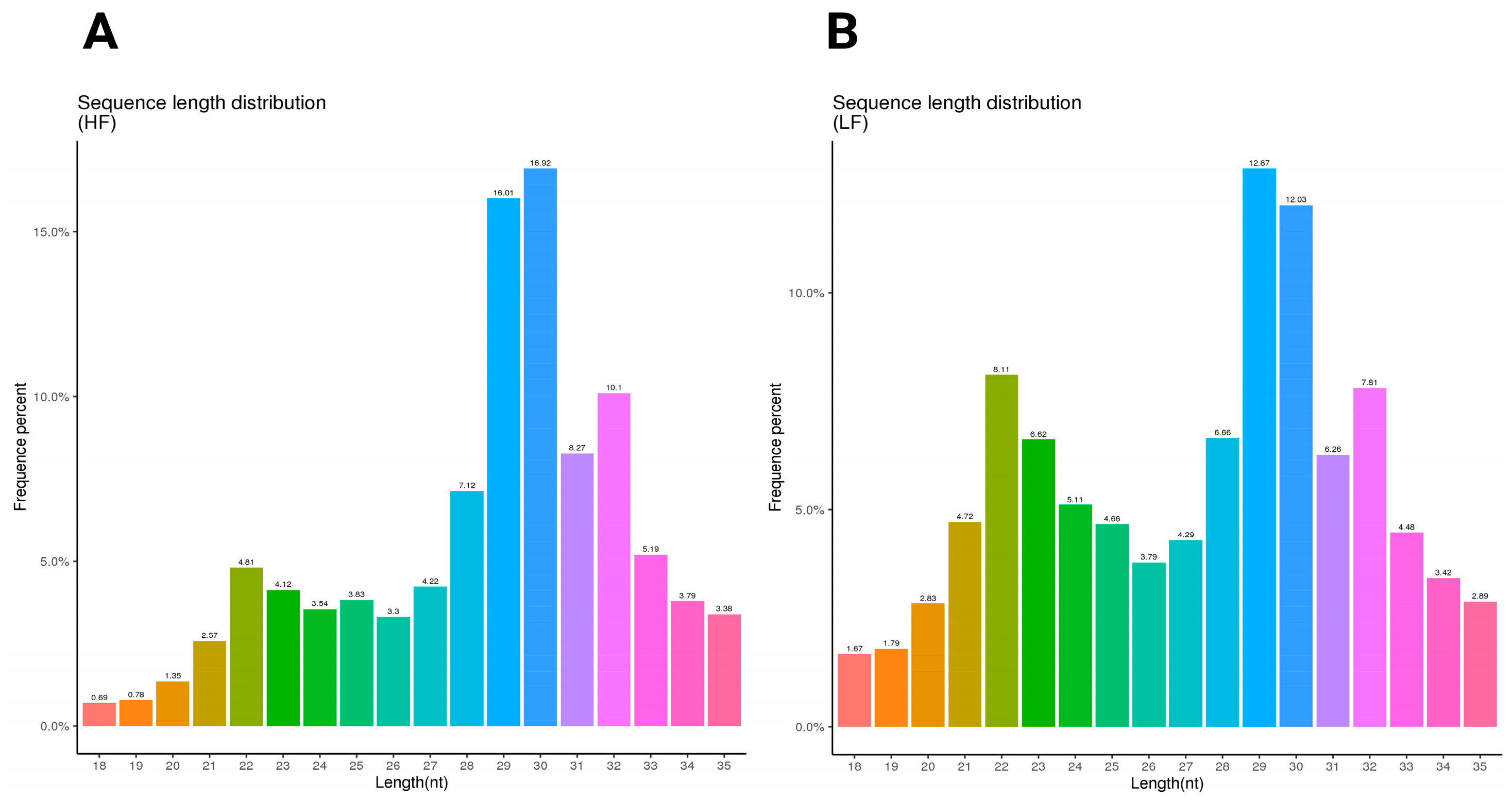
2.2. Length Distribution
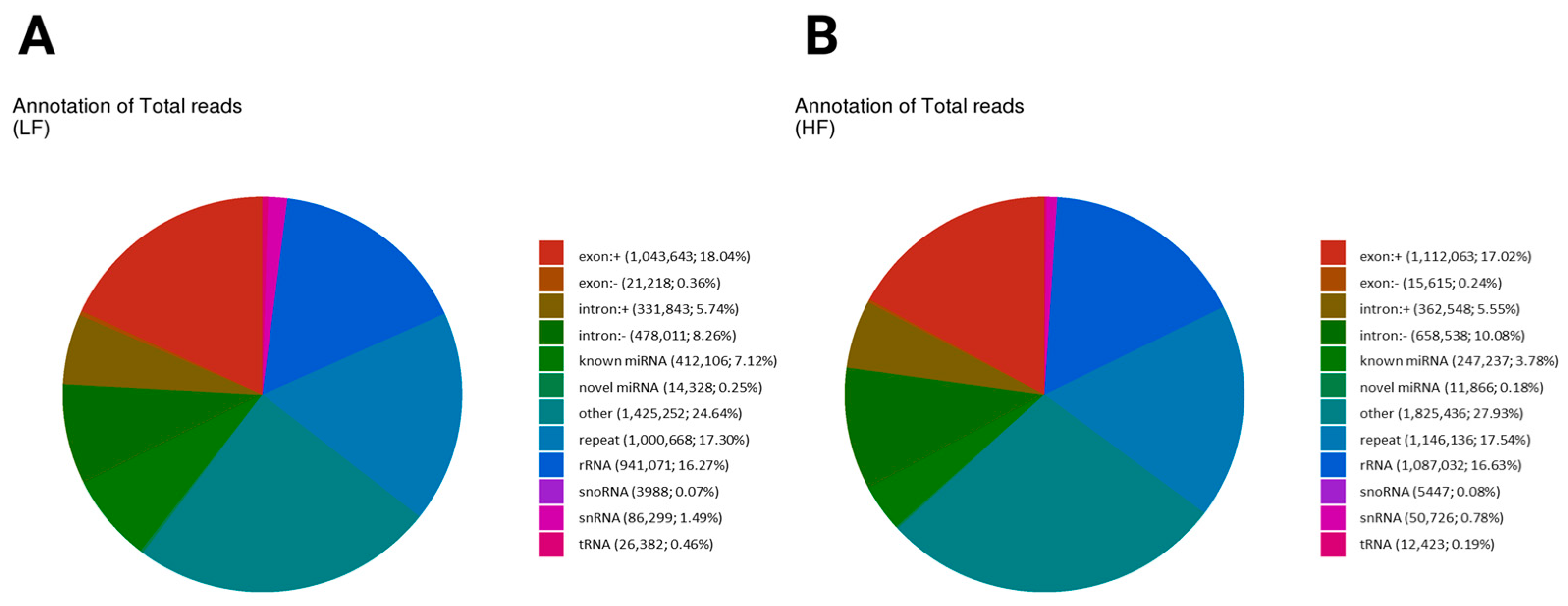
2.3. sncRNA Analysis and Annotation
2.4. Analysis of Known miRNA
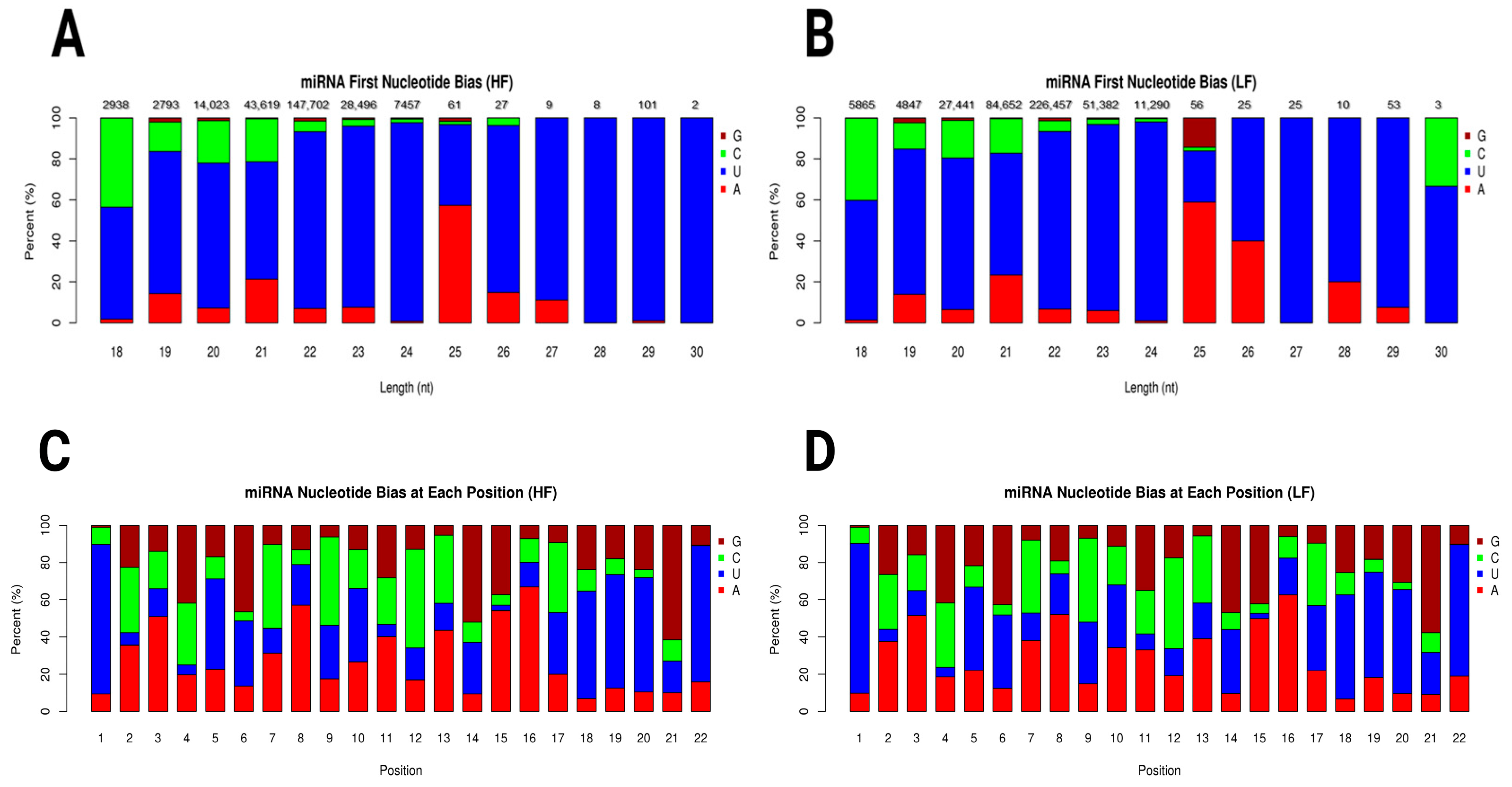
2.4.1. miRNA First Nucleotide Bias Across Length Variants in HF and LF Samples
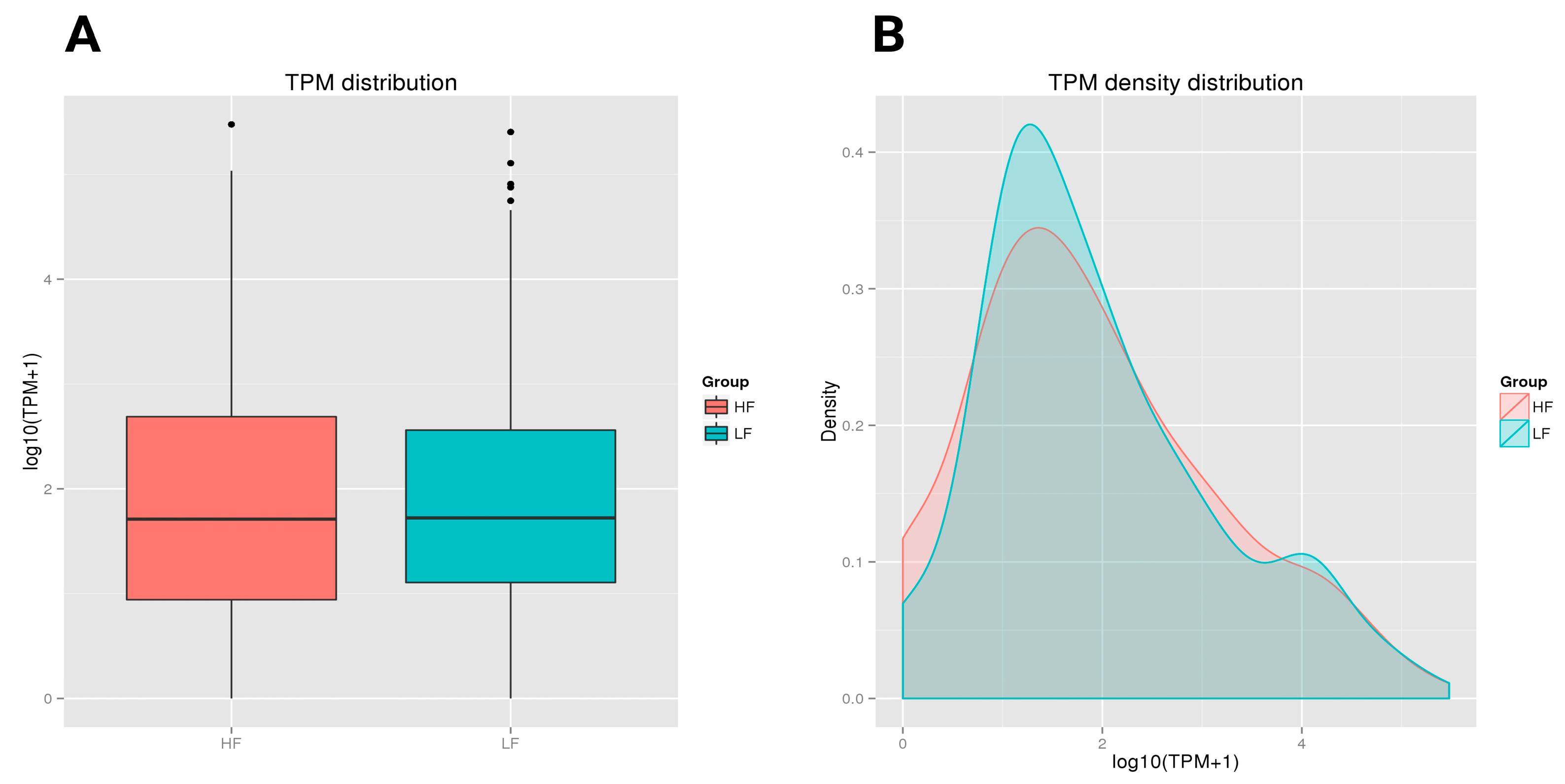
2.4.2. miRNA Expression
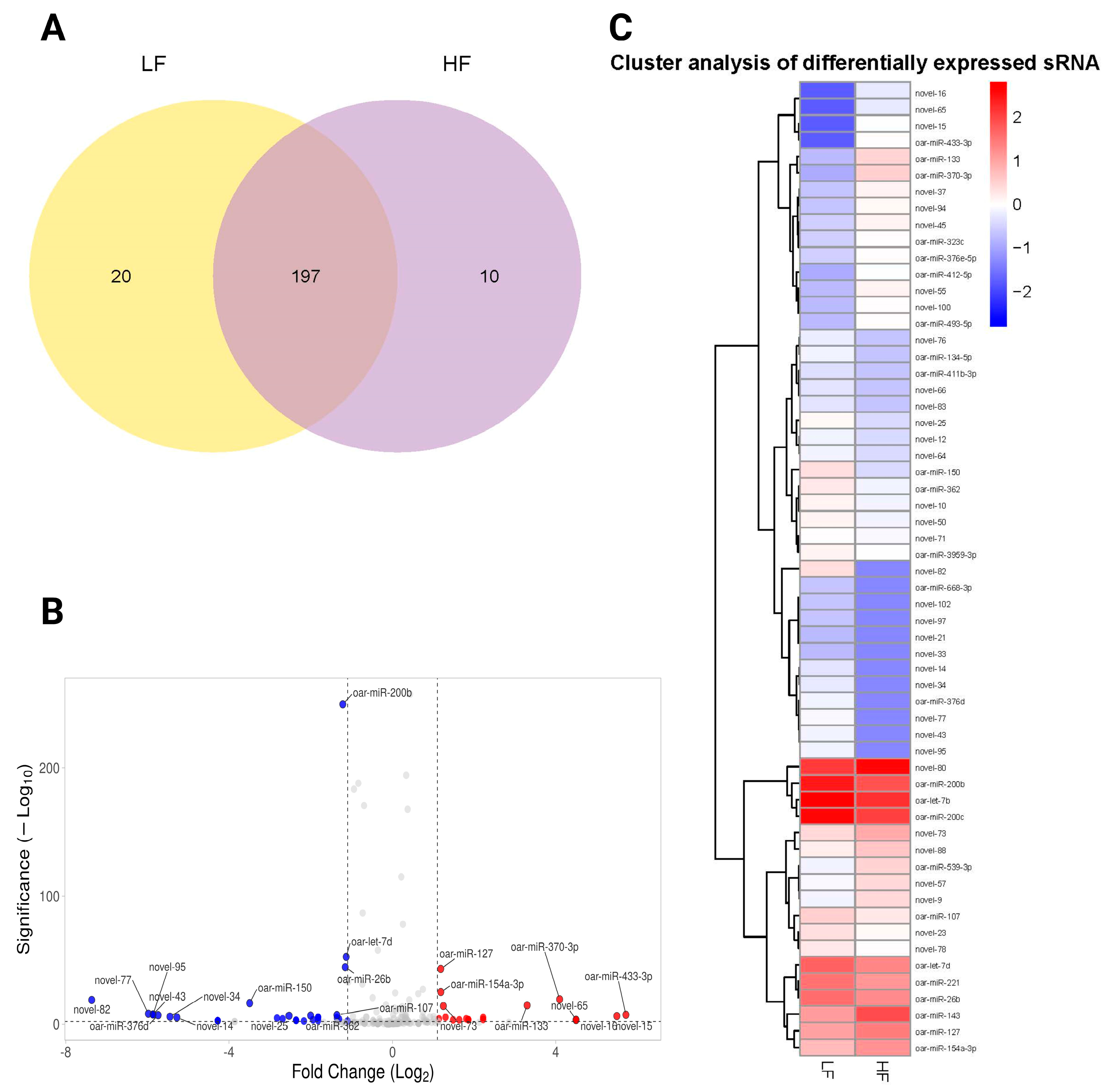
2.4.3. Differentially Expressed miRNAs Between LF and HF
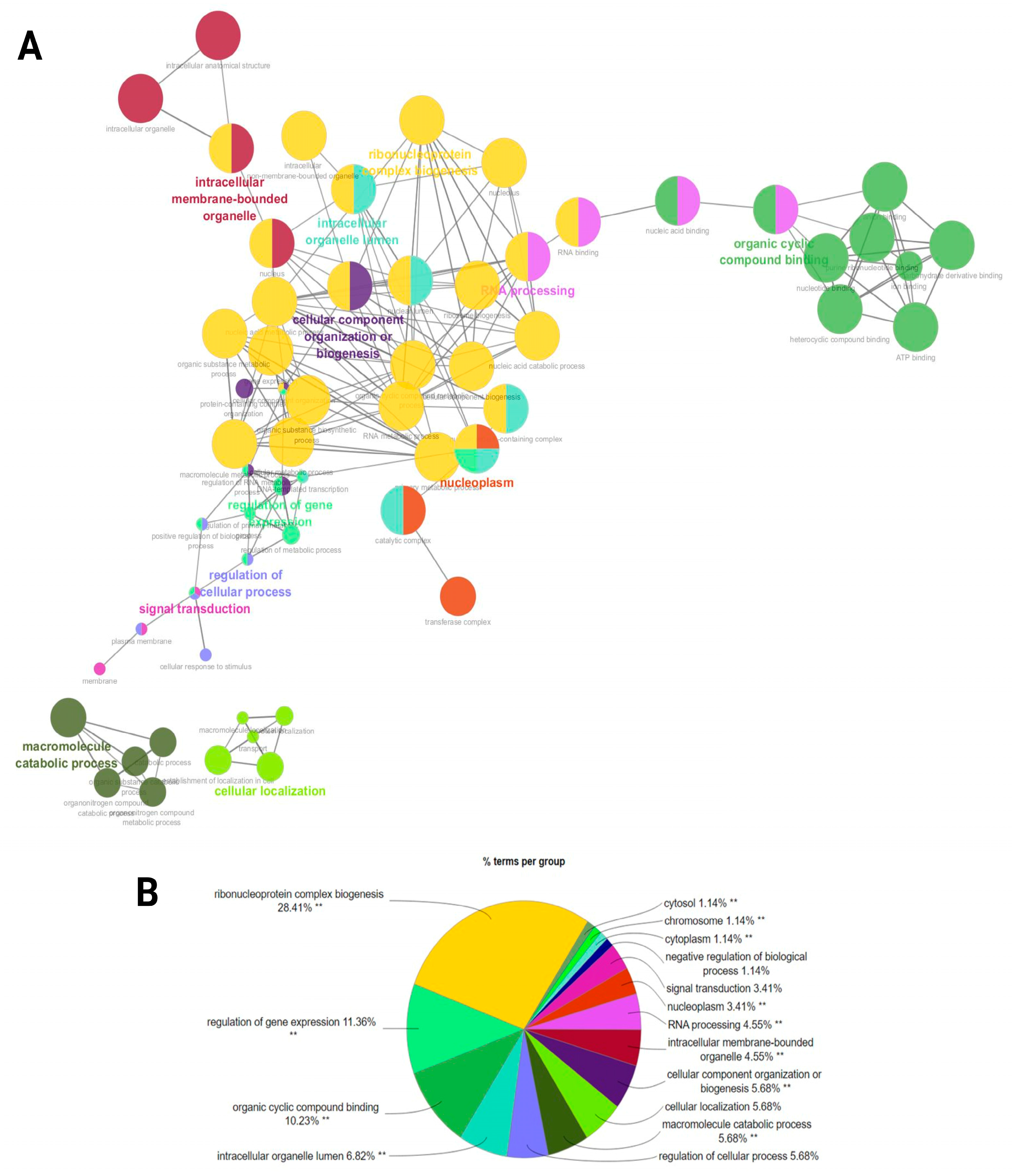
2.5. Functional Interaction Network Association Analysis of Target Genes in LF and HF Groups
3. Discussion
4. Materials and Methods
4.1. Ram Fertility Assessment and Experimental Design
4.2. Semen Collection
4.3. RNA Isolation
4.4. Library Preparation for sncRNA Sequencing
4.5. Alignment of RNA-Seq Reads and Assembly of Transcripts
4.6. miRNA and Analysis of Differentially Expressed microRNAs and Target Ene Prediction
4.7. Protein–Protein Interaction Network and Gene Ontology and Pathway Enrichment Analysis of Target Genes
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MacLaren, A.P.C. Ram fertility in South-West Scotland. Br. Vet. J. 1988, 144, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Hitit, M.; Memili, E. Sperm signatures of fertility and freezability. Anim. Reprod. Sci. 2022, 247, 107147. [Google Scholar] [CrossRef] [PubMed]
- Pardos, L.; Maza Rubio, M.T.; Fantova, E. The diversity of sheep production systems in Aragón (Spain): Characterisation and typification of meat sheep farms. Span. J. Agric. Res. 2008, 6, 497–507. [Google Scholar] [CrossRef]
- Sendler, E.; Johnson, G.D.; Mao, S.; Goodrich, R.J.; Diamond, M.P.; Hauser, R.; Krawetz, S.A. Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res. 2013, 41, 4104–4117. [Google Scholar] [CrossRef]
- Hitit, M.; Özbek, M.; Ayaz-Guner, S.; Guner, H.; Oztug, M.; Bodu, M.; Kirbas, M.; Bulbul, B.; Bucak, M.N.; Ataman, M.B.; et al. Proteomic fertility markers in ram sperm. Anim. Reprod. Sci. 2021, 235, 106882. [Google Scholar] [CrossRef]
- Kaya, A.; Dogan, S.; Vargovic, P.; Kutchy, N.A.; Ross, P.; Topper, E.; Oko, R.; van der Hoorn, F.; Sutovsky, P.; Memili, E. Sperm proteins ODF2 and PAWP as markers of fertility in breeding bulls. Cell Tissue Res. 2022, 387, 159–171. [Google Scholar] [CrossRef]
- Bodu, M.; Hitit, M.; Memili, E. Harnessing the value of fertility biomarkers in bull sperm for buck sperm. Anim. Reprod. Sci. 2025, 272, 107643. [Google Scholar] [CrossRef]
- Long, J.A. The ‘omics’ revolution: Use of genomic, transcriptomic, proteomic and metabolomic tools to predict male reproductive traits that impact fertility in livestock and poultry. Anim. Reprod. Sci. 2020, 220, 106354. [Google Scholar] [CrossRef]
- Carrell, D.T.; Aston, K.I.; Oliva, R.; Emery, B.R.; De Jonge, C.J. The “omics” of human male infertility: Integrating big data in a systems biology approach. Cell Tissue Res. 2016, 363, 295–312. [Google Scholar] [CrossRef]
- Krawetz, S.A.; Kruger, A.; Lalancette, C.; Tagett, R.; Anton, E.; Draghici, S.; Diamond, M.P. A survey of small RNAs in human sperm. Hum. Reprod. 2011, 26, 3401–3412. [Google Scholar] [CrossRef]
- Sharma, U. Paternal Contributions to Offspring Health: Role of Sperm Small RNAs in Intergenerational Transmission of Epigenetic Information. Front. Cell Dev. Biol. 2019, 7, 215. [Google Scholar] [CrossRef]
- Peng, H.; Shi, J.; Zhang, Y.; Zhang, H.; Liao, S.; Li, W.; Lei, L.; Han, C.; Ning, L.; Cao, Y.; et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012, 22, 1609–1612. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Lopes, S.M.C.d.S.; Tang, F.; Surani, M.A. Dynamic Equilibrium and Heterogeneity of Mouse Pluripotent Stem Cells with Distinct Functional and Epigenetic States. Cell Stem Cell 2008, 3, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.-F.; Hou, C.-C.; Yang, W.-X. Small non-coding RNAs and their associated proteins in spermatogenesis. Gene 2016, 578, 141–157. [Google Scholar] [CrossRef] [PubMed]
- García-López, J.; Alonso, L.; Cárdenas, D.B.; Artaza-Alvarez, H.; de Dios Hourcade, J.; Martínez, S.; Brieño-Enríquez, M.A.; Del Mazo, J. Diversity and functional convergence of small noncoding RNAs in male germ cell differentiation and fertilization. RNA 2015, 21, 946–962. [Google Scholar] [CrossRef]
- Liu, W.-M.; Pang, R.T.K.; Chiu, P.C.N.; Wong, B.P.C.; Lao, K.; Lee, K.-F.; Yeung, W.S.B. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc. Natl. Acad. Sci. USA 2012, 109, 490–494. [Google Scholar] [CrossRef]
- Yuan, S.; Schuster, A.; Tang, C.; Yu, T.; Ortogero, N.; Bao, J.; Zheng, H.; Yan, W. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 2016, 143, 635–647. [Google Scholar] [CrossRef]
- Chen, Q.; Yan, W.; Duan, E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat. Rev. Genet. 2016, 17, 733–743. [Google Scholar] [CrossRef]
- Hitit, M.; Kaya, A.; Memili, E. Sperm long non-coding RNAs as markers for ram fertility. Front. Vet. Sci. 2024, 11, 1337939. [Google Scholar] [CrossRef]
- Schuster, A.; Tang, C.; Xie, Y.; Ortogero, N.; Yuan, S.; Yan, W. SpermBase: A Database for Sperm-Borne RNA Contents. Biol. Reprod. 2016, 95, 1–12. [Google Scholar] [CrossRef]
- Pang, W.-K.; Amjad, S.; Ryu, D.-Y.; Adegoke, E.O.; Rahman, M.S.; Park, Y.-J.; Pang, M.-G. Establishment of a male fertility prediction model with sperm RNA markers in pigs as a translational animal model. J. Anim. Sci. Biotechnol. 2022, 13, 84. [Google Scholar] [CrossRef] [PubMed]
- Indriastuti, R.; Pardede, B.P.; Gunawan, A.; Ulum, M.F.; Arifiantini, R.I.; Purwantara, B. Sperm Transcriptome Analysis Accurately Reveals Male Fertility Potential in Livestock. Animals 2022, 12, 2955. [Google Scholar] [CrossRef] [PubMed]
- Donnellan, E.M.; Perrier, J.-P.; Keogh, K.; Štiavnická, M.; Collins, C.M.; Dunleavy, E.M.; Sellem, E.; Bernecic, N.C.; Lonergan, P.; Kenny, D.A.; et al. Identification of differentially expressed mRNAs and miRNAs in spermatozoa of bulls of varying fertility. Front. Vet. Sci. 2022, 9, 993561. [Google Scholar] [CrossRef] [PubMed]
- Ureña, I.; González, C.; Ramón, M.; Gòdia, M.; Clop, A.; Calvo, J.H.; Carabaño, M.J.; Serrano, M. Exploring the ovine sperm transcriptome by RNAseq techniques. I Effect of seasonal conditions on transcripts abundance. PLoS ONE 2022, 17, e0264978. [Google Scholar] [CrossRef]
- Sellem, E.; Jammes, H.; Schibler, L. Sperm-borne sncRNAs: Potential biomarkers for semen fertility? Reprod. Fertil. Dev. 2022, 34, 160–173. [Google Scholar] [CrossRef]
- Chen, C.; Wu, H.; Shen, D.; Wang, S.; Zhang, L.; Wang, X.; Gao, B.; Wu, T.; Li, B.; Li, K.; et al. Comparative profiling of small RNAs of pig seminal plasma and ejaculated and epididymal sperm. Reproduction 2017, 153, 785–796. [Google Scholar] [CrossRef]
- Werry, N. Sperm-Borne Small Non-Coding RNAs in Bovine Fertility. Doctoral Thesis, University of Guelph, Guelph, ON, Canada, 2023. [Google Scholar]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef]
- Sellem, E.; Marthey, S.; Rau, A.; Jouneau, L.; Bonnet, A.; Perrier, J.-P.; Fritz, S.; Le Danvic, C.; Boussaha, M.; Kiefer, H.; et al. A comprehensive overview of bull sperm-borne small non-coding RNAs and their diversity across breeds. Epigenetics Chromatin 2020, 13, 19. [Google Scholar] [CrossRef]
- Gòdia, M.; Estill, M.; Castelló, A.; Balasch, S.; Rodríguez-Gil, J.E.; Krawetz, S.A.; Sánchez, A.; Clop, A. A RNA-Seq Analysis to Describe the Boar Sperm Transcriptome and Its Seasonal Changes. Front. Genet. 2019, 10, 299. [Google Scholar] [CrossRef]
- Govindaraju, A.; Uzun, A.; Robertson, L.; Atli, M.O.; Kaya, A.; Topper, E.; Crate, E.A.; Padbury, J.; Perkins, A.; Memili, E. Dynamics of microRNAs in bull spermatozoa. Reprod. Biol. Endocrinol. 2012, 10, 82. [Google Scholar] [CrossRef]
- de Souza, L.P.; Domingues, W.B.; Blödorn, E.B.; da Silva Nunes, L.; Ortiz, H.G.; Komninou, E.R.; Campos, V.F. Expression of sperm microRNAs related to bull fertility: A systematic review. Res. Vet. Sci. 2024, 166, 105077. [Google Scholar] [CrossRef] [PubMed]
- Werry, N.; Russell, S.J.; Gillis, D.J.; Miller, S.; Hickey, K.; Larmer, S.; Lohuis, M.; Librach, C.; LaMarre, J. Characteristics of miRNAs Present in Bovine Sperm and Associations With Differences in Fertility. Front. Endocrinol. 2022, 13, 874371. [Google Scholar] [CrossRef] [PubMed]
- Keles, E.; Malama, E.; Bozukova, S.; Siuda, M.; Wyck, S.; Witschi, U.; Bauersachs, S.; Bollwein, H. The micro-RNA content of unsorted cryopreserved bovine sperm and its relation to the fertility of sperm after sex-sorting. BMC Genom. 2021, 22, 30. [Google Scholar] [CrossRef] [PubMed]
- Kasimanickam, V.; Kumar, N.; Kasimanickam, R. Investigation of Sperm and Seminal Plasma Candidate MicroRNAs of Bulls with Differing Fertility and In Silico Prediction of miRNA-mRNA Interaction Network of Reproductive Function. Animals 2022, 12, 2360. [Google Scholar] [CrossRef]
- Wu, C.; Wang, C.; Zhai, B.; Zhao, Y.; Zhao, Z.; Yuan, Z.; Zhang, M.; Tian, K.; Fu, X. Study of microRNA Expression Profile in Different Regions of Ram Epididymis. Reprod. Domest. Anim. 2021, 56, 1209–1219. [Google Scholar] [CrossRef]
- Xiong, S.; Ma, W.; Jing, J.; Zhang, J.; Dan, C.; Gui, J.-F.; Mei, J. An miR-200 Cluster on Chromosome 23 Regulates Sperm Motility in Zebrafish. Endocrinology 2018, 159, 1982–1991. [Google Scholar] [CrossRef]
- Xu, Z.; Xie, Y.; Zhou, C.; Hu, Q.; Gu, T.; Yang, J.; Zheng, E.; Huang, S.; Xu, Z.; Cai, G.; et al. Expression Pattern of Seminal Plasma Extracellular Vesicle Small RNAs in Boar Semen. Front. Vet. Sci. 2020, 7, 585276. [Google Scholar] [CrossRef]
- Abu-Halima, M.; Hammadeh, M.; Schmitt, J.; Leidinger, P.; Keller, A.; Meese, E.; Backes, C. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil. Steril. 2013, 99, 1249–1255.e16. [Google Scholar] [CrossRef]
- Huang, C.; Ji, X.R.; Huang, Z.H.; Liu, Q.; Wang, R.J.; Fan, L.Q.; Wu, H.L.; Bo, H.; Zhu, W.B. Long-term storage modifies the microRNA expression profile of cryopreserved human semen. Biomol. Biomed. 2024, 24, 51–60. [Google Scholar] [CrossRef]
- Li, L.; Li, H.; Tian, Y.; Hu, M.; Le, F.; Wang, L.; Liu, X.; Jin, F. Differential microRNAs expression in seminal plasma of normospermic patients with different sperm DNA fragmentation indexes. Reprod. Toxicol. 2020, 94, 8–12. [Google Scholar] [CrossRef]
- Afonso, J.; Lima, A.O.; de Sousa, M.A.P.; de Athayde, F.R.F.; Fortes, M.R.S. Transcription factors and miRNA act as contrary regulators of gene expression in the testis and epididymis of Bos indicus animals. Gene 2024, 899, 148133. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Song, R.; Ortogero, N.; Zheng, H.; Evanoff, R.; Small, C.L.; Griswold, M.D.; Namekawa, S.H.; Royo, H.; Turner, J.M.; et al. The RNase III Enzyme DROSHA Is Essential for MicroRNA Production and Spermatogenesis. J. Biol. Chem. 2012, 287, 25173–25190. [Google Scholar] [CrossRef] [PubMed]
- Tseden, K.; Topaloglu, Ö.; Meinhardt, A.; Dev, A.; Adham, I.; Müller, C.; Wolf, S.; Böhm, D.; Schlüter, G.; Engel, W.; et al. Premature translation of transition protein 2 mRNA causes sperm abnormalities and male infertility. Mol. Reprod. Dev. 2007, 74, 273–279. [Google Scholar] [CrossRef]
- Bedi, Y.S.; Roach, A.N.; Thomas, K.N.; Mehta, N.A.; Golding, M.C. Chromatin alterations during the epididymal maturation of mouse sperm refine the paternally inherited epigenome. Epigenet. Chromatin 2022, 15, 2. [Google Scholar] [CrossRef]
- Trigg, N.A.; Conine, C.C. Epididymal acquired sperm microRNAs modify post-fertilization embryonic gene expression. Cell Rep. 2024, 43, 114698. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011, 40, 37–52. [Google Scholar] [CrossRef]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef]
- Audic, S.; Claverie, J.-M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef]
- Baggerly, K.A.; Deng, L.; Morris, J.S.; Aldaz, C.M. Overdispersed logistic regression for SAGE: Modelling multiple groups and covariates. BMC Bioinform. 2004, 5, 144. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Bodu, M.; Hitit, M.; Memili, E. 163 Sperm small non-coding RNA biomarkers associated with ram fertility. In Proceedings of the 51st Annual Conference of the International Embryo Technology Society (IETS), Fort Worth, TX, USA, 18–22 January 2025. [Google Scholar]
| Sample | Reads | Bases | Error Rate | Q20 | Q30 | GC Content |
|---|---|---|---|---|---|---|
| LF | 14,962,876 | 0.75G | 0.01% | 98.98% | 96.62% | 53.11% |
| HF | 17,401,094 | 0.87G | 0.01% | 98.52% | 95.45% | 52.99% |
| Sample | Total Reads | Total Bases (bp) | Unique Reads | Unique Bases (bp) |
|---|---|---|---|---|
| LF | 9,851,512 | 269,292,004 | 1,550,984 | 40,934,899 |
| HF | 10,611,165 | 303,347,912 | 1,494,593 | 41,404,805 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bodu, M.; Hitit, M.; Donmez, H.; Kaya, A.; Ugur, M.R.; Memili, E. Exploration of Small Non-Coding RNAs as Molecular Markers of Ram Sperm Fertility. Int. J. Mol. Sci. 2025, 26, 2690. https://doi.org/10.3390/ijms26062690
Bodu M, Hitit M, Donmez H, Kaya A, Ugur MR, Memili E. Exploration of Small Non-Coding RNAs as Molecular Markers of Ram Sperm Fertility. International Journal of Molecular Sciences. 2025; 26(6):2690. https://doi.org/10.3390/ijms26062690
Chicago/Turabian StyleBodu, Mustafa, Mustafa Hitit, Huseyin Donmez, Abdullah Kaya, Muhammet Rasit Ugur, and Erdoğan Memili. 2025. "Exploration of Small Non-Coding RNAs as Molecular Markers of Ram Sperm Fertility" International Journal of Molecular Sciences 26, no. 6: 2690. https://doi.org/10.3390/ijms26062690
APA StyleBodu, M., Hitit, M., Donmez, H., Kaya, A., Ugur, M. R., & Memili, E. (2025). Exploration of Small Non-Coding RNAs as Molecular Markers of Ram Sperm Fertility. International Journal of Molecular Sciences, 26(6), 2690. https://doi.org/10.3390/ijms26062690








