Potential Role of Malassezia restricta in Pterygium Development
Abstract
:1. Introduction
2. Results
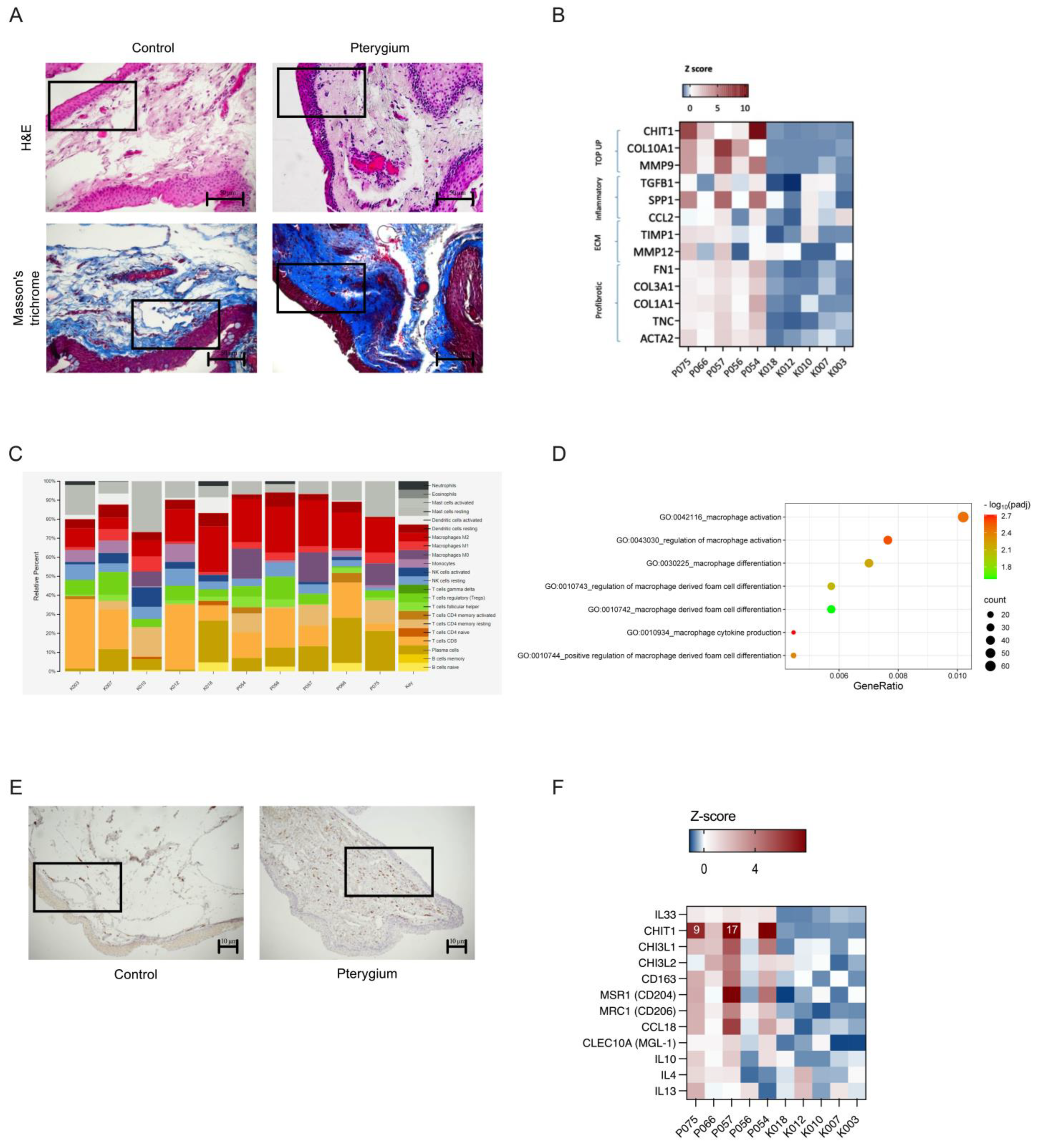
2.1. Experimental Design and Pterygium Characteristics
2.2. Metagenomic Analysis Revealed Higher Occurrence of Malassezia restricta in Pterygium Tissue, a Finding Corroborated by Fluorescent In Situ Hybridization (FISH)
2.3. Chitotriosidase (CHIT1), an Enzyme Critical for Breaking Down Chitin-Containing Pathogens like Malassezia, Showed the Highest Expression Levels in Pterygium Samples
2.4. Fibrosis and an Immunosuppressive State Enriched with M2 Macrophages—A Hallmark of Malassezia-Associated Infections—Were Present in the Pterygium Microenvironment
3. Discussion
4. Materials and Methods
4.1. Informed Consent
4.2. Patients
4.3. Tissue Samples
4.4. Histological and Staining Analysis
4.5. Immunohistochemistry
4.6. FISH
4.7. DNA and RNA Extraction
4.8. RNA Sequencing
4.9. Whole-Genome Microbiome Sequencing
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rezvan, F.; Khabazkhoob, M.; Hooshmand, E.; Yekta, A.; Saatchi, M.; Hashemi, H. Prevalence and Risk Factors of Pterygium: A Systematic Review and Meta-Analysis. Surv. Ophthalmol. 2018, 63, 719–735. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wu, J.; Geng, J.; Yuan, Z.; Huang, D. Geographical Prevalence and Risk Factors for Pterygium: A Systematic Review and Meta-Analysis. BMJ Open 2013, 3, e003787. [Google Scholar] [CrossRef] [PubMed]
- Chui, J.; Coroneo, M.T.; Tat, L.T.; Crouch, R.; Wakefield, D.; Di Girolamo, N. Ophthalmic Pterygium: A Stem Cell Disorder with Premalignant Features. Am. J. Pathol. 2011, 178, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Suarez, M.F.; Echenique, J.; López, J.M.; Medina, E.; Irós, M.; Serra, H.M.; Fini, M.E. Transcriptome Analysis of Pterygium and Pinguecula Reveals Evidence of Genomic Instability Associated with Chronic Inflammation. Int. J. Mol. Sci. 2021, 22, 12090. [Google Scholar] [CrossRef]
- Wanzeler, A.C.V.; Barbosa, I.A.F.; Duarte, B.; Borges, D.; Barbosa, E.B.; Kamiji, D.; Huarachi, D.R.G.; de Melo, M.B.; Alves, M. Mechanisms and Biomarker Candidates in Pterygium Development. Arq. Bras. Oftalmol. 2019, 82, 528–536. [Google Scholar] [CrossRef]
- Thompson, J.P.; Harbin, Z.; Das, H.; Deschner, L.A.; Seale, S.A.; Kheirkhah, A. Comparison of Pterygium Recurrence Rates Between Attending Physicians and Supervised Trainee Residents. Cornea 2022, 41, 12–15. [Google Scholar] [CrossRef]
- Aragona, P.; Baudouin, C.; Benitez Del Castillo, J.M.; Messmer, E.; Barabino, S.; Merayo-Lloves, J.; Brignole-Baudouin, F.; Inferrera, L.; Rolando, M.; Mencucci, R.; et al. The Ocular Microbiome and Microbiota and Their Effects on Ocular Surface Pathophysiology and Disorders. Surv. Ophthalmol. 2021, 66, 907–925. [Google Scholar] [CrossRef]
- Napolitano, P.; Filippelli, M.; Davinelli, S.; Bartollino, S.; dell’Omo, R.; Costagliola, C. Influence of Gut Microbiota on Eye Diseases: An Overview. Ann. Med. 2021, 53, 750–761. [Google Scholar] [CrossRef]
- Campagnoli, L.I.M.; Varesi, A.; Barbieri, A.; Marchesi, N.; Pascale, A. Targeting the Gut–Eye Axis: An Emerging Strategy to Face Ocular Diseases. Int. J. Mol. Sci. 2023, 24, 13338. [Google Scholar] [CrossRef]
- Leong, Y.; Tong, L. Barrier Function in the Ocular Surface: From Conventional Paradigms to New Opportunities. Ocul. Surf. 2015, 13, 103–109. [Google Scholar] [CrossRef]
- Nolan, J. Evaluation of Conjunctival and Nasal Bacterial Cultures before Intra-Ocular Operations. Br. J. Ophthalmol. 1967, 51, 483–485. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Brulc, J.M.; Iovieno, A.; Bates, B.; Garoutte, A.; Miller, D.; Revanna, K.V.; Gao, X.; Antonopoulos, D.A.; Slepak, V.Z.; et al. Diversity of Bacteria at Healthy Human Conjunctiva. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5408–5413. [Google Scholar] [CrossRef]
- Ozkan, J.; Coroneo, M.; Willcox, M.; Wemheuer, B.; Thomas, T. Identification and Visualization of a Distinct Microbiome in Ocular Surface Conjunctival Tissue. Investig. Ophthalmol. Vis. Sci. 2018, 59, 4268–4276. [Google Scholar] [CrossRef]
- Chalkia, A.K.; Spandidos, D.A.; Detorakis, E.T. Viral Involvement in the Pathogenesis and Clinical Features of Ophthalmic Pterygium (Review). Int. J. Mol. Med. 2013, 32, 539–543. [Google Scholar]
- Di Girolamo, N. Association of Human Papilloma Virus with Pterygia and Ocular-Surface Squamous Neoplasia. Eye 2012, 26, 202–211. [Google Scholar]
- Zhu, Y.; Yu, X.; Cheng, G. Human Skin Bacterial Microbiota Homeostasis: A Delicate Balance between Health and Disease. mLife 2023, 2, 107–120. [Google Scholar]
- De Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut Microbiome and Health: Mechanistic Insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef]
- Gomes, J.Á.P.; Frizon, L.; Demeda, V.F. Ocular Surface Microbiome in Health and Disease. Asia Pac. J. Ophthalmol. 2020, 9, 505–511. [Google Scholar] [CrossRef]
- Park, M.; Park, S.; Jung, W.H. Skin Commensal Fungus Malassezia and Its Lipases. J. Microbiol. Biotechnol. 2021, 31, 637–644. [Google Scholar] [CrossRef]
- Aykut, B.; Pushalkar, S.; Chen, R.; Li, Q.; Abengozar, R.; Kim, J.I.; Shadaloey, S.A.; Wu, D.; Preiss, P.; Verma, N.; et al. The Fungal Mycobiome Promotes Pancreatic Oncogenesis via Activation of MBL. Nature 2019, 574, 264–267. [Google Scholar] [CrossRef]
- Abdillah, A.; Ranque, S. Chronic Diseases Associated with Malassezia Yeast. J. Fungi 2021, 7, 855. [Google Scholar] [CrossRef] [PubMed]
- Soret, P.; Vandenborght, L.E.; Francis, F.; Coron, N.; Enaud, R.; Avalos, M.; Schaeverbeke, T.; Berger, P.; Fayon, M.; Thiebaut, R.; et al. Respiratory Mycobiome and Suggestion of Inter-Kingdom Network during Acute Pulmonary Exacerbation in Cystic Fibrosis. Sci. Rep. 2020, 10, 3589. [Google Scholar] [CrossRef]
- Limon, J.J.; Tang, J.; Li, D.; Wolf, A.J.; Michelsen, K.S.; Funari, V.; Gargus, M.; Nguyen, C.; Sharma, P.; Maymi, V.I.; et al. Malassezia Is Associated with Crohn’s Disease and Exacerbates Colitis in Mouse Models. Cell Host Microbe 2019, 25, 377–388.e6. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Levanduski, E.; Denz, P.; Villavicencio, H.S.; Bhatta, M.; Alhorebi, L.; Zhang, Y.; Gomez, E.C.; Morreale, B.; Senchanthisai, S.; et al. Fungal Mycobiome Drives IL-33 Secretion and Type 2 Immunity in Pancreatic Cancer. Cancer Cell 2022, 40, 153–167.e11. [Google Scholar] [CrossRef]
- Zhu, Y.; Herndon, J.M.; Sojka, D.K.; Kim, K.W.; Knolhoff, B.L.; Zuo, C.; Cullinan, D.R.; Luo, J.; Bearden, A.R.; Lavine, K.J.; et al. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity 2017, 47, 323–338.e6. [Google Scholar] [CrossRef]
- Faas, M.; Ipseiz, N.; Ackermann, J.; Culemann, S.; Grüneboom, A.; Schröder, F.; Rothe, T.; Scholtysek, C.; Eberhardt, M.; Böttcher, M.; et al. IL-33-Induced Metabolic Reprogramming Controls the Differentiation of Alternatively Activated Macrophages and the Resolution of Inflammation. Immunity 2021, 54, 2531–2546.e5. [Google Scholar] [CrossRef]
- Hassanshahi, A.; Moradzad, M.; Ghalamkari, S.; Fadaei, M.; Cowin, A.J.; Hassanshahi, M. Macrophage-Mediated Inflammation in Skin Wound Healing. Cells 2022, 11, 2953. [Google Scholar] [CrossRef]
- Jiang, Y.; Cai, R.; Huang, Y.; Zhu, L.; Xiao, L.; Wang, C.; Wang, L. Macrophages in Organ Fibrosis: From Pathogenesis to Therapeutic Targets. Cell Death Discov. 2024, 10, 487. [Google Scholar]
- Vannella, K.M.; Ramalingam, T.R.; Hart, K.M.; De Queiroz Prado, R.; Sciurba, J.; Barron, L.; Borthwick, L.A.; Smith, A.D.; Mentink-Kane, M.; White, S.; et al. Acidic Chitinase Primes the Protective Immune Response to Gastrointestinal Nematodes. Nat. Immunol. 2016, 17, 538–544. [Google Scholar] [CrossRef]
- Anwar, W.; Amin, H.; Khan, H.A.A.; Akhter, A.; Bashir, U.; Anjum, T.; Kalsoom, R.; Javed, M.A.; Zohaib, K.A. Chitinase of Trichoderma Longibrachiatum for Control of Aphis Gossypii in Cotton Plants. Sci. Rep. 2023, 13, 13181. [Google Scholar] [CrossRef]
- Choudhary, V.; Choudhary, M.; Bollag, W.B. Exploring Skin Wound Healing Models and the Impact of Natural Lipids on the Healing Process. Int. J. Mol. Sci. 2024, 25, 3790. [Google Scholar] [CrossRef] [PubMed]
- Summer, R.; Mora, A.L. Lipid Metabolism: A New Player in the Conundrum of Lung Fibrosis. Am. J. Respir. Cell Mol. Biol. 2019, 61, 669–670. [Google Scholar] [PubMed]
- Zhang, X.; Han, P.; Qiu, J.; Huang, F.; Luo, Q.; Cheng, J.; Shan, K.; Yang, Y.; Zhang, C. Single-Cell RNA Sequencing Reveals the Complex Cellular Niche of Pterygium. Ocul. Surf. 2024, 32, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Mayslich, C.; Grange, P.A.; Dupin, N.; Brüggemann, H. Cutibacterium acnes as an Opportunistic Pathogen: An Update of Its Virulence-Associated Factors. Microorganisms 2021, 9, 303. [Google Scholar] [CrossRef]
- Lalitha, P.; Prajna, N.V.; Sikha, M.; Gunasekaran, R.; Hinterwirth, A.; Worden, L.; Chen, C.; Zhong, L.; Liu, Z.; Lietman, T.M.; et al. Evaluation of Metagenomic Deep Sequencing as a Diagnostic Test for Infectious Keratitis. Ophthalmology 2021, 128, 473–475. [Google Scholar] [CrossRef]
- Ramírez-Soto, M.C.; Bonifaz, A. Ocular Fungal Infections. J. Fungi 2022, 8, 1078. [Google Scholar] [CrossRef]
- Lešin, M.; Paradžik, M.; Marin Lovria, J.; Olujia, I.; Ljubia, A.; Vuainovia, A.; Buaan, K.; Puljak, L. Cauterisation versus Fibrin Glue for Conjunctival Autografting in Primary Pterygium Surgery (CAGE CUP): Study Protocol of a Randomised Controlled Trial. BMJ Open 2018, 8, e020714. [Google Scholar] [CrossRef]
- Kurt, A.; Kılıç, R.; Tad, M.; Polat, O.A. YKL-40 Expression in Pterygium: A Potential Role in the Pathogenesis. Int. Ophthalmol. 2019, 39, 1445–1450. [Google Scholar] [CrossRef]
- Lee, K.; Zhang, I.; Kyman, S.; Kask, O.; Kathryn, E. Co-Infection of Malassezia sympodialis with Bacterial Pathobionts Pseudomonas aeruginosa or Staphylococcus aureus Leads to Distinct Sinonasal Inflammatory Responses in a Murine Acute Sinusitis Model. Front. Cell Infect. Microbiol. 2020, 10, 472. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining Cell Type Abundance and Expression from Bulk Tissues with Digital Cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Chen, S. Ultrafast One-Pass FASTQ Data Preprocessing, Quality Control, and Deduplication Using Fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and Sensitive Classification of Metagenomic Sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [PubMed]
- Martí, J.M. Recentrifuge: Robust Comparative Analysis and Contamination Removal for Metagenomics. PLoS Comput. Biol. 2019, 15, e1006967. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a Package of R Functions for Community Ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar]




| Parameter | Pterygium Group (N = 46) | Control Group (N = 22) | p |
|---|---|---|---|
| Sex—N (%) | |||
| Male | 23 (50%) | 9 (41%) | 0.482 |
| Female | 23 (50%) | 13 (59%) | |
| Age (yr) | 64.3 (36.48–87.69) | 71.64 (56.05–83.31) | 0.015 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paradzik Simunovic, M.; Degoricija, M.; Korac-Prlic, J.; Lesin, M.; Stanic, R.; Puljak, L.; Olujic, I.; Marin Lovric, J.; Vucinovic, A.; Ljubic, Z.; et al. Potential Role of Malassezia restricta in Pterygium Development. Int. J. Mol. Sci. 2025, 26, 2976. https://doi.org/10.3390/ijms26072976
Paradzik Simunovic M, Degoricija M, Korac-Prlic J, Lesin M, Stanic R, Puljak L, Olujic I, Marin Lovric J, Vucinovic A, Ljubic Z, et al. Potential Role of Malassezia restricta in Pterygium Development. International Journal of Molecular Sciences. 2025; 26(7):2976. https://doi.org/10.3390/ijms26072976
Chicago/Turabian StyleParadzik Simunovic, Martina, Marina Degoricija, Jelena Korac-Prlic, Mladen Lesin, Robert Stanic, Livia Puljak, Ivana Olujic, Josipa Marin Lovric, Ana Vucinovic, Zana Ljubic, and et al. 2025. "Potential Role of Malassezia restricta in Pterygium Development" International Journal of Molecular Sciences 26, no. 7: 2976. https://doi.org/10.3390/ijms26072976
APA StyleParadzik Simunovic, M., Degoricija, M., Korac-Prlic, J., Lesin, M., Stanic, R., Puljak, L., Olujic, I., Marin Lovric, J., Vucinovic, A., Ljubic, Z., Thissen, J., Reen Kok, C., Jaing, C., Bucan, K., & Terzic, J. (2025). Potential Role of Malassezia restricta in Pterygium Development. International Journal of Molecular Sciences, 26(7), 2976. https://doi.org/10.3390/ijms26072976






