Alterations in Gene Expression and Alternative Splicing Induced by Plasmid-Mediated Overexpression of GFP and P2RY12 Within the A549 Cell Line
Abstract
:1. Introduction
2. Results
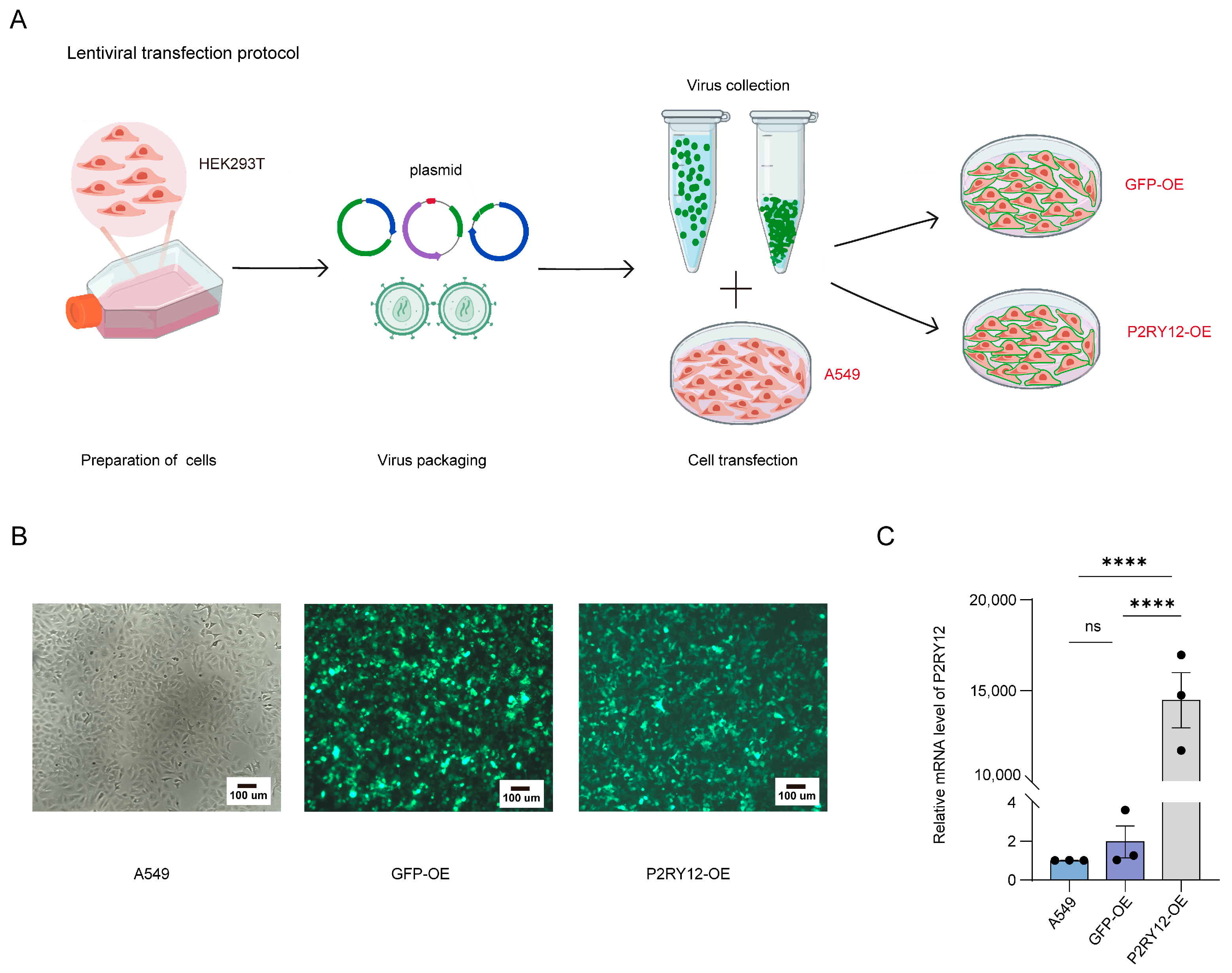
2.1. Establishment of A549, GFP-OE, and P2RY12-OE Cell Lines
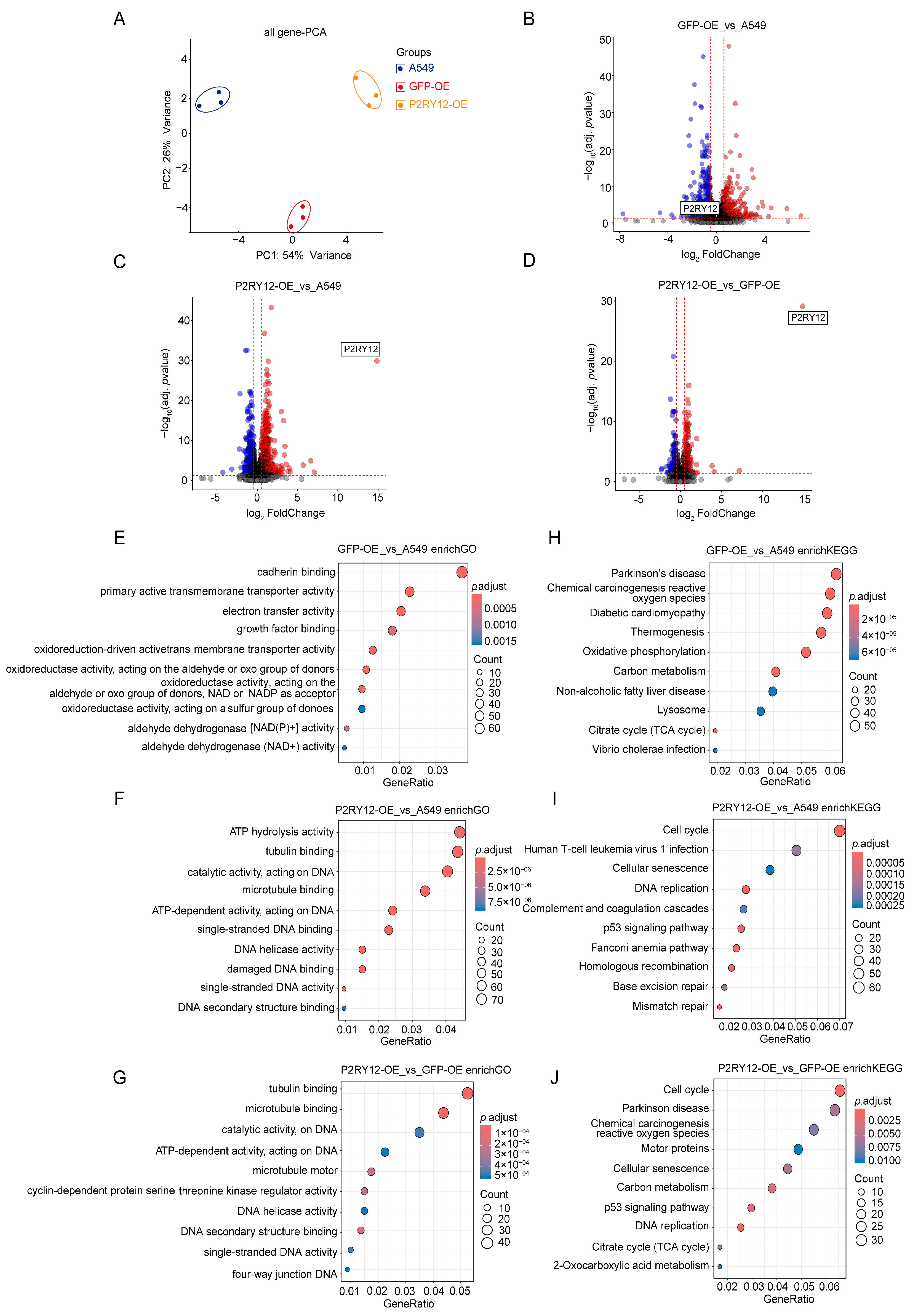
2.2. Gene Expression Differences Between A549, GFP-OE, and P2RY12-OE
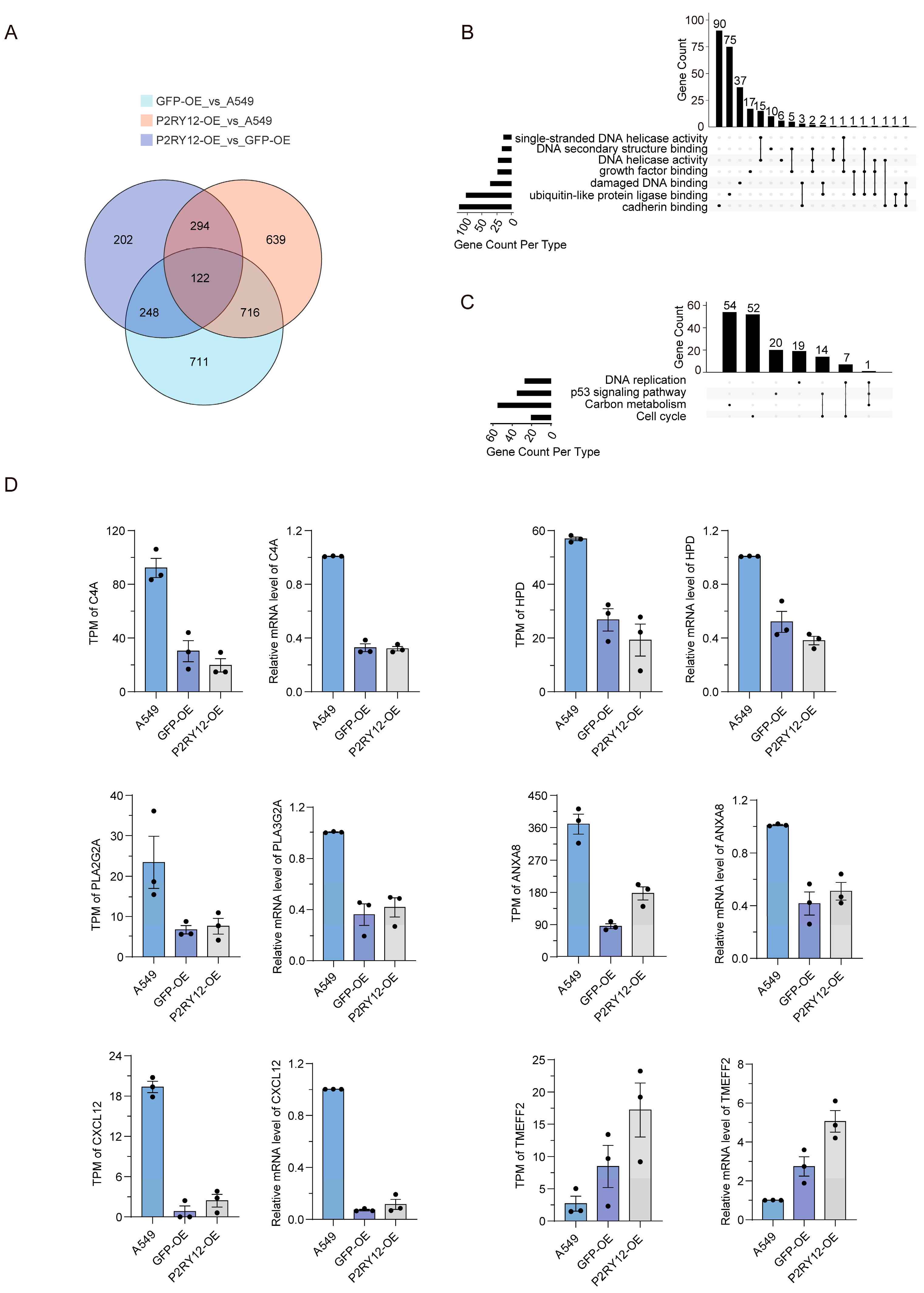
2.3. Overlap and Validation of DEGs Between A549, GFP-OE, and P2RY12-OE
2.4. Identification of Alternative Splicing Events in GFP-OE and P2RY12-OE
2.5. Overlap Between DEGs and gDTUs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Plasmid Transfection
4.3. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.4. RNA-Seq Data Analysis and Validation
4.5. Identification and Quantification of Alternative Splicing Events
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DEGs | Differentially expressed genes |
| gDTUs | Genes with differential transcript usage |
| GFP-OE | GFP over-expression |
| P2RY12-OE | P2RY12 over-expression |
| AS | Alternative splicing |
| AL | Alternative last exon |
| AF | Alternative first exon |
| A5 | Alternative 5′ splice site |
| RI | Retained intron |
| ES | Exon skipping |
| ME | Mutually exclusive exons |
| A3 | Alternative 3′ splice site |
| LUAD | Lung adenocarcinoma |
| PCA | Principal component analysis |
| GO | Gene Ontology |
| FBL | Fibrillarin |
| C4A | Complement C4A |
| TMEFF2 | Transmembrane protein with an EGF-like and two Follistatin-like domains 2 |
| ANXA8 | Annexin A8 |
| HPD | 4-hydroxyphenylpyruvate dioxygenase |
| PLA2G2A | Phospholipase A2, Group IIA |
| CXCL12 | C-X-C motif chemokine ligand 12 |
| NSCLC | Non-small cell lung cancer |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
References
- Lederberg, J. Cell genetics and hereditary symbiosis. Physiol. Rev. 1952, 32, 403–430. [Google Scholar] [CrossRef]
- Cohen, S.N.; Chang, A.C.; Boyer, H.W.; Helling, R.B. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. USA 1973, 70, 3240–3244. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gao, Y.; Wang, M.; Liu, Y.; Wang, F.; Shi, J.; Wang, Z.; Li, R. Adaptive evolution of plasmid and chromosome contributes to the fitness of a blaNDM-bearing cointegrate plasmid in Escherichia coli. ISME J. 2024, 18, wrae037. [Google Scholar] [CrossRef]
- Wein, T.; Dagan, T. Plasmid evolution. Curr. Biol. 2020, 30, R1158–R1163. [Google Scholar] [CrossRef]
- Hinnebusch, A.G.; Ivanov, I.P.; Sonenberg, N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 2016, 352, 1413–1416. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, N.; Gao, G. Quantifying the regulatory potential of genetic variants via a hybrid sequence-oriented model with SVEN. Nat. Commun. 2024, 15, 10917. [Google Scholar] [CrossRef] [PubMed]
- Chong, Z.X.; Yeap, S.K.; Ho, W.Y. Transfection types, methods and strategies: A technical review. PeerJ 2021, 9, e11165. [Google Scholar] [CrossRef]
- Miles, L.B.; Calcinotto, V.; Oveissi, S.; Serrano, R.J.; Sonntag, C.; Mulia, O.; Lee, C.; Bryson-Richardson, R.J. CRIMP: A CRISPR/Cas9 insertional mutagenesis protocol and toolkit. Nat. Commun. 2024, 15, 5011. [Google Scholar] [CrossRef]
- Szarzanowicz, M.J.; Waldburger, L.M.; Busche, M.; Geiselman, G.M.; Kirkpatrick, L.D.; Kehl, A.J.; Tahmin, C.; Kuo, R.C.; McCauley, J.; Pannu, H.; et al. Binary vector copy number engineering improves Agrobacterium-mediated transformation. Nat. Biotechnol. 2024, 1–9. [Google Scholar] [CrossRef]
- Yu, L.; Cao, S.; Li, J.; Han, B.; Zhong, H.; Zhong, R. Prognostic value and immune infiltration of a novel stromal/immune score-related P2RY12 in lung adenocarcinoma microenvironment. Int. Immunopharmacol. 2021, 98, 107734. [Google Scholar] [CrossRef]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef] [PubMed]
- Giepmans, B.N.; Adams, S.R.; Ellisman, M.H.; Tsien, R.Y. The fluorescent toolbox for assessing protein location and function. Science 2006, 312, 217–224. [Google Scholar] [CrossRef]
- Verma, S.; Moreno, I.Y.; Gesteira, T.F.; Coulson-Thomas, V.J. Toxicity of nuclear-localized GFP in reporter mice. Sci. Rep. 2024, 14, 24642. [Google Scholar] [CrossRef] [PubMed]
- Harla, I.; Pawluś, W.; Zarębski, M.; Dobrucki, J.W. Induction of DNA single- and double-strand breaks by excited intra- or extracellular green fluorescent protein. J. Photochem. Photobiol. B 2024, 259, 113001. [Google Scholar] [CrossRef]
- Abdelaal, A.S.; Yazdani, S.S. A genetic toolkit for co-expression of multiple proteins of diverse physiological implication. Biotechnol. Rep. 2021, 32, e00692. [Google Scholar] [CrossRef]
- Yang, H.; Wang, Y.; Liu, W.; He, T.; Liao, J.; Qian, Z.; Zhao, J.; Cong, Z.; Sun, D.; Liu, Z.; et al. Genome-wide pan-GPCR cell libraries accelerate drug discovery. Acta Pharm. Sin. B 2024, 14, 4296–4311. [Google Scholar] [CrossRef]
- Bi, Z.; Li, H.; Liang, Y.; Sun, D.; Liu, S.; Chen, W.; Leng, L.; Song, C.; Zhang, S.; Cong, Z.; et al. Emerging paradigms for target discovery of traditional medicines: A genome-wide pan-GPCR perspective. Innovation 2025, 6, 100774. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.M.; Doyle, M.V.; Mark, D.F. Quantitation of mRNA by the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 1989, 86, 9717–9721. [Google Scholar] [CrossRef]
- Tsuyuzaki, K.; Sato, H.; Sato, K.; Nikaido, I. Benchmarking principal component analysis for large-scale single-cell RNA-sequencing. Genome Biol. 2020, 21, 9. [Google Scholar] [CrossRef]
- Yin, H.; Duo, H.; Li, S.; Qin, D.; Xie, L.; Xiao, Y.; Sun, J.; Tao, J.; Zhang, X.; Li, Y.; et al. Unlocking biological insights from differentially expressed genes: Concepts, methods, and future perspectives. J. Adv. Res. 2024. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Levine, A.J. p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer 2020, 20, 471–480. [Google Scholar] [CrossRef]
- Kudo, S.; Caaveiro, J.M.; Tsumoto, K. Adhesive Dimerization of Human P-Cadherin Catalyzed by a Chaperone-like Mechanism. Structure 2016, 24, 1523–1536. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dong, J.; Wang, W.; Yu, D.; Fan, X.; Hui, Y.C.; Lee, C.S.K.; Lam, W.H.; Alary, N.; Yang, Y.; et al. The human pre-replication complex is an open complex. Cell 2023, 186, 98–111.e21. [Google Scholar] [CrossRef] [PubMed]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Xu, X.; Yuan, Y.; Zhang, B.; Li, Z.; Xie, Y.; Yan, R.; Zheng, Z.; Ji, J.; et al. The intra-S phase checkpoint directly regulates replication elongation to preserve the integrity of stalled replisomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2019183118. [Google Scholar] [CrossRef]
- Cui, L.; Zheng, Y.; Xu, R.; Lin, Y.; Zheng, J.; Lin, P.; Guo, B.; Sun, S.; Zhao, X. Alternative pre-mRNA splicing in stem cell function and therapeutic potential: A critical review of current evidence. Int. J. Biol. Macromol. 2024, 268, 131781. [Google Scholar] [CrossRef]
- Kjer-Hansen, P.; Weatheritt, R.J. The function of alternative splicing in the proteome: Rewiring protein interactomes to put old functions into new contexts. Nat. Struct. Mol. Biol. 2023, 30, 1844–1856. [Google Scholar] [CrossRef]
- Trincado, J.L.; Entizne, J.C.; Hysenaj, G.; Singh, B.; Skalic, M.; Elliott, D.J.; Eyras, E. SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018, 19, 40. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef] [PubMed]
- Martinez, N.M.; Pan, Q.; Cole, B.S.; Yarosh, C.A.; Babcock, G.A.; Heyd, F.; Zhu, W.; Ajith, S.; Blencowe, B.J.; Lynch, K.W. Alternative splicing networks regulated by signaling in human T cells. Rna 2012, 18, 1029–1040. [Google Scholar] [CrossRef]
- Funk, J.S.; Klimovich, M.; Drangenstein, D.; Pielhoop, O.; Hunold, P.; Borowek, A.; Noeparast, M.; Pavlakis, E.; Neumann, M.; Balourdas, D.I.; et al. Deep CRISPR mutagenesis characterizes the functional diversity of TP53 mutations. Nat. Genet. 2025, 57, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Cockcroft, S. Expanding functions of the phosphatidylinositol/phosphatidate lipid transporter, PITPNC1 in physiology and in pathology. Adv. Biol. Regul. 2024, 95, 101056. [Google Scholar] [CrossRef]
- Sun, X.; Gao, C.; Xu, X.; Li, M.; Zhao, X.; Wang, Y.; Wang, Y.; Zhang, S.; Yan, Z.; Liu, X.; et al. FBL promotes cancer cell resistance to DNA damage and BRCA1 transcription via YBX1. EMBO Rep. 2023, 24, e56230. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Matsuyama, M.; Iwamiya, T. Novel and effective plasmid transfection protocols for functional analysis of genetic elements in human cardiac fibroblasts. PLoS ONE 2024, 19, e0309566. [Google Scholar] [CrossRef]
- Maurer, A.C.; Benyamini, B.; Fan, V.B.; Whitney, O.N.; Dailey, G.M.; Darzacq, X.; Weitzman, M.D.; Tjian, R. Double-Strand Break Repair Pathways Differentially Affect Processing and Transduction by Dual AAV Vectors. bioRxiv 2023. [Google Scholar] [CrossRef]
- Son, H.I.; Hamrick, G.S.; Shende, A.R.; Kim, K.; Yang, K.; Huang, T.J.; You, L. Population-level amplification of gene regulation by programmable gene transfer. Nat. Chem. Biol. 2025, 1–10. [Google Scholar] [CrossRef]
- Yuan, S.; Zhou, G.; Xu, G. Translation machinery: The basis of translational control. J. Genet. Genom. 2024, 51, 367–378. [Google Scholar] [CrossRef]
- Bhatter, N.; Dmitriev, S.E.; Ivanov, P. Cell death or survival: Insights into the role of mRNA translational control. Semin. Cell Dev. Biol. 2024, 154, 138–154. [Google Scholar] [CrossRef] [PubMed]
- Wachter, A. Gene regulation by structured mRNA elements. Trends Genet. 2014, 30, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Gilbert, W. Why genes in pieces? Nature 1978, 271, 501. [Google Scholar] [CrossRef]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef]
- Blencowe, B.J. Alternative splicing: New insights from global analyses. Cell 2006, 126, 37–47. [Google Scholar] [CrossRef]
- Kim, E.; Magen, A.; Ast, G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007, 35, 125–131. [Google Scholar] [CrossRef]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef]
- Bradley, R.K.; Anczuków, O. RNA splicing dysregulation and the hallmarks of cancer. Nat. Rev. Cancer 2023, 23, 135–155. [Google Scholar] [CrossRef]
- Blázquez-Encinas, R.; García-Vioque, V.; Caro-Cuenca, T.; Moreno-Montilla, M.T.; Mangili, F.; Alors-Pérez, E.; Ventura, S.; Herrera-Martínez, A.D.; Moreno-Casado, P.; Calzado, M.A.; et al. Altered splicing machinery in lung carcinoids unveils NOVA1, PRPF8 and SRSF10 as novel candidates to understand tumor biology and expand biomarker discovery. J. Transl. Med. 2023, 21, 879. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, T.; Koujin, T.; Shindo, T.; Bilir, Ş.; Osakada, H.; Nishimura, K.; Hirano, Y.; Asakawa, H.; Mori, C.; Kobayashi, S.; et al. Transfected plasmid DNA is incorporated into the nucleus via nuclear envelope reformation at telophase. Commun. Biol. 2022, 5, 78. [Google Scholar] [CrossRef]
- Delenda, C. Lentiviral vectors: Optimization of packaging, transduction and gene expression. J. Gene Med. 2004, 6 (Suppl. S1), S125–S138. [Google Scholar] [CrossRef] [PubMed]
- Chey, Y.C.J.; Gierus, L.; Lushington, C.; Arudkumar, J.C.; Geiger, A.B.; Staker, L.G.; Robertson, L.J.; Pfitzner, C.; Kennedy, J.G.; Lee, R.H.B.; et al. Optimal SpCas9- and SaCas9-mediated gene editing by enhancing gRNA transcript levels through scaffold poly-T tract reduction. BMC Genom. 2025, 26, 138. [Google Scholar] [CrossRef]
- Huang, W.Y.; Aramburu, J.; Douglas, P.S.; Izumo, S. Transgenic expression of green fluorescence protein can cause dilated cardiomyopathy. Nat. Med. 2000, 6, 482–483. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.K.; Joyce, P.B.; Molinete, M.; Halban, P.A.; Gorr, S.U. Oligomerization of green fluorescent protein in the secretory pathway of endocrine cells. Biochem. J. 2001, 360, 645–649. [Google Scholar] [CrossRef]
- Ganini, D.; Leinisch, F.; Kumar, A.; Jiang, J.; Tokar, E.J.; Malone, C.C.; Petrovich, R.M.; Mason, R.P. Fluorescent proteins such as eGFP lead to catalytic oxidative stress in cells. Redox Biol. 2017, 12, 462–468. [Google Scholar] [CrossRef]
- Mahnert, C.; Oyarzún, D.A.; Berrios, J. Multiscale modelling of bioprocess dynamics and cellular growth. Microb. Cell Fact. 2024, 23, 315. [Google Scholar] [CrossRef]
- Figueroa-Bossi, N.; Fernández-Fernández, R.; Kerboriou, P.; Bouloc, P.; Casadesús, J.; Sánchez-Romero, M.A.; Bossi, L. Transcription-driven DNA supercoiling counteracts H-NS-mediated gene silencing in bacterial chromatin. Nat. Commun. 2024, 15, 2787. [Google Scholar] [CrossRef]
- Gharaati-Far, N.; Tohidkia, M.R.; Dehnad, A.; Omidi, Y. Efficiency and cytotoxicity analysis of cationic lipids-mediated gene transfection into AGS gastric cancer cells. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1001–1008. [Google Scholar] [CrossRef]
- Zhu, C.; Kros, J.M.; van der Weiden, M.; Zheng, P.; Cheng, C.; Mustafa, D.A. Expression site of P2RY12 in residential microglial cells in astrocytomas correlates with M1 and M2 marker expression and tumor grade. Acta Neuropathol. Commun. 2017, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Kidmose, R.T.; Laursen, N.S.; Dobó, J.; Kjaer, T.R.; Sirotkina, S.; Yatime, L.; Sottrup-Jensen, L.; Thiel, S.; Gál, P.; Andersen, G.R. Structural basis for activation of the complement system by component C4 cleavage. Proc. Natl. Acad. Sci. USA 2012, 109, 15425–15430. [Google Scholar] [CrossRef] [PubMed]
- Cheung, D.; Hassan, M.A.; Huynh, T.; Feng, X.; Wang, H. Shedding light on the role of complement C4 activation in cancer. Hum. Immunol. 2025, 86, 111226. [Google Scholar] [CrossRef]
- Zhou, G.Z.; Sun, Y.H.; Shi, Y.Y.; Zhang, Q.; Zhang, L.; Cui, L.Q.; Sun, G.C. ANXA8 Regulates Proliferation of Human Non-Small Lung Cancer Cells A549 via EGFR-AKT-mTOR Signaling Pathway. Mol. Biol. 2021, 55, 870–880. [Google Scholar] [CrossRef]
- Teicher, B.A.; Fricker, S.P. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin. Cancer Res. 2010, 16, 2927–2931. [Google Scholar] [CrossRef]
- Wagner, P.L.; Hyjek, E.; Vazquez, M.F.; Meherally, D.; Liu, Y.F.; Chadwick, P.A.; Rengifo, T.; Sica, G.L.; Port, J.L.; Lee, P.C.; et al. CXCL12 and CXCR4 in adenocarcinoma of the lung: Association with metastasis and survival. J. Thorac. Cardiovasc. Surg. 2009, 137, 615–621. [Google Scholar] [CrossRef]
- Liu, Z.; Cui, L.; Wang, J.; Zhao, W.; Teng, Y. Aspirin boosts the synergistic effect of EGFR/p53 inhibitors on lung cancer cells by regulating AKT/mTOR and p53 pathways. Cell Biochem. Funct. 2024, 42, e3902. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, J.; Wang, C.; Wang, T.; Zeng, Y.; Li, X.; Zuo, Y.; Chen, H.; Zhang, C.; Cao, Y.; et al. Aptamer-functionalized triptolide with release controllability as a promising targeted therapy against triple-negative breast cancer. J. Exp. Clin. Cancer Res. 2024, 43, 207. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, L.; Fang, Z.; Liu, D.; Shao, M.; Liu, Y.; Liao, B.; Jin, Y. PRPF8 controls alternative splicing of PIRH2 to modulate the p53 pathway and survival of human ESCs. J. Cell Physiol. 2023, 238, 1909–1920. [Google Scholar] [CrossRef]
- Jung, J.H.; Lee, H.; Cao, B.; Liao, P.; Zeng, S.X.; Lu, H. RNA-binding motif protein 10 induces apoptosis and suppresses proliferation by activating p53. Oncogene 2020, 39, 1031–1040. [Google Scholar] [CrossRef]
- Kim, T.K.; Eberwine, J.H. Mammalian cell transfection: The present and the future. Anal. Bioanal. Chem. 2010, 397, 3173–3178. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G. Thirteen years of clusterProfiler. Innovation 2024, 5, 100722. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Liu, Z.; Qian, Y.; Wu, M.; Mo, J.; Wang, C.; Xu, G.; Leng, L.; Zhang, S. Alterations in Gene Expression and Alternative Splicing Induced by Plasmid-Mediated Overexpression of GFP and P2RY12 Within the A549 Cell Line. Int. J. Mol. Sci. 2025, 26, 2973. https://doi.org/10.3390/ijms26072973
Liu Q, Liu Z, Qian Y, Wu M, Mo J, Wang C, Xu G, Leng L, Zhang S. Alterations in Gene Expression and Alternative Splicing Induced by Plasmid-Mediated Overexpression of GFP and P2RY12 Within the A549 Cell Line. International Journal of Molecular Sciences. 2025; 26(7):2973. https://doi.org/10.3390/ijms26072973
Chicago/Turabian StyleLiu, Qingqing, Zhaoyu Liu, Yongqi Qian, Mingxu Wu, Jing Mo, Can Wang, Guoqing Xu, Liang Leng, and Sanyin Zhang. 2025. "Alterations in Gene Expression and Alternative Splicing Induced by Plasmid-Mediated Overexpression of GFP and P2RY12 Within the A549 Cell Line" International Journal of Molecular Sciences 26, no. 7: 2973. https://doi.org/10.3390/ijms26072973
APA StyleLiu, Q., Liu, Z., Qian, Y., Wu, M., Mo, J., Wang, C., Xu, G., Leng, L., & Zhang, S. (2025). Alterations in Gene Expression and Alternative Splicing Induced by Plasmid-Mediated Overexpression of GFP and P2RY12 Within the A549 Cell Line. International Journal of Molecular Sciences, 26(7), 2973. https://doi.org/10.3390/ijms26072973





