Cell Wall–Based Machine Learning Models to Predict Plant Growth Using Onion Epidermis
Abstract
1. Introduction
2. Results
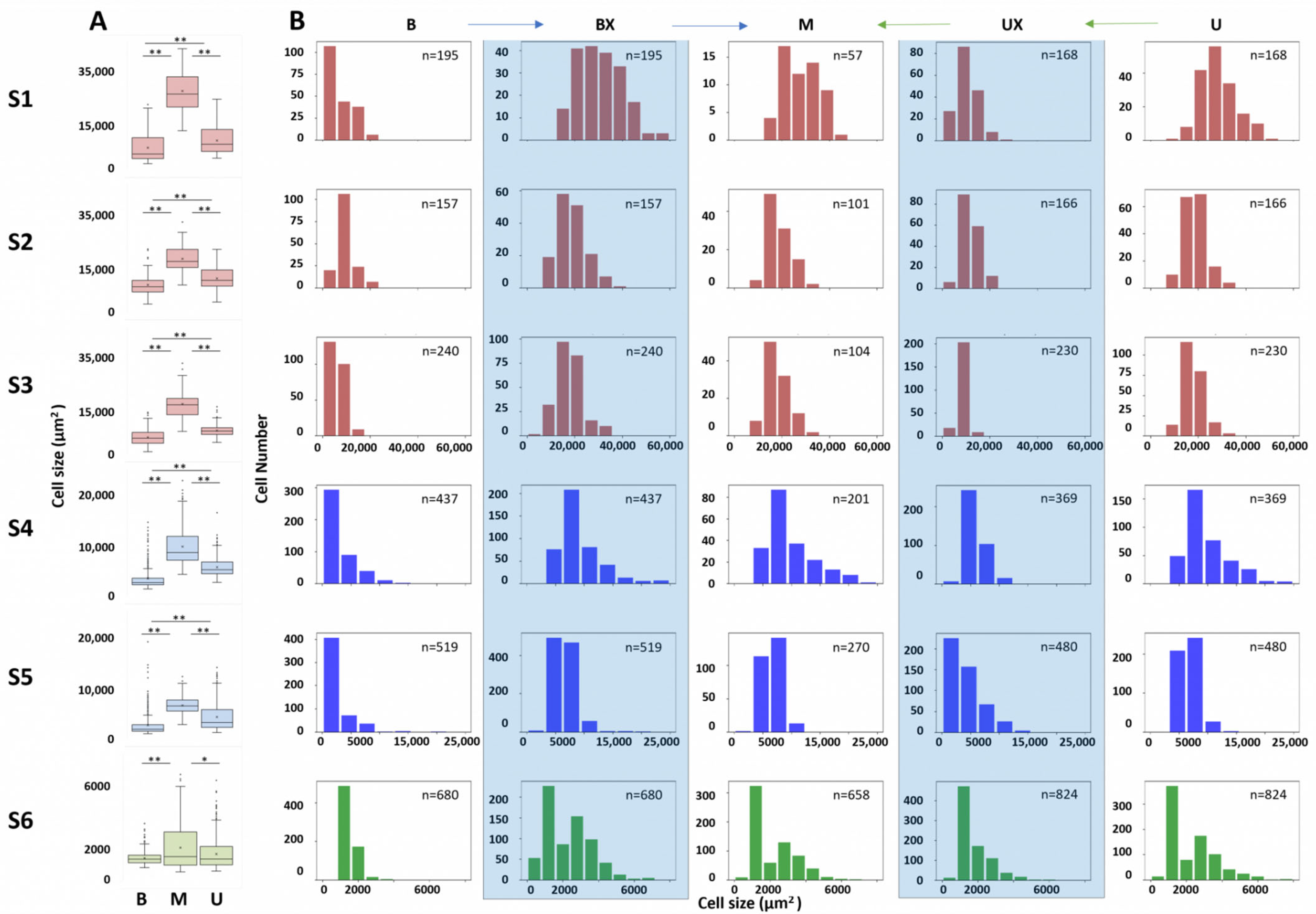
2.1. Analysis of Adaxial Epidermal Cell Size Using Cell Wall Staining
2.2. FTIR and Biochemical Spectroscopic Analysis
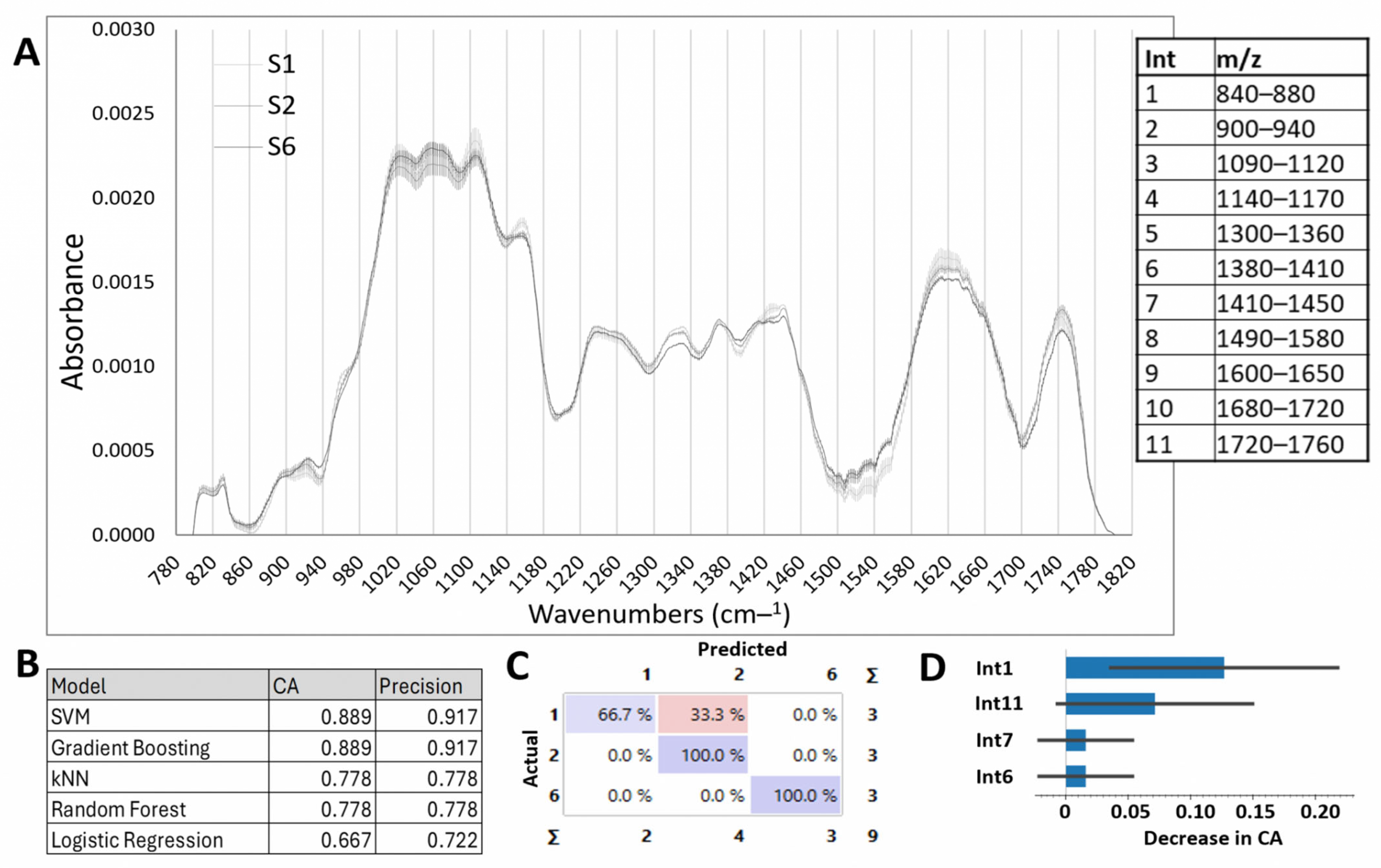
2.2.1. Cell Wall Compounds: FTIR Analysis
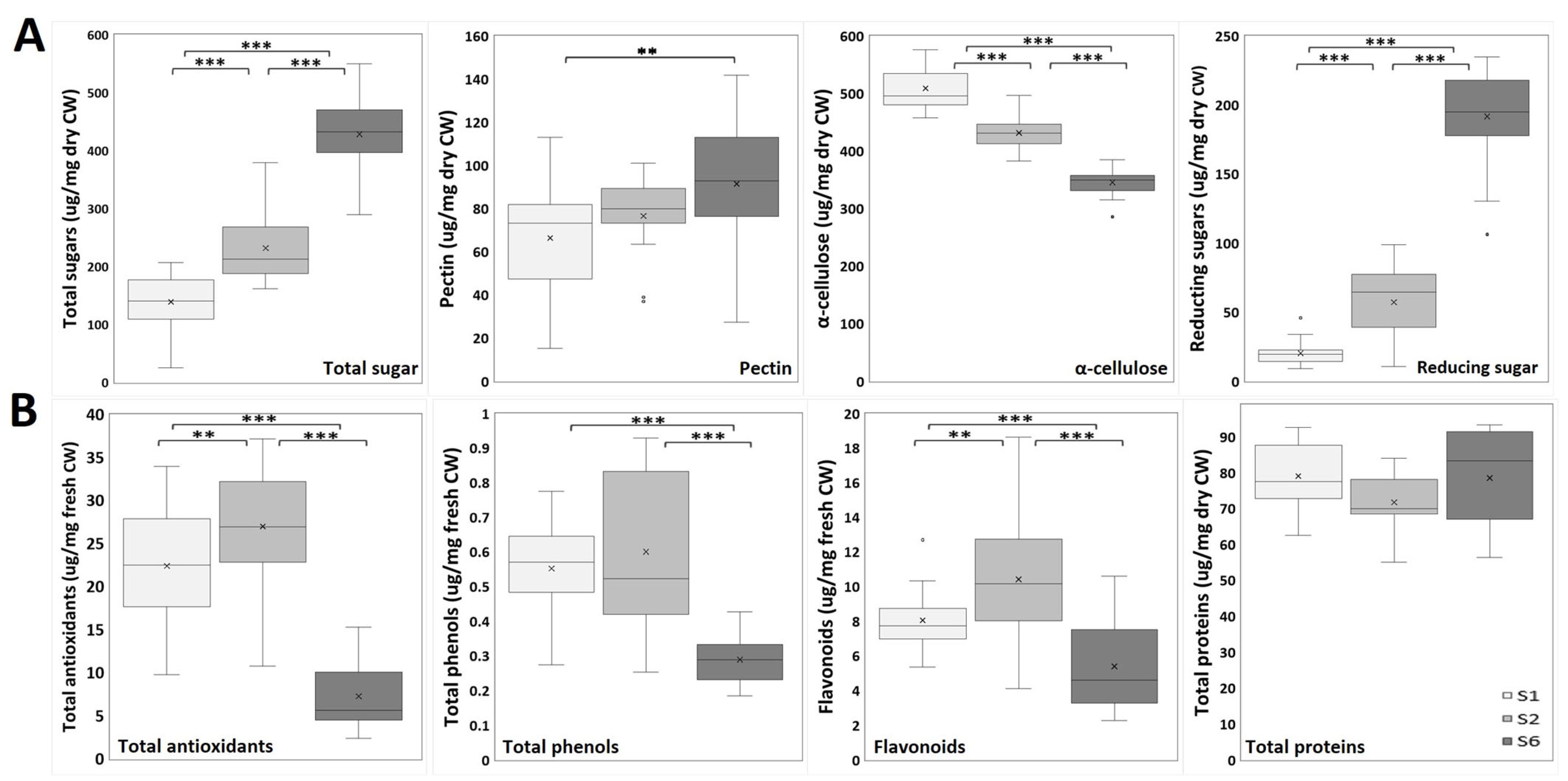
2.2.2. Biochemical Analysis: Cell Wall and Nutraceutical Compounds
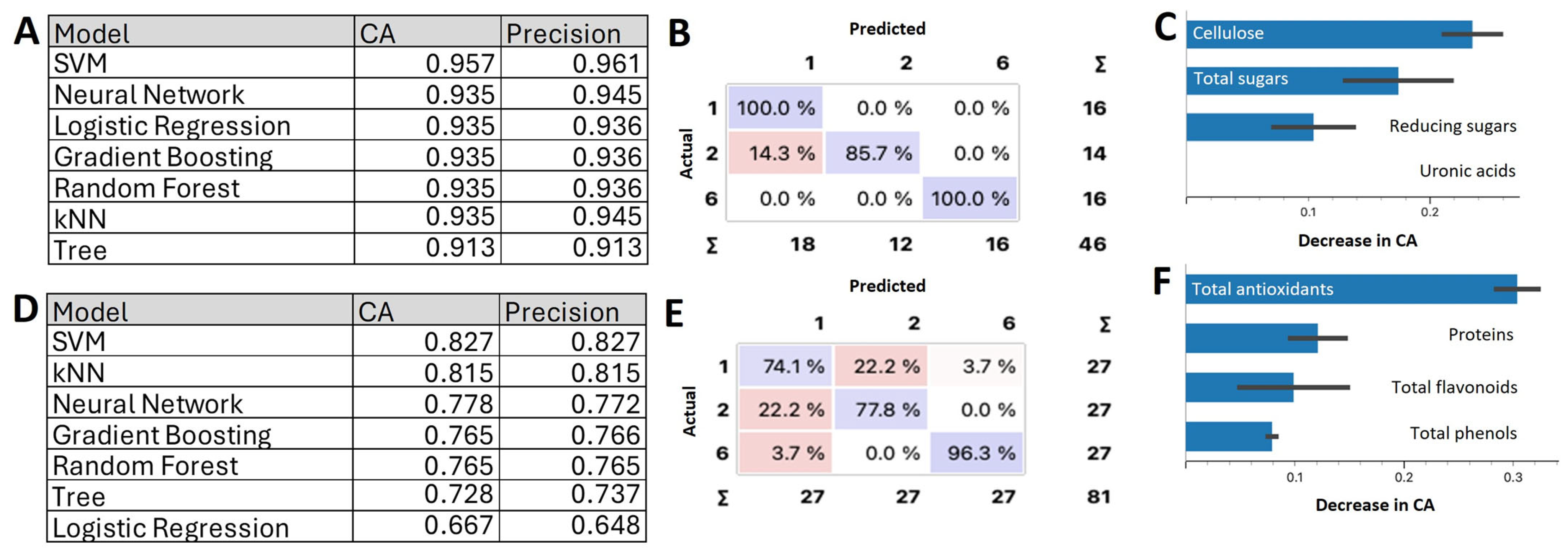
2.2.3. Machine Learning Models for Layer Classification Using the CWC and NUTC Data
2.3. Gene Expression Analysis of Cell Wall Enzymes by RT-qPCR
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Microscopy Analysis of Cell Morphology
4.3. Spectrophotometric and Biochemical Analysis
4.3.1. Cell Wall Compound (CWC) Analysis
4.3.2. Nutraceutical Analysis
4.3.3. Total Protein Quantification
4.4. CWE Gene Expression
4.4.1. CWE Gene Selection
4.4.2. RNA Extraction
4.4.3. Gene Expression Analysis Using RT-qPCR
4.5. Supervised Analysis and Predictive Models
4.5.1. FTIR Analysis
4.5.2. CWC and NUTC Analysis
4.5.3. Correlational Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hilty, J.; Muller, B.; Pantin, F.; Leuzinger, S. Plant growth: The What, the How, and the Why. New Phytol. 2021, 232, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Plant cell wall extensibility: Connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 2016, 67, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Huang, S.; Zhou, J.; Bai, Y.; Liu, Y.; Shi, L.; Liu, S.; Hu, Z.; Tang, Y. miR397-LACs mediated cadmium stress tolerance in Arabidopsis thaliana. Plant Mol. Biol. 2023, 113, 415–430. [Google Scholar] [CrossRef]
- Höfte, H.; Voxeur, A. Plant cell walls. Curr. Biol. 2017, 27, R865–R870. [Google Scholar] [CrossRef]
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T.; et al. Toward a Systems Approach to Understanding Plant Cell Walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef]
- Timm, M.; Offringa, L.C.; Van Klinken, B.J.W.; Slavin, J. Beyond Insoluble Dietary Fiber: Bioactive Compounds in Plant Foods. Nutrients 2023, 15, 4138. [Google Scholar] [CrossRef] [PubMed]
- Alsulami, F.J.; Shaheed, S.U. Role of Natural Antioxidants in Cancer. In Nutrition and Dietary Interventions in Cancer; Saeed, R.F., Shaheed, S.U., Eds.; Springer: Cham, Switzerland, 2024; Volume 191, pp. 95–117. [Google Scholar] [CrossRef]
- Flieger, J.; Flieger, W.; Baj, J.; Maciejewski, R. Antioxidants: Classification, Natural Sources, Activity/Capacity Measurements, and Usefulness for the Synthesis of Nanoparticles. Materials 2021, 14, 4135. [Google Scholar] [CrossRef]
- Liu, R.H. Health-promoting components of fruits and vegetables in the diet. Adv. Nutr. 2013, 4, 384S–392S. [Google Scholar] [CrossRef]
- Le Gall, H.; Philippe, F.; Domon, J.M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell Wall Metabolism in Response to Abiotic Stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Xu, J.; Yan, J.; Li, W.; Wang, Q.; Wang, C.; Guo, J.; Geng, D.; Guan, Q.; Ma, F. Integrative analyses of widely targeted metabolic profiling and transcriptome data reveals molecular insight into metabolomic variations during apple (Malus domestica) fruit development and ripening. Int. J. Mol. Sci. 2020, 21, 4797. [Google Scholar] [CrossRef]
- Ibragimova, N.; Mokshina, N.; Ageeva, M.; Gurjanov, O.; Mikshina, P. Rearrangement of the Cellulose-Enriched Cell Wall in Flax Phloem Fibers over the Course of the Gravitropic Reaction. Int. J. Mol. Sci. 2020, 21, 5322. [Google Scholar] [CrossRef] [PubMed]
- Saffer, A.M.; Baskin, T.I.; Verma, A.; Stanislas, T.; Oldenbourg, R.; Irish, V.F. Cellulose assembles into helical bundles of uniform handedness in cell walls with abnormal pectin composition. Plant J. 2023, 116, 855–870. [Google Scholar] [CrossRef] [PubMed]
- Tian, L. Recent advances in understanding carotenoid-derived signaling molecules in regulating plant growth and development. Front. Plant Sci. 2015, 6, 790. [Google Scholar] [CrossRef]
- Liu, X.; Le Bourvellec, C.; Renard, C.M.G.C. Interactions between cell wall polysaccharides and polyphenols: Effect of molecular internal structure. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3574–3617. [Google Scholar] [CrossRef]
- Van Sandt, V.S.T.; Suslov, D.; Verbelen, J.P.; Vissenberg, K. Xyloglucan Endotransglucosylase Activity Loosens a Plant Cell Wall. Ann. Bot. 2007, 100, 1467–1473. [Google Scholar] [CrossRef]
- Cosgrove, D.J.; Li, L.C.; Cho, H.T.; Hoffmann-Benning, S.; Moore, R.C.; Blecker, D. The Growing World of Expansins. Plant Cell Physiol. 2002, 43, 1436–1444. [Google Scholar] [CrossRef]
- Chandrasekar, B.; van der Hoorn, R.A.L. Beta galactosidases in Arabidopsis and tomato—A mini review. Biochem. Soc. Trans. 2016, 44, 150–158. [Google Scholar] [CrossRef]
- Verhage, L. Get in shape—How a polygalacturonase affects plant morphology. Plant J. 2021, 106, 1491–1492. [Google Scholar] [CrossRef]
- Wang, D.; Yeats, T.H.; Uluisik, S.; Rose, J.K.C.; Seymour, G.B. Fruit softening: Revisiting the role of pectin. Trends Plant Sci. 2018, 23, 302–310. [Google Scholar] [CrossRef]
- Guo, S.; Wang, M.; Song, X.; Zhou, G.; Kong, Y. The evolving views of the simplest pectic polysaccharides: Homogalacturonan. Plant Cell Rep. 2022, 41, 2111–2123. [Google Scholar] [CrossRef]
- Röckel, N.; Wolf, S.; Kost, B.; Rausch, T.; Greiner, S. Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis and reflects the distribution of esterified and de-esterified pectins. Plant J. 2008, 53, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Pogorelko, G.; Fursova, O.; Lin, M.; Pyle, E.; Jass, J.; Zabotina, O.A. Post-synthetic modification of plant cell walls by expression of microbial hydrolases in the apoplast. Plant Mol. Biol. 2011, 77, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Perrot, T.; Pauly, M.; Ramírez, V. Emerging roles of β-glucanases in plant development and adaptative responses. Plants 2022, 11, 1119. [Google Scholar] [CrossRef]
- Mendonça, M.; Barroca, M.; Collins, T. Endo-1,4-β-xylanase-containing glycoside hydrolase families: Characteristics, singularities, and similarities. Biotechnol. Adv. 2023, 65, 108148. [Google Scholar] [CrossRef]
- Fry, S.C. Primary cell wall metabolism: Tracking the careers of wall polymers in living plant cells. New Phytol. 2004, 161, 641–675. [Google Scholar] [CrossRef]
- Wu, S.; Ning, F.; Wu, X.; Wang, W. Proteomic Characterization of Differential Abundant Proteins Accumulated between Lower and Upper Epidermises of Fleshy Scales in Onion (Allium cepa L.) Bulbs. PLoS ONE 2016, 11, e0168959. [Google Scholar] [CrossRef]
- Krivanek, J.; Soldatov, R.A.; Kastriti, M.E.; Chontorotzea, T.; Herdina, A.N.; Petersen, J.; Szarowska, B.; Landova, M.; Matejova, V.K.; Holla, L.I.; et al. Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth. Nat. Commun. 2020, 11, 4816. [Google Scholar] [CrossRef]
- Suslov, D.; Verbelen, J.P.; Vissenberg, K. Onion epidermis as a new model to study the control of growth anisotropy in higher plants. J. Exp. Bot. 2009, 60, 4175–4187. [Google Scholar] [CrossRef]
- Nicolas, W.J.; Fäßler, F.; Dutka, P.; Schur, F.K.M.; Jensen, G.; Meyerowitz, E. Cryo-electron tomography of the onion cell wall shows bimodally oriented cellulose fibers and reticulated homogalacturonan networks. Curr. Biol. 2022, 32, 2375–2389.e6. [Google Scholar] [CrossRef]
- Haswell, E.S.; Dixit, R. Counting what counts: The importance of quantitative approaches to studying plant cell biology. Curr. Opin. Plant Biol. 2018, 46, A1–A3. [Google Scholar] [CrossRef]
- Araújo, S.O.; Peres, R.S.; Ramalho, J.C.; Lidon, F.; Barata, J. Machine Learning Applications in Agriculture: Current Trends, Challenges, and Future Perspectives. Agronomy 2023, 13, 2976. [Google Scholar] [CrossRef]
- Civantos-Gómez, I.; Rubio Teso, M.L.; Galeano, J.; Rubiales, D.; Iriondo, J.M.; García-Algarra, J. Climate change conditions the selection of rust-resistant candidate wild lentil populations for in situ conservation. Front. Plant Sci. 2022, 13, 1010799. [Google Scholar] [CrossRef]
- Van Dijk, A.D.J.; Kootstra, G.; Kruijer, W.; De Ridder, D. Machine learning in plant science and plant breeding. iScience 2021, 24, 101890. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Ko, J.; Shin, T.; Yeom, J.M. Incorporation of machine learning and deep neural network approaches into a remote sensing-integrated crop model for the simulation of rice growth. Sci. Rep. 2022, 12, 9030. [Google Scholar] [CrossRef]
- Sakeef, N.; Scandola, S.; Kennedy, C.; Lummer, C.; Chang, J.; Uhrig, R.G.; Lin, G. Machine learning classification of plant genotypes grown under different light conditions through the integration of multi-scale time-series data. Comput. Struct. Biotechnol. J. 2023, 21, 3183–3195. [Google Scholar] [CrossRef]
- Yoosefzadeh Najafabadi, M.; Hesami, M.; Eskandari, M. Machine Learning-Assisted Approaches in Modernized Plant Breeding Programs. Genes 2023, 14, 777. [Google Scholar] [CrossRef]
- Zakieva, A.; Cerrone, L.; Greb, T. Deep machine learning for cell segmentation and quantitative analysis of radial plant growth. Cells Dev. 2023, 174, 203842. [Google Scholar] [CrossRef]
- Silva, J.C.F.; Teixeira, R.M.; Silva, F.F.; Brommonschenkel, S.H.; Fontes, E.P.B. Machine learning approaches and their current application in plant molecular biology: A systematic review. Plant Sci. 2019, 284, 37–47. [Google Scholar] [CrossRef]
- Miedes, E.; Zarra, I.; Hoson, T.; Herbers, K.; Sonnewald, U.; Lorences, E.P. Xyloglucan endotransglucosylase and cell wall extensibility. J. Plant Physiol. 2011, 168, 196–203. [Google Scholar] [CrossRef]
- Miedes, E.; Suslov, D.; Vandenbussche, F.; Kenobi, K.; Ivakov, A.; Van Der Straeten, D.; Lorences, E.P.; Mellerowicz, E.J.; Verbelen, J.P.; Vissenberg, K. Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. J. Exp. Bot. 2013, 64, 2481–2497. [Google Scholar] [CrossRef]
- Sablowski, R.; Gutiérrez, C. Cycling in a crowd: Coordination of plant cell division, growth, and cell fate. Plant Cell 2022, 34, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Le Gloanec, C.; Collet, L.; Silveira, S.R.; Wang, B.; Routier-Kierzkowska, A.-L.; Kierzkowski, D. Cell type-specific dynamics underlie cellular growth variability in plants. Development 2022, 149, 200783. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Yang, J.; Zabotina, O.A.; Yu, C. Surface-enhanced Raman spectroscopic chemical imaging reveals distribution of pectin and its co-localization with xyloglucan inside onion epidermal cell wall. PLoS ONE 2021, 16, e0250650. [Google Scholar] [CrossRef]
- Xiang, M.; Yuan, S.; Zhang, Q.; Liu, X.; Li, Q.; Leng, Z.; Sha, J.; Anderson, C.T.; Xiao, C. Galactosylation of xyloglucan is essential for the stabilization of the actin cytoskeleton and endomembrane system through the proper assembly of cell walls. J. Exp. Bot. 2023, 74, 5104–5123. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Structure and growth of plant cell walls. Nat. Rev. Mol. Cell Biol. 2024, 25, 340–358. [Google Scholar] [CrossRef]
- Moussu, S.; Lee, H.K.; Haas, K.T.; Broyart, C.; Rathgeb, U.; De Bellis, D.; Levasseur, T.; Schoenaers, S.; Fernandez, G.S.; Grossniklaus, U.; et al. Plant cell wall patterning and expansion mediated by protein-peptide-polysaccharide interaction. Science 2023, 382, 719–725. [Google Scholar] [CrossRef]
- Zhou, Y.; Gao, Y.H.; Zhang, B.C.; Yang, H.L.; Tian, Y.B.; Huang, Y.H.; Yin, C.C.; Tao, J.J.; Wei, W.; Zhang, W.K.; et al. CELLULOSE SYNTHASE-LIKE C proteins modulate cell wall establishment during ethylene-mediated root growth inhibition in rice. Plant Cell 2024, 36, 3751–3769. [Google Scholar] [CrossRef]
- Wilson, M.H.; Holman, T.J.; Sørensen, I.; Cancho Sánchez, E.; Wells, D.M.; Swarup, R.; Knox, J.P.; Willats, W.G.; Ubeda Tomás, S.; Holdsworth, M. Multi-omics analysis identifies genes mediating the extension of cell walls in the Arabidopsis thaliana root elongation zone. Front. Cell Dev. Biol. 2015, 3, 10. [Google Scholar] [CrossRef]
- Cheddadi, I.; Génard, M.; Bertin, N.; Godin, C. Coupling water fluxes with cell wall mechanics in a multicellular model of plant development. PLoS Comput. Biol. 2019, 15, e1007121. [Google Scholar] [CrossRef]
- Kaur, H.; Manna, M.; Thakur, T.; Gautam, V.; Salvi, P. Imperative role of sugar signaling and transport during drought stress responses in plants. Physiol. Plant 2021, 171, 833–848. [Google Scholar] [CrossRef]
- Ng, A.; Parker, M.; Parr, A.; Saunders, P.K.; Smith, A.; Waldron, K. Physicochemical characteristics of onion (Allium cepa L.) tissues. J. Agric. Food Chem. 2000, 48, 5612–5617. [Google Scholar] [CrossRef] [PubMed]
- Coolong, T.; Randle, W.; Wicker, L. Structural and chemical differences in the cell wall regions in relation to scale firmness of three onion (Allium cepa L.) selections at harvest and during storage. J. Sci. Food Agric. 2008, 88, 1277–1286. [Google Scholar] [CrossRef]
- Zhang, T.; Mahgsoudy-Louyeh, S.; Tittmann, B.; Cosgrove, D. Visualization of the nanoscale pattern of recently-deposited cellulose microfibrils and matrix materials in never-dried primary walls of the onion epidermis. Cellulose 2014, 21, 853–862. [Google Scholar] [CrossRef]
- Pérez-Gregorio, R.; García-Falcón, M.S.; Simal-Gándara, J.; Rodrigues, A.; Almeida, D. Identification and quantification of flavonoids in traditional cultivars of red and white onions at harvest. J. Food Compos. Anal. 2010, 23, 592–598. [Google Scholar] [CrossRef]
- Sharma, K.; Assefa, A.D.; Kim, S.; Ko, E.; Lee, E.T.; Park, S.W. Evaluation of total phenolics, flavonoids, and antioxidant activity of 18 Korean onion cultivars: A comparative study. J. Sci. Food Agric. 2014, 94, 1521–1529. [Google Scholar] [CrossRef]
- Tedesco, I.; Carbone, V.; Spagnuolo, C.; Minasi, P.; Russo, G. Identification and quantification of flavonoids from two southern Italian cultivars of Allium cepa L., Tropea (Red Onion) and Montoro (Copper Onion), and their capacity to protect human erythrocytes from oxidative stress. J. Agric. Food Chem. 2015, 63, 5229–5238. [Google Scholar] [CrossRef]
- Rodov, V.; Tietel, Z.; Vinokur, Y.; Horev, B.; Eshel, D. Ultraviolet light stimulates flavonol accumulation in peeled onions and controls microorganisms on their surface. J. Agric. Food Chem. 2010, 58, 9071–9076. [Google Scholar] [CrossRef]
- Milea, Ș.A.; Vasile, M.A.; Crăciunescu, O.; Prelipcean, A.-M.; Bahrim, G.E.; Râpeanu, G.; Oancea, A.; Stănciuc, N. Co-microencapsulation of flavonoids from yellow onion skins and lactic acid bacteria lead to multifunctional ingredients for nutraceutical and pharmaceutical applications. Pharmaceutics 2020, 12, 1053. [Google Scholar] [CrossRef]
- Liakos, K.G.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine Learning in Agriculture: A Review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef]
- Wolf, S.; Greiner, S. Growth control by cell wall pectins. Protoplasma 2012, 249, 169–175. [Google Scholar] [CrossRef]
- Moneo-Sánchez, M.; Izquierdo, L.; Martín, I.; Hernández-Nistal, J.; Albornos, L.; Dopico, B.; Labrador, E. Knockout mutants of Arabidopsis thaliana β-galactosidase: Modifications in the cell wall saccharides and enzymatic activities. Biol. Plant 2018, 62, 80–88. [Google Scholar] [CrossRef]
- Sampedro, J.; Gianzo, C.; Iglesias, N.; Guitián, E.; Revilla, G.; Zarra, I. AtBGAL10 is the main xyloglucan β-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiol. 2012, 158, 1146–1157. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, L. Plant cell wall loosening factors. Chin. Plant Physiol. J. 2011, 47, 925–935. [Google Scholar]
- Behar, H.; Graham, S.; Brumer, H. Comprehensive cross-genome survey and phylogeny of glycoside hydrolase family 16 members reveals the evolutionary origin of EG16 and XTH proteins in plant lineages. Plant J. 2018, 95, 1114–1128. [Google Scholar] [CrossRef]
- Shinohara, N.; Nishitani, K. Cryogenian origin and subsequent diversification of the plant cell-wall enzyme XTH family. Plant Cell Physiol. 2021, 62, 1874–1889. [Google Scholar] [CrossRef]
- Miedes, E.; Lorences, E.P. Changes in cell wall pectin and pectinase activity in apple and tomato fruits during Penicillium expansum infection. J. Sci. Food Agric. 2006, 86, 1359–1364. [Google Scholar] [CrossRef]
- Maris, A.; Suslov, D.; Fry, S.; Verbelen, J.; Vissenberg, K. Enzymic characterization of two recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis and their effect on root growth and cell wall extension. J. Exp. Bot. 2009, 60, 3959–3972. [Google Scholar] [CrossRef]
- Farkaš, V.; Ait-Mohand, F.; Stratilová, E. Sensitive detection of transglycosylating activity of xyloglucan endotransglycosylase/hydrolase (XTH) after isoelectric focusing in polyacrylamide gels. Plant Physiol. Biochem. 2005, 43, 431–435. [Google Scholar] [CrossRef]
- Sampedro, J.; Pardo, B.; Gianzo, C.; Guitián, E.; Revilla, G.; Zarra, I. Lack of α-xylosidase activity in Arabidopsis alters xyloglucan composition and results in growth defects. Plant Physiol. 2010, 154, 1105–1115. [Google Scholar] [CrossRef]
- Wolf, S.; Mouille, G.; Pelloux, J. Homogalacturonan methyl-esterification and plant development. Mol. Plant 2009, 2, 851–860. [Google Scholar] [CrossRef]
- Hongo, S.; Sato, K.; Yokoyama, R.; Nishitani, K. Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. Plant Cell 2012, 24, 2624–2634. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yu, Y.; Liang, Y.; Anderson, C.T.; Cao, J. A profusion of molecular scissors for pectins: Classification, expression, and functions of plant polygalacturonases. Front. Plant Sci. 2018, 9, 1208. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.; Xiao, C.; Yi, H.; Kandemir, B.; Wang, J.Z.; Puri, V.; Anderson, C.T. POLYGALACTURONASE INVOLVED IN EXPANSION3 functions in seedling development, rosette growth, and stomatal dynamics in Arabidopsis thaliana. Plant Cell 2017, 29, 2413–2432. [Google Scholar] [CrossRef]
- Leonard, R.; Pabst, M.; Bondili, J.S.; Chambat, G.; Veit, C.; Strasser, R.; Altmann, F. Identification of an Arabidopsis gene encoding a GH95 α1,2-fucosidase active on xyloglucan oligo- and polysaccharides. Phytochemistry 2008, 69, 1983–1988. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Peaucelle, A.; Braybrook, S.A.; Le Guillou, L.; Bron, E.; Kuhlemeier, C.; Höfte, H. Pectin-Induced Changes in Cell Wall Mechanics Underlie Organ Initiation in Arabidopsis. Curr. Biol. 2011, 21, 1720–1726. [Google Scholar] [CrossRef]
- Günl, M.; Neumetzler, L.; Krämer, F.; de Souza, A.; Schultink, A.; Peña, M.; York, W.S.; Pauly, M. AXY8 encodes an α-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 2011, 23, 4025–4040. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Pattathil, S.; Hahn, M.G.; Molina, A.; Miedes, E. Characterization of Plant Cell Wall Damage-Associated Molecular Patterns Regulating Immune Responses. In Plant Pattern Recognition Receptors; Shan, L., He, P., Eds.; Springer: New York, NY, USA, 2017; Volume 1578, pp. 13–23. [Google Scholar] [CrossRef]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Blumenkrantz, N.; Asboe-Hansen, G. New method for quantitative determination of uronic acids. Anal. Biochem. 1973, 54, 484–489. [Google Scholar] [CrossRef]
- Liao, H.; Dong, W.; Shi, X.; Liu, H.; Yuan, K. Analysis and comparison of the active components and antioxidant activities of extracts from Abelmoschus esculentus L. Phcog. Mag. 2012, 8, 156. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Demšar, J.; Curk, T.; Erjavec, A.; Gorup, Č.; Hočevar, T.; Milutinovič, M.; Možina, M.; Polajnar, M.; Toplak, M.; Starič, A.; et al. Orange: Data Mining Toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Walters, S.J.; Campbell, M.J. The use of bootstrap methods for analysing Health-Related Quality of Life outcomes (particularly the SF-36). Health Qual. Life Outcomes 2004, 2, 70. [Google Scholar] [CrossRef] [PubMed]


| Pearson Coefficient | Total Sugar | Uronic Acid | α-Cellulose | Reducing Sugar | Total Antioxidants | Total Phenols | Flavonoids | Total Protein |
|---|---|---|---|---|---|---|---|---|
| XTH | 0.0740 | 0.0543 | 0.0793 | 0.1834 | −0.3271 | −0.2462 | −0.3853 | 0.2013 |
| Glucanase | 0.7180 | 0.0660 | −0.5599 | 0.7575 | −0.8687 | −0.7716 | −0.7780 | 0.3506 |
| Xylanase | −0.4785 | −0.0111 | 0.4158 | −0.4027 | 0.2773 | 0.2604 | 0.1591 | −0.0376 |
| Expansin | 0.3501 | −0.1167 | −0.4240 | 0.2851 | −0.0803 | −0.0824 | 0.0091 | −0.0138 |
| Galactosidase | −0.7815 | −0.0727 | 0.7560 | −0.7537 | 0.5477 | 0.6119 | 0.3442 | −0.0613 |
| Fucosidase | 0.9577 | 0.1292 | −0.8385 | 0.9662 | −0.9134 | −0.8780 | −0.6842 | 0.1053 |
| PG | 0.2823 | 0.0370 | −0.2317 | 0.3200 | −0.5384 | −0.4106 | −0.4796 | 0.2992 |
| PLL18 | 0.5125 | −0.0807 | −0.4253 | 0.6115 | −0.6310 | −0.5103 | −0.5362 | 0.2233 |
| PME | −0.9467 | 0.0734 | 0.8774 | −0.9482 | 0.7665 | 0.8163 | 0.5503 | −0.1846 |
| Xylosidase | −0.1705 | 0.0094 | 0.1314 | −0.0698 | −0.1008 | −0.1306 | −0.3869 | 0.1985 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khoulali, C.; Pastor, J.M.; Galeano, J.; Vissenberg, K.; Miedes, E. Cell Wall–Based Machine Learning Models to Predict Plant Growth Using Onion Epidermis. Int. J. Mol. Sci. 2025, 26, 2946. https://doi.org/10.3390/ijms26072946
Khoulali C, Pastor JM, Galeano J, Vissenberg K, Miedes E. Cell Wall–Based Machine Learning Models to Predict Plant Growth Using Onion Epidermis. International Journal of Molecular Sciences. 2025; 26(7):2946. https://doi.org/10.3390/ijms26072946
Chicago/Turabian StyleKhoulali, Celia, Juan Manuel Pastor, Javier Galeano, Kris Vissenberg, and Eva Miedes. 2025. "Cell Wall–Based Machine Learning Models to Predict Plant Growth Using Onion Epidermis" International Journal of Molecular Sciences 26, no. 7: 2946. https://doi.org/10.3390/ijms26072946
APA StyleKhoulali, C., Pastor, J. M., Galeano, J., Vissenberg, K., & Miedes, E. (2025). Cell Wall–Based Machine Learning Models to Predict Plant Growth Using Onion Epidermis. International Journal of Molecular Sciences, 26(7), 2946. https://doi.org/10.3390/ijms26072946







