The Sequence [RRKLPVGRS] Is a Nuclear Localization Signal for Importin 8 Binding (NLS8): A Chemical Biology and Bioinformatics Study
Abstract
:1. Introduction
2. Results
2.1. In Silico Characterization of Importin 8 NLSs
| Cargo Protein | AlphaFold Code | References | Interacting Amino Acids | ΔG (Kcal/mol) |
|---|---|---|---|---|
| AGO1 | AF-Q9UL18-F1 | [69] | 181SPPEGYYHPLGGGREVWFGF200 | −442.91 |
| 228VIEFMCEVLDIRN240 | −298.03 | |||
| 336YLPLEVCNIVAGQRC350 | −590.03 | |||
| AGO2 | AF-Q9UKV8-F1 | [69,70] | 166MRHLPSMRYTPVGRSFFT183 | −1106.31 |
| 185SEGCSNPLGGGREVWF200 | −470.15 | |||
| 380SKLMRSASFNTDPYVRE396 | −974.05 | |||
| AGO3 | AF-Q9H9G7-F1 | [69] | 186PEGYDHPLGGGREVW200 | −302.12 |
| 359DNQTSTMIKAT369 | −446.97 | |||
| 373APDRQEEISRLVRSA387 | −592.07 | |||
| AGO4 | AF-Q9HCK5-F1 | [69] | 162MRYTPVGRSF171 | −855.04 |
| 175PEGYYHPLGGG185 | −350.66 | |||
| 362APDRQEEISRLVKSNSMVGGPDPYLKE388 | −628.82 | |||
| RPL23A | AF-P62750-F1 | [71] | 10PAPPKAEAKAKALKAKKAVLKGVHSHKKKK39 | −2127.57 |
| SMAD1 | AF-Q15797-F1 | [72] | 187SPNSSYPNSPGSSS199 | −609.18 |
| SMAD3 | AF-P84022-F1 | [72] | 251SQPSMTVDGFTDPSNSERFCLGL273 | −352.31 |
| SRP19 | AF-P09132-F1 | [71] | 45TATEIQDVCSA55 | −226.35 |
| 63EKNKMYSREWNRDVQYRGRVRV84 | −1307.71 | |||
| 111MIPKLKTRTQKTGGAD126 | −1125.31 | |||
| TFE3 | AF-P19532-F1 | [73] | 173PREVLKVQTHLENPTRYHLQQARRQQVKQYLSTTLGPKLASQ214 | −1319.95 |
| WT1 | AF-P19544-F1 | [74] | 1MGSDVRDLN9 | −380.46 |
| 58PAPPPPPPPPPHSFIK73 | −661.45 | |||
| 365SRSDQLKRHQRRHT378 | −1123.92 | |||
| ZFP2 | AF-Q6ZN57-F1 | [74] | 322GVKPFECNECGKAFSKNSSLTQHRRIHTGEKPYECMVCGKHFTGRSSLTVHQVIHTGEKPYECNECGKAFSQ393 | −1439.80 |
| 418AFIKNSSLTV427 | −655.69 | |||
| ZNF264 | AF-O43296-F1 | [74] | 169SRIGQEQVSPGDRVRSH185 | −714.25 |
| 201NNFKCSECGKVFNKKHLLAGHEKIHSGVKPYEC233 | −1203.69 | |||
| 236CGKTFIKSTHLLQHH250 | −853.09 | |||
| 327RPGFLRHYVVHS338 | −922.59 | |||
| ZNF774 | AF-Q6NX45-F1 | [74] | 261KPYACLECHKS271 | −779.05 |
| 280THQRTHTGVKPY291 | −865.15 | |||
| 373RPFKCENC380 | −724.34 |

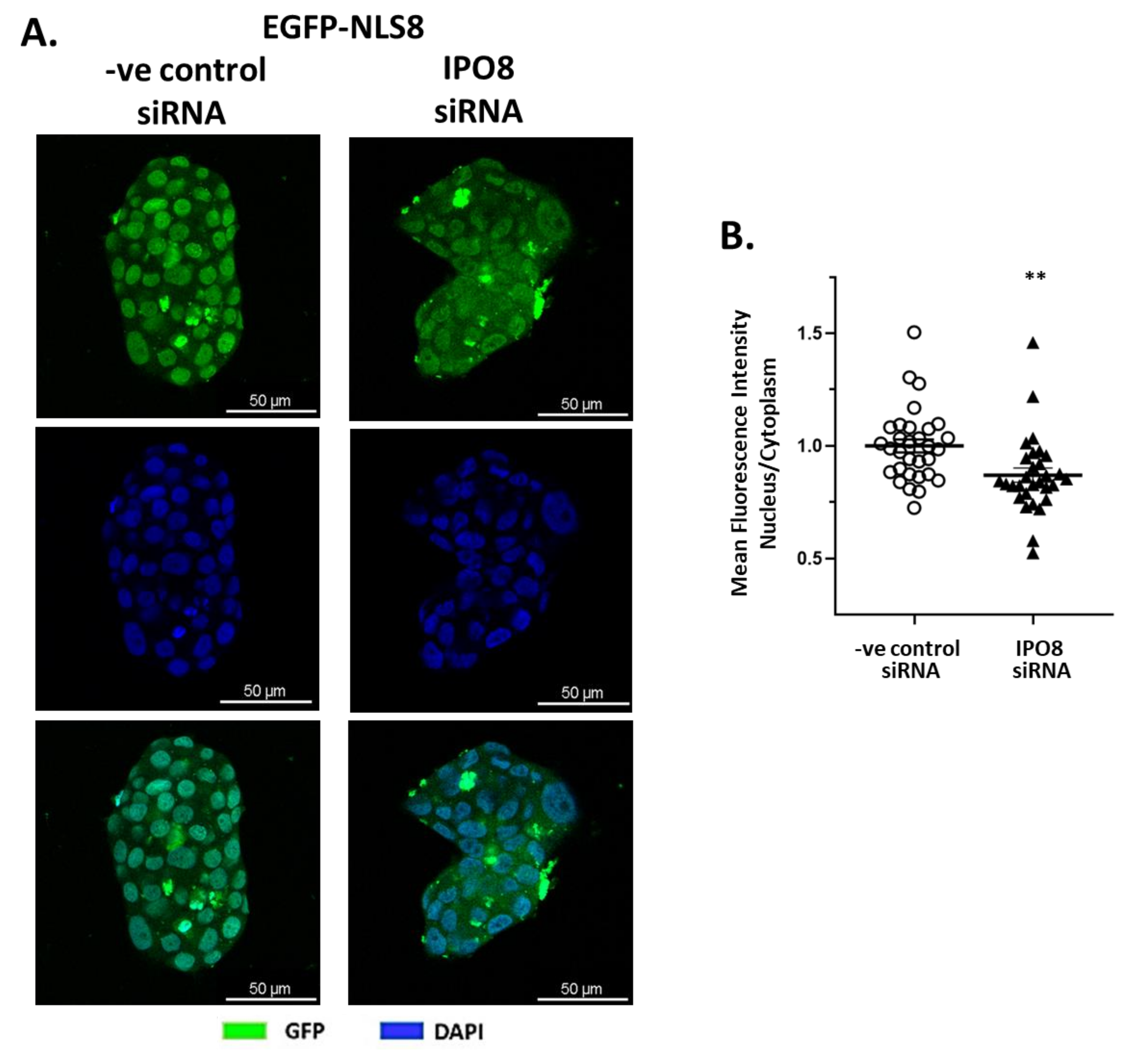
2.2. In Vitro Validation of RRKLPVGRS Sequence as the Importin 8 NLS
3. Discussion
4. Materials and Methods
4.1. Bioinformatics Methods for Identification of the Ιmportin 8 NLS Sequence
4.1.1. Selection and Generation of PDB Files
4.1.2. Flexible Refined Solutions of Proteins Models
4.1.3. Protein–Protein Binding Simulations
4.1.4. Peptide Sequence Alignment
4.2. In Vitro Methods for Validation of Ιmportin 8 NLS Sequence
4.2.1. Cell Culture and Materials
4.2.2. Preparation of GFP-NLS8 Plasmid
4.2.3. Cell Transfection for GFP-NLS8 and IPO8 Silencing
4.2.4. RNA Isolation and Real-Time PCR
4.2.5. Quantification of Nuclear Translocation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Çağatay, T.; Chook, Y.M. Karyopherins in cancer. Curr. Opin. Cell Biol. 2018, 52, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Görlich, D.; Mattaj, I.W. Nucleocytoplasmic Transport. Science 1996, 271, 1513–1519. [Google Scholar] [CrossRef]
- Fontes, M.R.M.; Teh, T.; Kobe, B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α11Edited by K. Nagai. J. Mol. Biol. 2000, 297, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Conti, E. Structures of importins. Results Probl. Cell Differ. 2002, 35, 93–113. [Google Scholar] [CrossRef] [PubMed]
- Güttler, T.; Görlich, D. Ran-dependent nuclear export mediators: A structural perspective. EMBO J. 2011, 30, 3457–3474. [Google Scholar] [CrossRef]
- Lonhienne, T.G.; Forwood, J.K.; Marfori, M.; Robin, G.; Kobe, B.; Carroll, B.J. Importin-β Is a GDP-to-GTP Exchange Factor of Ran: Implications for the Mechanism of Nuclear Import. J. Biol. Chem. 2009, 284, 22549–22558. [Google Scholar] [CrossRef]
- Moore, M.S.; Blobel, G. A G protein involved in nucleocytoplasmic transport: The role of Ran. Trends Biochem. Sci. 1994, 19, 211–216. [Google Scholar] [CrossRef]
- Soniat, M.; Chook, Y.M. Nuclear localization signals for four distinct karyopherin-β nuclear import systems. Biochem. J. 2015, 468, 353–362. [Google Scholar] [CrossRef]
- Chook, Y.M.; Süel, K.E. Nuclear import by karyopherin-βs: Recognition and inhibition. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2011, 1813, 1593–1606. [Google Scholar] [CrossRef]
- Kalderon, D.; Richardson, W.D.; Markham, A.F.; Smith, A.E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 1984, 311, 33–38. [Google Scholar] [CrossRef]
- Kalderon, D.; Roberts, B.L.; Richardson, W.D.; Smith, A.E. A short amino acid sequence able to specify nuclear location. Cell 1984, 39, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Iijima, M.; Suzuki, M.; Tanabe, A.; Nishimura, A.; Yamada, M. Two motifs essential for nuclear import of the hnRNP A1 nucleocytoplasmic shuttling sequence M9 core. FEBS Lett. 2006, 580, 1365–1370. [Google Scholar] [CrossRef]
- Izaurralde, E.; Jarmolowski, A.; Beisel, C.; Mattaj, I.W.; Dreyfuss, G.; Fischer, U. A Role for the M9 Transport Signal of hnRNP A1 in mRNA Nuclear Export. J. Cell Biol. 1997, 137, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Bogerd, H.P.; Benson, R.E.; Truant, R.; Herold, A.; Phingbodhipakkiya, M.; Cullen, B.R. Definition of a Consensus Transportin-specific Nucleocytoplasmic Transport Signal*. J. Biol. Chem. 1999, 274, 9771–9777. [Google Scholar] [CrossRef]
- Terry, L.J.; Shows, E.B.; Wente, S.R. Crossing the Nuclear Envelope: Hierarchical Regulation of Nucleocytoplasmic Transport. Science 2007, 318, 1412–1416. [Google Scholar] [CrossRef] [PubMed]
- Robbins, J.; Dilwortht, S.M.; Laskey, R.A.; Dingwall, C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell 1991, 64, 615–623. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Yamada, K.; Yoneda, Y. Importin α: A key molecule in nuclear transport and non-transport functions. J. Biochem. 2016, 160, 69–75. [Google Scholar] [CrossRef]
- Freitas, N.; Cunha, C. Mechanisms and Signals for the Nuclear Import of Proteins. Curr. Genom. 2009, 10, 550–557. [Google Scholar] [CrossRef]
- Marfori, M.; Mynott, A.; Ellis, J.J.; Mehdi, A.M.; Saunders, N.F.W.; Curmi, P.M.; Forwood, J.K.; Bodén, M.; Kobe, B. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2011, 1813, 1562–1577. [Google Scholar] [CrossRef]
- Adam, S.A. Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr. Opin. Cell Biol. 1999, 11, 402–406. [Google Scholar] [CrossRef]
- Seidel, M.; Romanov, N.; Obarska-Kosinska, A.; Becker, A.; Trevisan Doimo de Azevedo, N.; Provaznik, J.; Nagaraja, S.R.; Landry, J.J.M.; Benes, V.; Beck, M. Co-translational binding of importins to nascent proteins. Nat. Commun. 2023, 14, 3418. [Google Scholar] [CrossRef] [PubMed]
- Dickson, C.F.; Hertel, S.; Tuckwell, A.J.; Li, N.; Ruan, J.; Al-Izzi, S.C.; Ariotti, N.; Sierecki, E.; Gambin, Y.; Morris, R.G.; et al. The HIV capsid mimics karyopherin engagement of FG-nucleoporins. Nature 2024, 626, 836–842. [Google Scholar] [CrossRef]
- Vogel, O.A.; Forwood, J.K.; Leung, D.W.; Amarasinghe, G.K.; Basler, C.F. Viral Targeting of Importin Alpha-Mediated Nuclear Import to Block Innate Immunity. Cells 2023, 13, 71. [Google Scholar] [CrossRef]
- Bhambid, M.; Dey, V.; Walunj, S.; Patankar, S. Toxoplasma Gondii Importin α Shows Weak Auto-Inhibition. Protein J. 2023, 42, 327–342. [Google Scholar] [CrossRef] [PubMed]
- Panagiotopoulos, A.A.; Polioudaki, C.; Ntallis, S.G.; Dellis, D.; Notas, G.; Panagiotidis, C.A.; Theodoropoulos, P.A.; Castanas, E.; Kampa, M. The sequence [EKRKI(E/R)(K/L/R/S/T)] is a nuclear localization signal for importin 7 binding (NLS7). Biochim Biophys. Acta Gen. Subj. 2021, 1865, 129851. [Google Scholar] [CrossRef]
- Panagiotopoulos, A.A.; Kalyvianaki, K.; Tsodoulou, P.K.; Darivianaki, M.N.; Dellis, D.; Notas, G.; Daskalakis, V.; Theodoropoulos, P.A.; Panagiotidis, C.A.; Castanas, E.; et al. Recognition motifs for importin 4 [(L)PPRS(G/P)P] and importin 5 [KP(K/Y)LV] binding, identified by bio-informatic simulation and experimental in vitro validation. Comput. Struct. Biotechnol. J. 2022, 20, 5952–5961. [Google Scholar] [CrossRef]
- Van Gucht, I.; Meester, J.A.N.; Bento, J.R.; Bastiaansen, M.; Bastianen, J.; Luyckx, I.; Van Den Heuvel, L.; Neutel, C.H.G.; Guns, P.J.; Vermont, M.; et al. A human importin-β-related disorder: Syndromic thoracic aortic aneurysm caused by bi-allelic loss-of-function variants in IPO8. Am. J. Hum. Genet. 2021, 108, 1115–1125. [Google Scholar] [CrossRef]
- Moradimotlagh, A.; Chen, S.; Koohbor, S.; Moon, K.M.; Foster, L.J.; Reiner, N.; Nandan, D. Leishmania infection upregulates and engages host macrophage Argonaute 1, and system-wide proteomics reveals Argonaute 1-dependent host response. Front. Immunol. 2023, 14, 1287539. [Google Scholar] [CrossRef]
- Li, W.; Liu, M.; Feng, Y.; Xu, Y.F.; Che, J.P.; Wang, G.C.; Zheng, J.H.; Gao, H.J. Evaluation of Argonaute protein as a predictive marker for human clear cell renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2013, 6, 1086–1094. [Google Scholar]
- Kim, D.H.; Villeneuve, L.M.; Morris, K.V.; Rossi, J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006, 13, 793–797. [Google Scholar] [CrossRef]
- Zhang, Q.; Higginbotham, J.N.; Jeppesen, D.K.; Yang, Y.P.; Li, W.; McKinley, E.T.; Graves-Deal, R.; Ping, J.; Britain, C.M.; Dorsett, K.A.; et al. Transfer of Functional Cargo in Exomeres. Cell Rep. 2019, 27, 940–954.e946. [Google Scholar] [CrossRef]
- Gómez Acuña, L.I.; Nazer, E.; Rodríguez-Seguí, S.A.; Pozzi, B.; Buggiano, V.; Marasco, L.E.; Agirre, E.; He, C.; Alló, M.; Kornblihtt, A.R. Nuclear role for human Argonaute-1 as an estrogen-dependent transcription coactivator. J. Cell Biol. 2020, 219, e201908097. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e418. [Google Scholar] [CrossRef] [PubMed]
- Mori, R.; Tanaka, K.; Shimokawa, I. Identification and functional analysis of inflammation-related miRNAs in skin wound repair. Dev. Growth Differ. 2018, 60, 306–315. [Google Scholar] [CrossRef]
- Elkayam, E.; Kuhn, C.D.; Tocilj, A.; Haase, A.D.; Greene, E.M.; Hannon, G.J.; Joshua-Tor, L. The structure of human argonaute-2 in complex with miR-20a. Cell 2012, 150, 100–110. [Google Scholar] [CrossRef]
- Schmidt, M.F.; Korb, O.; Abell, C. MicroRNA-specific argonaute 2 protein inhibitors. ACS Chem. Biol. 2013, 8, 2122–2126. [Google Scholar] [CrossRef]
- Ferreira, R.; Santos, T.; Amar, A.; Gong, A.; Chen, T.C.; Tahara, S.M.; Giannotta, S.L.; Hofman, F.M. Argonaute-2 promotes miR-18a entry in human brain endothelial cells. J. Am. Heart Assoc. 2014, 3, e000968. [Google Scholar] [CrossRef]
- Lopez-Orozco, J.; Fayad, N.; Khan, J.Q.; Felix-Lopez, A.; Elaish, M.; Rohamare, M.; Sharma, M.; Falzarano, D.; Pelletier, J.; Wilson, J.; et al. The RNA Interference Effector Protein Argonaute 2 Functions as a Restriction Factor Against SARS-CoV-2. J Mol Biol 2023, 435, 168170. [Google Scholar] [CrossRef]
- Zoulikha, M.; Xiao, Q.; Boafo, G.F.; Sallam, M.A.; Chen, Z.; He, W. Pulmonary delivery of siRNA against acute lung injury/acute respiratory distress syndrome. Acta Pharm. Sinica. B 2022, 12, 600–620. [Google Scholar] [CrossRef]
- Alizada, A.; Hannon, G.J.; Nicholson, B.C. Transcriptional regulation of the piRNA pathway by Ovo in animal ovarian germ cells. Genes Dev. 2025, 3, 221–241. [Google Scholar] [CrossRef]
- Winter, J.; Diederichs, S. Argonaute-3 activates the let-7a passenger strand microRNA. RNA Biol. 2013, 10, 1631–1643. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Xu, C.; Mei, J.; Chen, Y.; Wang, D. Argonaute 3 (AGO3) promotes malignancy potential of cervical cancer via regulation of Wnt/β-catenin signaling pathway. Reprod. Biol. 2021, 21, 100479. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, W.; Xu, W.; Zhang, H.; Liu, X.; Chen, X.; Chen, H. Genome-Wide Association Study and Identification of Candidate Genes Associated with Seed Number per Pod in Soybean. Int. J. Mol. Sci. 2024, 25, 2536. [Google Scholar] [CrossRef]
- Till, S.; Ladurner, A.G. RNA Pol IV plays catch with Argonaute 4. Cell 2007, 131, 643–645. [Google Scholar] [CrossRef] [PubMed]
- Potenza, N.; Papa, U.; Russo, A. Differential expression of Dicer and Argonaute genes during the differentiation of human neuroblastoma cells. Cell Biol. Int. 2009, 33, 734–738. [Google Scholar] [CrossRef]
- Chalertpet, K.; Pin-On, P.; Aporntewan, C.; Patchsung, M.; Ingrungruanglert, P.; Israsena, N.; Mutirangura, A. Argonaute 4 as an Effector Protein in RNA-Directed DNA Methylation in Human Cells. Front. Genet. 2019, 10, 645. [Google Scholar] [CrossRef]
- Jiang, H.; Lin, J.J.; Tao, J.; Fisher, P.B. Suppression of human ribosomal protein L23A expression during cell growth inhibition by interferon-beta. Oncogene 1997, 14, 473–480. [Google Scholar] [CrossRef]
- Ramos-Sáenz, A.; González-Álvarez, D.; Rodríguez-Galán, O.; Rodríguez-Gil, A.; Gaspar, S.G.; Villalobo, E.; Dosil, M.; de la Cruz, J. Pol5 is an essential ribosome biogenesis factor required for 60S ribosomal subunit maturation in Saccharomyces cerevisiae. RNA 2019, 25, 1561–1575. [Google Scholar] [CrossRef]
- Humeres, C.; Venugopal, H.; Frangogiannis, N.G. Smad-dependent pathways in the infarcted and failing heart. Curr. Opin. Pharmacol. 2022, 64, 102207. [Google Scholar] [CrossRef]
- Nickel, J.; Mueller, T.D. Specification of BMP Signaling. Cells 2019, 8, 1579. [Google Scholar] [CrossRef]
- Chen, D.; Zhao, M.; Mundy, G.R. Bone morphogenetic proteins. Growth Factors 2004, 22, 233–241. [Google Scholar] [CrossRef]
- Wu, W.; Wang, X.; Yu, X.; Lan, H.Y. Smad3 Signatures in Renal Inflammation and Fibrosis. Int. J. Biol. Sci. 2022, 18, 2795–2806. [Google Scholar] [CrossRef]
- Jeon, H.Y.; Pornour, M.; Ryu, H.; Khadka, S.; Xu, R.; Jang, J.; Li, D.; Chen, H.; Hussain, A.; Fazli, L.; et al. SMAD3 promotes expression and activity of the androgen receptor in prostate cancer. Nucleic Acids Res 2023, 51, 2655–2670. [Google Scholar] [CrossRef]
- Habeeb, O.; Korty, K.E.; Azzato, E.M.; Astbury, C.; Farkas, D.H.; Ko, J.S.; Billings, S.D. EWSR1-SMAD3 rearranged fibroblastic tumor: Case series and review. J. Cutan. Pathol. 2021, 48, 255–262. [Google Scholar] [CrossRef]
- Linder, M.I.; Mizoguchi, Y.; Hesse, S.; Csaba, G.; Tatematsu, M.; Łyszkiewicz, M.; Ziȩtara, N.; Jeske, T.; Hastreiter, M.; Rohlfs, M.; et al. Human genetic defects in SRP19 and SRPRA cause severe congenital neutropenia with distinctive proteome changes. Blood 2023, 141, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Morita, S.; Tabata, K.; Nakamura, K.; Kawai, G. Solution structure of a SRP19 binding domain in human SRP RNA. J. Biochem. 2002, 132, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Faoro, C.; Ataide, S.F. Noncanonical Functions and Cellular Dynamics of the Mammalian Signal Recognition Particle Components. Front. Mol. Biosci. 2021, 8, 679584. [Google Scholar] [CrossRef] [PubMed]
- Di Malta, C.; Zampelli, A.; Granieri, L.; Vilardo, C.; De Cegli, R.; Cinque, L.; Nusco, E.; Pece, S.; Tosoni, D.; Sanguedolce, F.; et al. TFEB and TFE3 drive kidney cystogenesis and tumorigenesis. EMBO Mol. Med. 2023, 15, e16877. [Google Scholar] [CrossRef]
- Takamatsu, D.; Kohashi, K.; Kiyozawa, D.; Kinoshita, F.; Ieiri, K.; Baba, M.; Eto, M.; Oda, Y. TFE3-immunopositive papillary renal cell carcinoma: A clinicopathological, immunohistochemical, and genetic study. Pathol. Res. Pract. 2023, 242, 154313. [Google Scholar] [CrossRef]
- Kmeid, M.; Akgul, M. TFE3 Rearrangement and Expression in Renal Cell Carcinoma. Int. J. Surg. Pathol. 2023, 31, 509–520. [Google Scholar] [CrossRef]
- Zhou, K.; Zheng, Z.; Li, Y.; Han, W.; Zhang, J.; Mao, Y.; Chen, H.; Zhang, W.; Liu, M.; Xie, L.; et al. TFE3, a potential therapeutic target for Spinal Cord Injury via augmenting autophagy flux and alleviating ER stress. Theranostics 2020, 10, 9280–9302. [Google Scholar] [CrossRef]
- Salvatorelli, L.; Parenti, R.; Leone, G.; Musumeci, G.; Vasquez, E.; Magro, G. Wilms tumor 1 (WT1) protein: Diagnostic utility in pediatric tumors. Acta Histochem. 2015, 117, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Petrosyan, A.; Villani, V.; Aguiari, P.; Thornton, M.E.; Wang, Y.; Rajewski, A.; Zhou, S.; Cravedi, P.; Grubbs, B.H.; De Filippo, R.E.; et al. Identification and Characterization of the Wilms Tumor Cancer Stem Cell. Adv. Sci. 2023, 10, e2206787. [Google Scholar] [CrossRef]
- Ogasawara, M. Wilms’ tumor 1-targeting cancer vaccine: Recent advancements and future perspectives. Hum. Vaccines Immunother. 2024, 20, 2296735. [Google Scholar] [CrossRef]
- Mörking, P.A.; Rampazzo Rde, C.; Walrad, P.; Probst, C.M.; Soares, M.J.; Gradia, D.F.; Pavoni, D.P.; Krieger, M.A.; Matthews, K.; Goldenberg, S.; et al. The zinc finger protein TcZFP2 binds target mRNAs enriched during Trypanosoma cruzi metacyclogenesis. Mem. Inst. Oswaldo Cruz 2012, 107, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.M.; Guo, Y.; Yu, H.; Guo, T. RNA editing affects cis-regulatory elements and predicts adverse cancer survival. Cancer Med. 2021, 10, 6114–6127. [Google Scholar] [CrossRef]
- Guan, C.; He, L.; Chang, Z.; Gu, X.; Liang, J.; Liu, R. ZNF774 is a potent suppressor of hepatocarcinogenesis through dampening the NOTCH2 signaling. Oncogene 2020, 39, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Weinmann, L.; Höck, J.; Ivacevic, T.; Ohrt, T.; Mütze, J.; Schwille, P.; Kremmer, E.; Benes, V.; Urlaub, H.; Meister, G. Importin 8 Is a Gene Silencing Factor that Targets Argonaute Proteins to Distinct mRNAs. Cell 2009, 136, 496–507. [Google Scholar] [CrossRef]
- Höck, J.; Weinmann, L.; Ender, C.; Rüdel, S.; Kremmer, E.; Raabe, M.; Urlaub, H.; Meister, G. Proteomic and functional analysis of Argonaute-containing mRNA–protein complexes in human cells. EMBO Rep. 2007, 8, 1052–1060. [Google Scholar] [CrossRef]
- Dean, K.A.; von Ahsen, O.; Görlich, D.; Fried, H.M. Signal recognition particle protein 19 is imported into the nucleus by importin 8 (RanBP8) and transportin. J. Cell Sci. 2001, 114, 3479–3485. [Google Scholar] [CrossRef]
- Xu, L.; Yao, X.; Chen, X.; Lu, P.; Zhang, B.; Ip, Y.T. Msk is required for nuclear import of TGF-β/BMP-activated Smads. J. Cell Biol. 2007, 178, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Perera, R.M.; Stoykova, S.; Nicolay, B.N.; Ross, K.N.; Fitamant, J.; Boukhali, M.; Lengrand, J.; Deshpande, V.; Selig, M.K.; Ferrone, C.R.; et al. Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism. Nature 2015, 524, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Luck, K.; Kim, D.-K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef]
- Woong-Hee, S.; Gyu Rie, L.; Lim, H.; Hasup, L.; Chaok, S. Prediction of Protein Structure and Interaction by GALAXY Protein Modeling Programs. Bio Des. 2014, 2, 1–11. [Google Scholar]
- Ko, J.; Park, H.; Heo, L.; Seok, C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012, 40, W294–W297. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.R.; Heo, L.; Seok, C. Effective protein model structure refinement by loop modeling and overall relaxation. Proteins Struct. Funct. Bioinform. 2016, 84, 293–301. [Google Scholar] [CrossRef]
- Ritchie, D.W.; Grudinin, S. Spherical polar Fourier assembly of protein complexes with arbitrary point group symmetry. J. Appl. Crystallogr. 2016, 49, 158–167. [Google Scholar] [CrossRef]
- Ritchie, D.W. Evaluation of protein docking predictions using Hex 3.1 in CAPRI rounds 1 and 2. Proteins Struct. Funct. Bioinform. 2003, 52, 98–106. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Lin, K.C.; Lin, M.W.; Hsu, M.N.; Yu-Chen, G.; Chao, Y.C.; Tuan, H.Y.; Chiang, C.S.; Hu, Y.C. Graphene oxide sensitizes cancer cells to chemotherapeutics by inducing early autophagy events, promoting nuclear trafficking and necrosis. Theranostics 2018, 8, 2477–2487. [Google Scholar] [CrossRef] [PubMed]
- Conforti, F.; Zhang, X.; Rao, G.; De Pas, T.; Yonemori, Y.; Rodriguez, J.A.; McCutcheon, J.N.; Rahhal, R.; Alberobello, A.T.; Wang, Y.; et al. Therapeutic Effects of XPO1 Inhibition in Thymic Epithelial Tumors. Cancer Res. 2017, 77, 5614–5627. [Google Scholar] [CrossRef] [PubMed]
- Conforti, F.; Wang, Y.; Rodriguez, J.A.; Alberobello, A.T.; Zhang, Y.W.; Giaccone, G. Molecular Pathways: Anticancer Activity by Inhibition of Nucleocytoplasmic Shuttling. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 4508–4513. [Google Scholar] [CrossRef]
- Saenz-Ponce, N.; Pillay, R.; de Long, L.M.; Kashyap, T.; Argueta, C.; Landesman, Y.; Hazar-Rethinam, M.; Boros, S.; Panizza, B.; Jacquemyn, M.; et al. Targeting the XPO1-dependent nuclear export of E2F7 reverses anthracycline resistance in head and neck squamous cell carcinomas. Sci. Transl. Med. 2018, 10, 1–12. [Google Scholar] [CrossRef]
- García-García, M.; Sánchez-Perales, S.; Jarabo, P.; Calvo, E.; Huyton, T.; Fu, L.; Ng, S.C.; Sotodosos-Alonso, L.; Vázquez, J.; Casas-Tintó, S.; et al. Mechanical control of nuclear import by Importin-7 is regulated by its dominant cargo YAP. Nat. Commun. 2022, 13, 1174. [Google Scholar] [CrossRef]
- Nganou, G.; Silva, C.G.; Gladwyn-Ng, I.; Engel, D.; Coumans, B.; Delgado-Escueta, A.V.; Tanaka, M.; Nguyen, L.; Grisar, T.; de Nijs, L.; et al. Importin-8 Modulates Division of Apical Progenitors, Dendritogenesis and Tangential Migration During Development of Mouse Cortex. Front. Mol. Neurosci. 2018, 11, 234. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, L.; Chen, L.; Gong, B.; Jia, D.; Sun, Q. Nuclear transport proteins: Structure, function, and disease relevance. Signal Transduct. Target. Ther. 2023, 8, 425. [Google Scholar] [CrossRef]
- Wei, Y.; Li, L.; Wang, D.; Zhang, C.Y.; Zen, K. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J. Biol. Chem. 2014, 289, 10270–10275. [Google Scholar] [CrossRef]
- Kobayashi, H.; Tomari, Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta 2016, 1859, 71–81. [Google Scholar] [CrossRef]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Nowak, I.; Sarshad, A.A. Argonaute Proteins Take Center Stage in Cancers. Cancers 2021, 13, 788. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Cao, H.; Xu, J.; Xu, M.; He, W.; Zhang, W.; Dong, L.; Chen, D. Circ_RPL23A acts as a miR-1233 sponge to suppress the progression of clear cell renal cell carcinoma by promoting ACAT2. J. Bioenerg. Biomembr. 2021, 53, 415–428. [Google Scholar] [CrossRef]
- Meng, X.M.; Nikolic-Paterson, D.J.; Lan, H.Y. TGF-β: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef]
- Lai, L.Y.S.; Gracie, N.P.; Gowripalan, A.; Howell, L.M.; Newsome, T.P. SMAD proteins: Mediators of diverse outcomes during infection. Eur. J. Cell Biol. 2022, 101, 151204. [Google Scholar] [CrossRef] [PubMed]
- Padmakumar, D.; Chandraprabha, V.R.; Gopinath, P.; Vimala Devi, A.R.T.; Anitha, G.R.J.; Sreelatha, M.M.; Padmakumar, A.; Sreedharan, H. A concise review on the molecular genetics of acute myeloid leukemia. Leuk. Res. 2021, 111, 106727. [Google Scholar] [CrossRef]
- Jiang, Y.; Lv, X.; Ge, X.; Qu, H.; Zhang, Q.; Lu, K.; Lu, Y.; Xue, C.; Zhang, L.; Wang, X. Wilms tumor gent 1 (WT1)-specific adoptive immunotherapy in hematologic diseases. Int. Immunopharmacol. 2021, 94, 107504. [Google Scholar] [CrossRef]
- Stroggilos, R.; Frantzi, M.; Zoidakis, J.; Mokou, M.; Moulavasilis, N.; Mavrogeorgis, E.; Melidi, A.; Makridakis, M.; Stravodimos, K.; Roubelakis, M.G.; et al. Gene Expression Monotonicity across Bladder Cancer Stages Informs on the Molecular Pathogenesis and Identifies a Prognostic Eight-Gene Signature. Cancers 2022, 14, 2542. [Google Scholar] [CrossRef]
- Bahramy, A.; Zafari, N.; Izadi, P.; Soleymani, F.; Kavousi, S.; Noruzinia, M. The Role of miRNAs 340-5p, 92a-3p, and 381-3p in Patients with Endometriosis: A Plasma and Mesenchymal Stem-Like Cell Study. BioMed Res. Int. 2021, 2021, 5298006. [Google Scholar] [CrossRef]
- Dahlberg, J.E.; Lund, E.; Goodwin, E.B. Nuclear translation: What is the evidence? RNA 2003, 9, 1–8. [Google Scholar] [CrossRef]
- Nagai, M.; Yoneda, Y. Small GTPase Ran and Ran-binding proteins. Biomol. Concepts 2012, 3, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Görlich, D.; Dabrowski, M.; Bischoff, F.R.; Kutay, U.; Bork, P.; Hartmann, E.; Prehn, S.; Izaurralde, E. A novel class of RanGTP binding proteins. J. Cell Biol. 1997, 138, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2005, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef]
- Baek, M.; Shin, W.-H.; Chung, H.W.; Seok, C. GalaxyDock BP2 score: A hybrid scoring function for accurate protein–ligand docking. J. Comput.-Aided Mol. Des. 2017, 31, 653–666. [Google Scholar] [CrossRef]
- Shin, W.-H.; Kim, J.-K.; Kim, D.-S.; Seok, C. GalaxyDock2: Protein–ligand docking using beta-complex and global optimization. J. Comput. Chem. 2013, 34, 2647–2656. [Google Scholar] [CrossRef]
- Park, T.; Won, J.; Baek, M.; Seok, C. GalaxyHeteromer: Protein heterodimer structure prediction by template-based and ab initio docking. Nucleic Acids Res. 2021, 49, W237–W241. [Google Scholar] [CrossRef]
- Malamos, P.; Kalyvianaki, K.; Panagiotopoulos, A.A.; Vogiatzoglou, A.P.; Tsikalaki, A.A.; Katifori, A.; Polioudaki, H.; Darivianaki, M.N.; Theodoropoulos, P.A.; Panagiotidis, C.A.; et al. Nuclear translocation of the membrane oxoeicosanoid/androgen receptor, OXER1: Possible mechanisms involved. Mol. Cell. Endocrinol. 2024, 594, 112357. [Google Scholar] [CrossRef]
- Kalyvianaki, K.; Drosou, I.; Notas, G.; Castanas, E.; Kampa, M. Enhanced OXER1 expression is indispensable for human cancer cell migration. Biochem. Biophys. Res. Commun. 2021, 584, 95–100. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panagiotopoulos, A.A.; Kalyvianaki, K.; Angelidaki, A.; Dellis, D.; Panagiotidis, C.A.; Kampa, M.; Castanas, E. The Sequence [RRKLPVGRS] Is a Nuclear Localization Signal for Importin 8 Binding (NLS8): A Chemical Biology and Bioinformatics Study. Int. J. Mol. Sci. 2025, 26, 2814. https://doi.org/10.3390/ijms26062814
Panagiotopoulos AA, Kalyvianaki K, Angelidaki A, Dellis D, Panagiotidis CA, Kampa M, Castanas E. The Sequence [RRKLPVGRS] Is a Nuclear Localization Signal for Importin 8 Binding (NLS8): A Chemical Biology and Bioinformatics Study. International Journal of Molecular Sciences. 2025; 26(6):2814. https://doi.org/10.3390/ijms26062814
Chicago/Turabian StylePanagiotopoulos, Athanasios A., Konstantina Kalyvianaki, Aikaterini Angelidaki, Dimitris Dellis, Christos A. Panagiotidis, Marilena Kampa, and Elias Castanas. 2025. "The Sequence [RRKLPVGRS] Is a Nuclear Localization Signal for Importin 8 Binding (NLS8): A Chemical Biology and Bioinformatics Study" International Journal of Molecular Sciences 26, no. 6: 2814. https://doi.org/10.3390/ijms26062814
APA StylePanagiotopoulos, A. A., Kalyvianaki, K., Angelidaki, A., Dellis, D., Panagiotidis, C. A., Kampa, M., & Castanas, E. (2025). The Sequence [RRKLPVGRS] Is a Nuclear Localization Signal for Importin 8 Binding (NLS8): A Chemical Biology and Bioinformatics Study. International Journal of Molecular Sciences, 26(6), 2814. https://doi.org/10.3390/ijms26062814








