Comprehensive Transcriptomic Analysis of the Isolated Candida tropicalis with Enhanced Tolerance of Furfural Inhibitor
Abstract
:1. Introduction
2. Results and Discussion
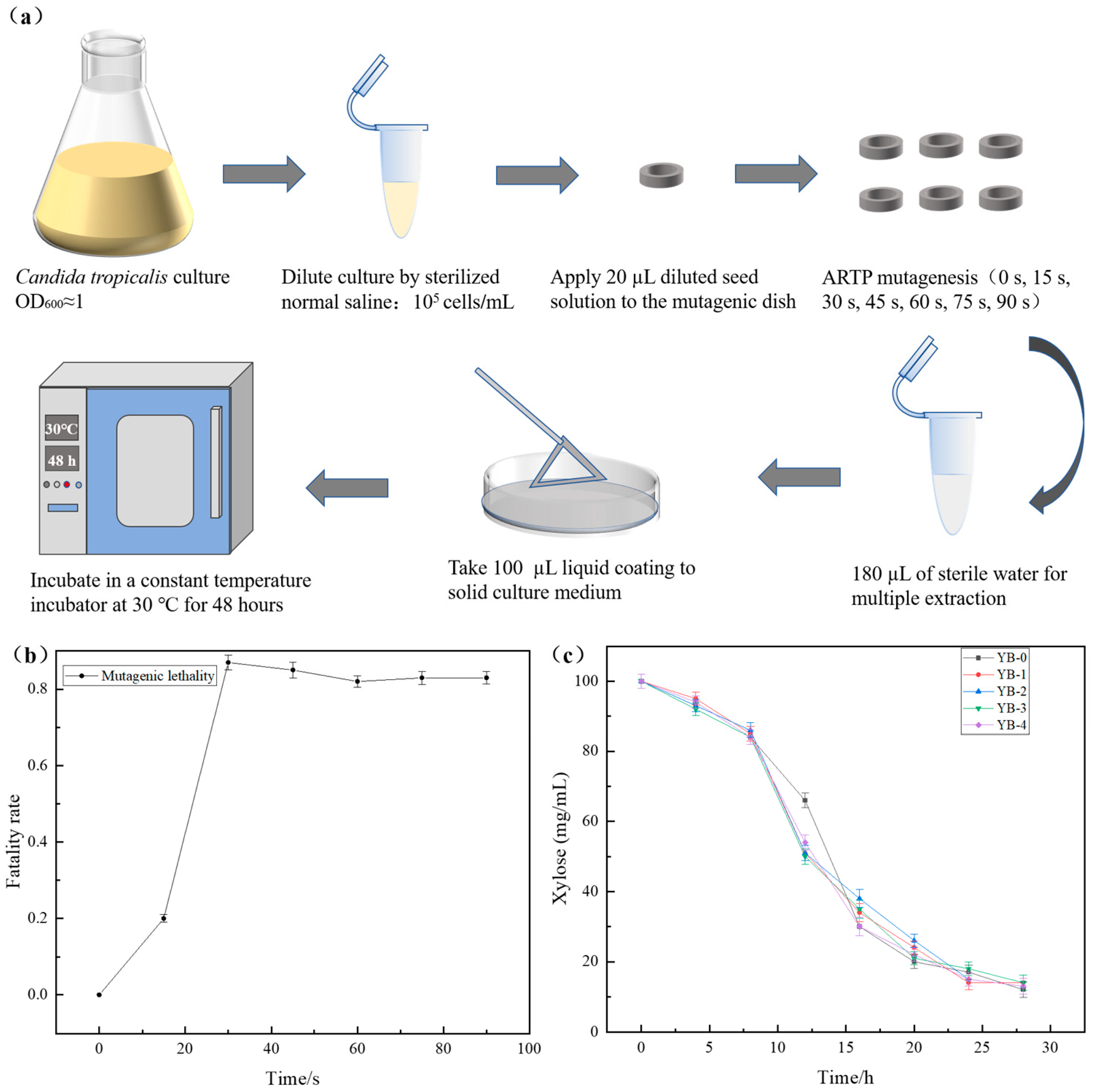
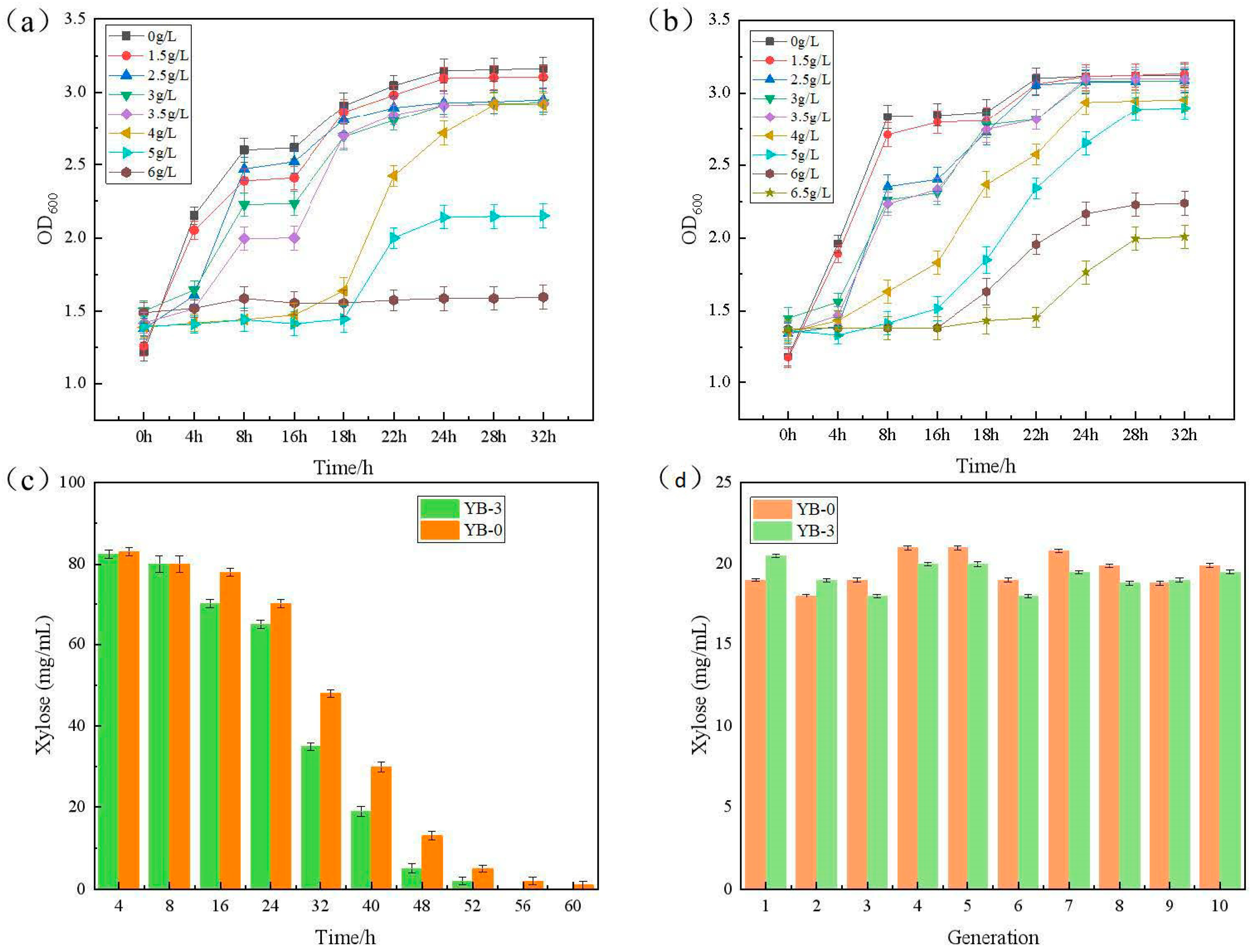
2.1. Enhanced Furfural Tolerance with Mutant Strains by the ARTP Mutagenesis
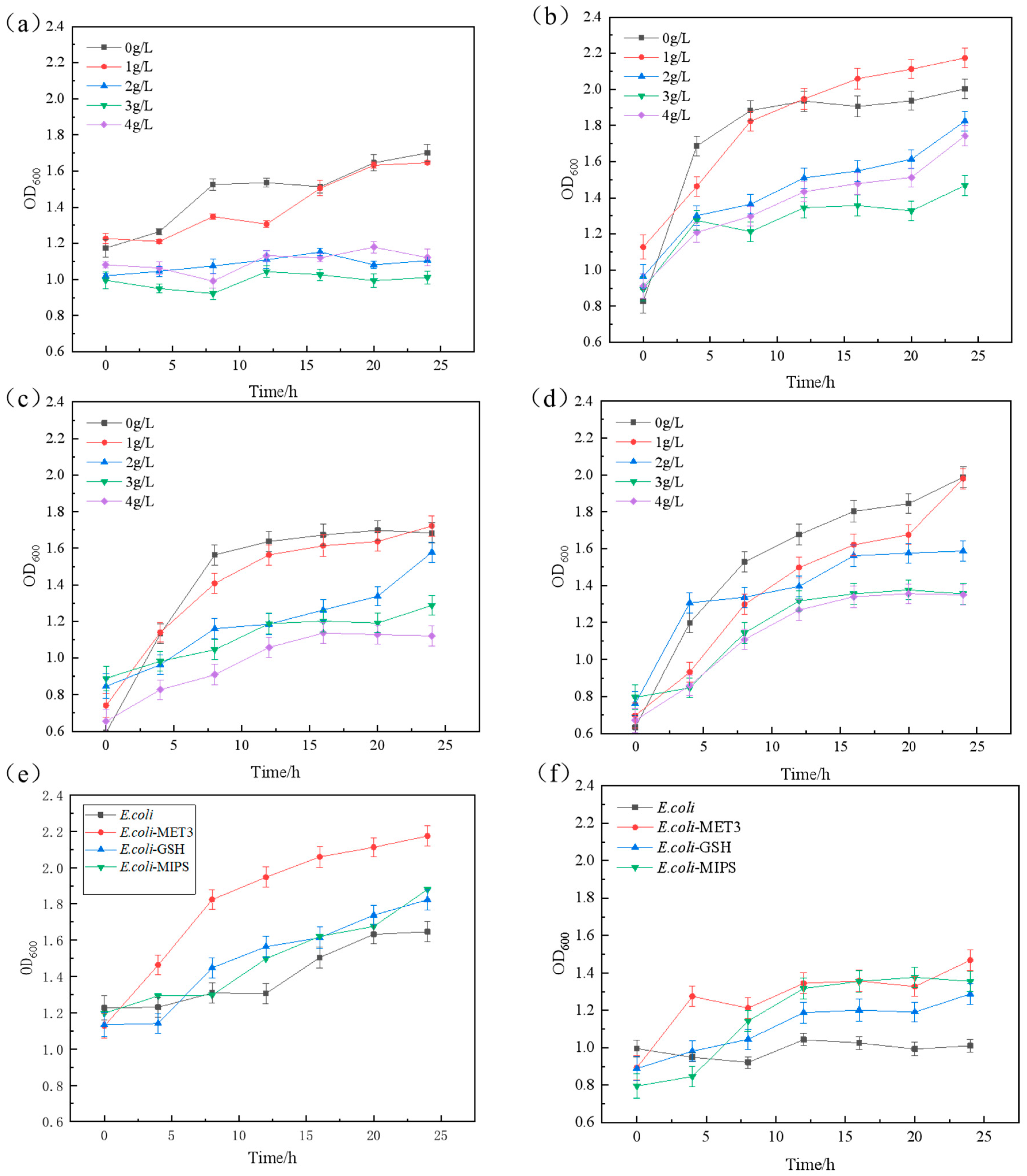
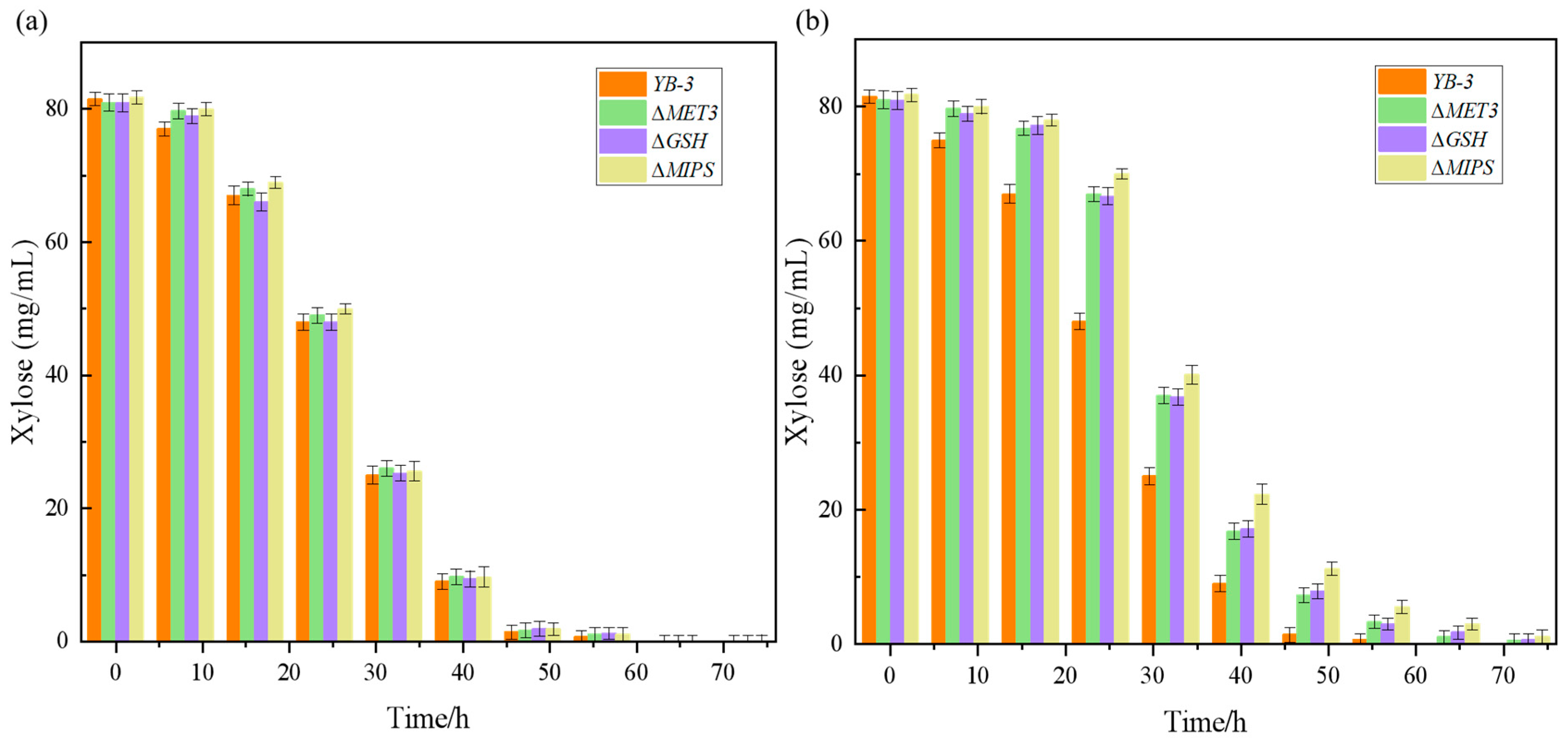
2.2. Mutagenic Growth Fermentation and Stability Under the Furfural Stress
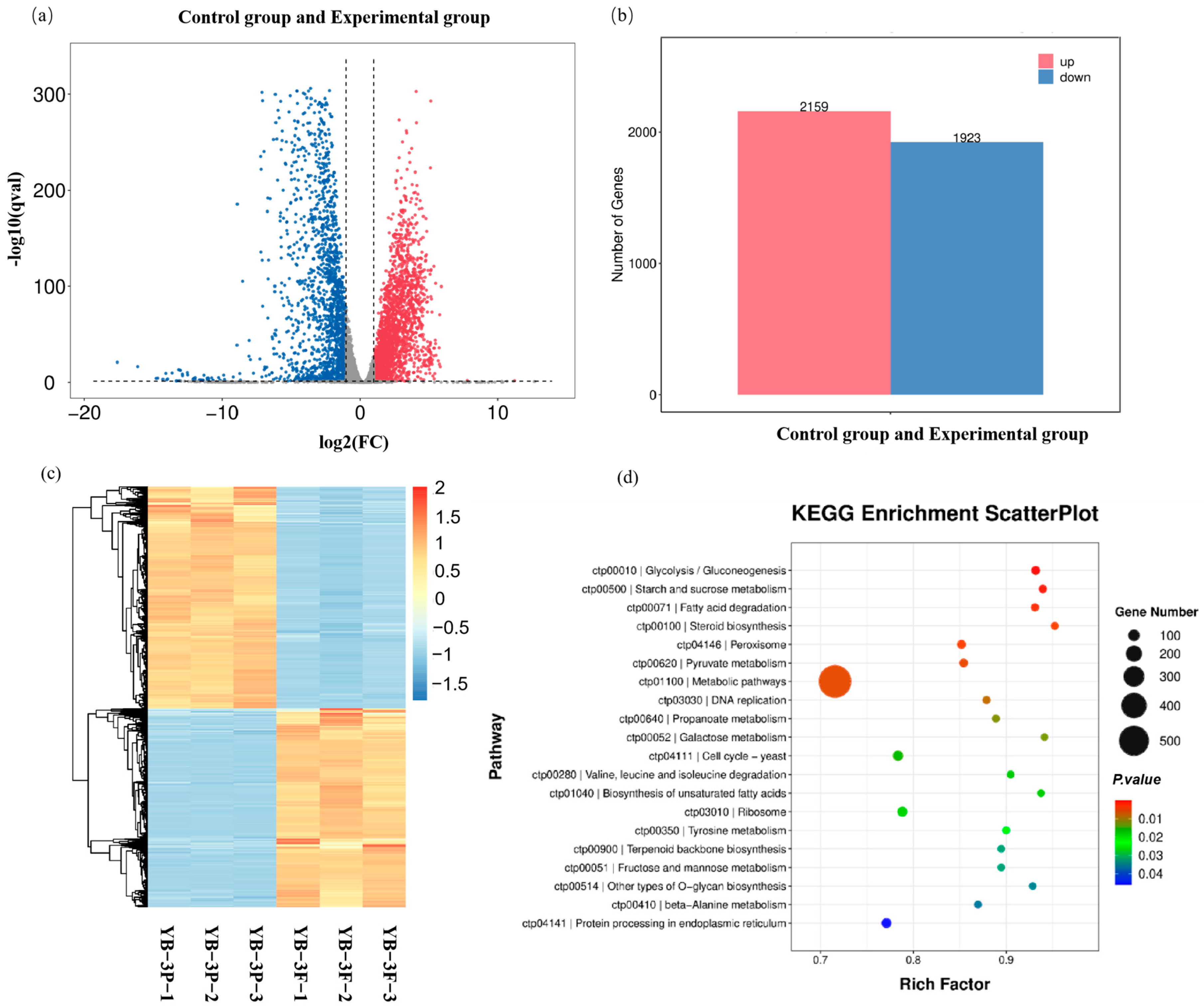
2.3. Transcriptome Analysis and Identification of Furfural Tolerance-Related Genes
2.4. Identification of Furfural Tolerance Gene Knockout Verification
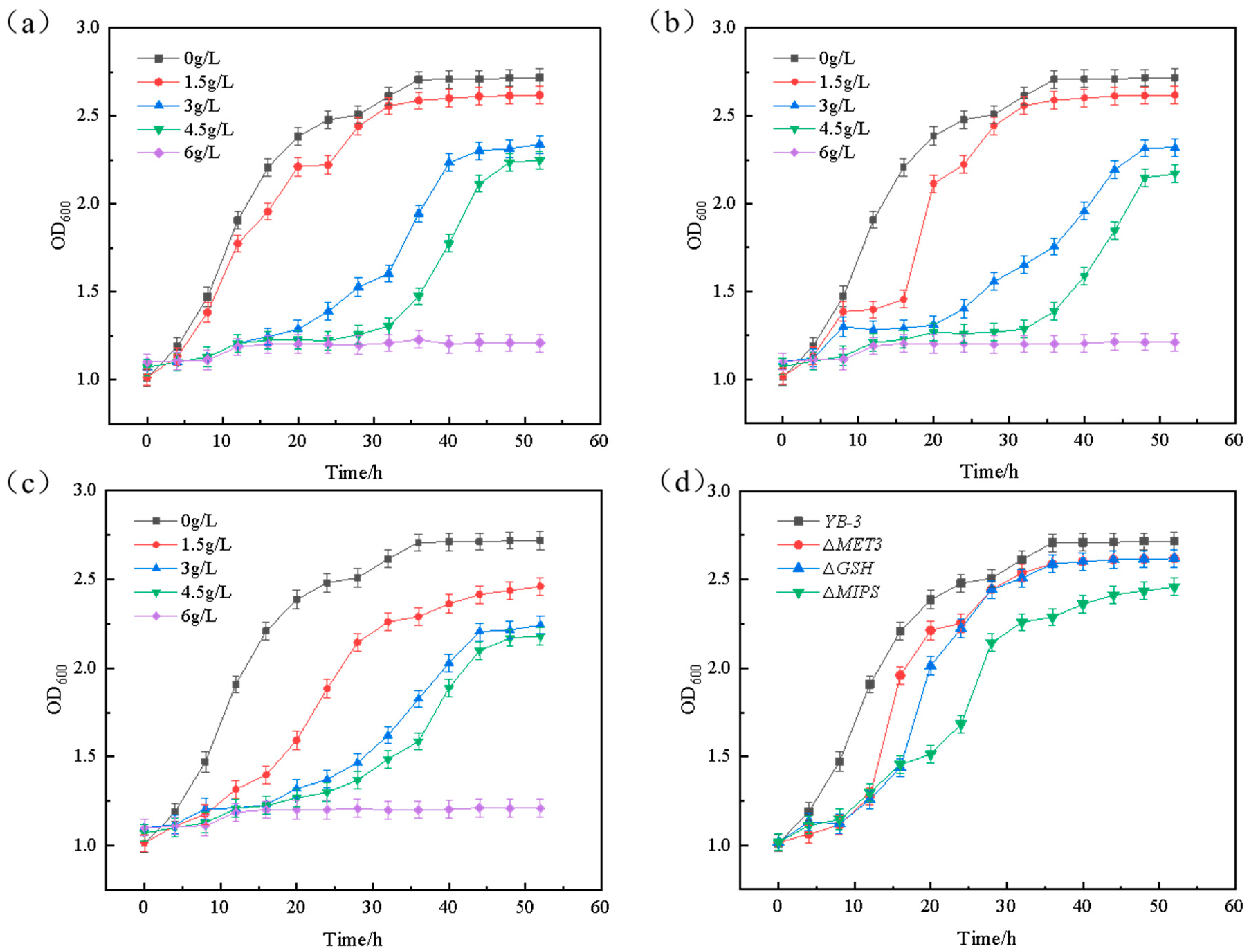
2.5. The Exogenous Verifications of Genes to the Furfural Tolerance
3. Materials and Methods
3.1. Strains and Plasmids
3.2. Mutagenesis Experiment by the ARTP
3.3. The Growth Curve, Fermentation Performance, and Stability Experiments of Mutant Strains Under Furfural Stress
3.4. Samples Preparation and Transcriptome Analysis
3.5. RT-qPCR Analysis
3.6. Knockout Verification of Furfural Tolerance-Related Genes
3.7. Exogenous Verification of Furfural Tolerance Genes MET3, MIPS, and GSH Genes
3.8. Analytical Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Robak, K.; Balcerek, M. Current state-of-the-art in ethanol production from lignocellulosic feedstocks. Microbiol. Res. 2020, 240, 126534. [Google Scholar] [CrossRef]
- Hachimi Alaoui, C.; Réthoré, G.; Weiss, P.; Fatimi, A. Sustainable Biomass Lignin-Based Hydrogels: A Review on Properties, Formulation, and Biomedical Applications. Int. J. Mol. Sci. 2023, 24, 13493. [Google Scholar] [CrossRef] [PubMed]
- Parsin, S.; Kaltschmitt, M. Processing of hemicellulose in wheat straw by steaming and ultrafiltration—A novel approach. Bioresour. Technol. 2024, 393, 130071. [Google Scholar] [CrossRef]
- Jin, T.; Xing, X.; Xie, Y.; Sun, Y.; Bian, S.; Liu, L.; Chen, G.; Wang, X.; Yu, X.; Su, Y. Evaluation of Preparation and Detoxification of Hemicellulose Hydrolysate for Improved Xylitol Production from Quinoa Straw. Int. J. Mol. Sci. 2022, 24, 516. [Google Scholar] [CrossRef] [PubMed]
- Hoang, A.T.; Nizetic, S.; Ong, H.C.; Chong, C.T.; Atabani, A.E.; Pham, V.V. Acid-based lignocellulosic biomass biorefinery for bioenergy production: Advantages, application constraints, and perspectives. J. Environ. Manag. 2021, 296, 113194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, X.; Yong, Q.; Yang, S.; Ouyang, J.; Yu, S. Impacts of lignocellulose-derived inhibitors on l-lactic acid fermentation by Rhizopus oryzae. Bioresour. Technol. 2016, 203, 173–180. [Google Scholar] [CrossRef]
- Contreras, F.; Pramanik, S.; Rozhkova, A.M.; Zorov, I.N.; Korotkova, O.; Sinitsyn, A.P.; Schwaneberg, U.; Davari, M.D. Engineering Robust Cellulases for Tailored Lignocellulosic Degradation Cocktails. Int. J. Mol. Sci. 2020, 21, 1589. [Google Scholar] [CrossRef]
- Fan, X.; Cheng, G.; Zhang, H.; Li, M.; Wang, S.; Yuan, Q. Effects of acid impregnated steam explosion process on xylose recovery and enzymatic conversion of cellulose in corncob. Carbohydr. Polym. 2014, 114, 21–26. [Google Scholar] [CrossRef]
- Zhang, H.; Fan, X.; Qiu, X.; Zhang, Q.; Wang, W.; Li, S.; Deng, L.; Koffas, M.A.G.; Wei, D.; Yuan, Q. A novel cleaning process for industrial production of xylose in pilot scale from corncob by using screw-steam-explosive extruder. Bioprocess Biosyst. Eng. 2014, 37, 2425–2436. [Google Scholar] [CrossRef]
- Fan, X.; Li, M.; Zhang, J.; Tang, P.; Yuan, Q. Optimization of SO2-catalyzed hydrolysis of corncob for xylose and xylitol production. J. Chem. Technol. Biot. 2014, 89, 1720–1726. [Google Scholar] [CrossRef]
- Wang, L.; Yang, M.; Fan, X.; Zhu, X.; Xu, T.; Yuan, Q. An environmentally friendly and efficient method for xylitol bioconversion with high-temperature-steaming corncob hydrolysate by adapted Candida tropicalis. Process Biochem. 2011, 46, 1619–1626. [Google Scholar]
- Rasmussen, H.; Sørensen, H.R.; Meyer, A.S. Formation of degradation compounds from lignocellulosic biomass in the biorefinery: Sugar reaction mechanisms. Carbohydr. Res. 2014, 385, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Ujor, V.C.; Okonkwo, C.C. Microbial detoxification of lignocellulosic biomass hydrolysates: Biochemical and molecular aspects, challenges, exploits and future perspectives. Front. Bioeng. Biotechnol. 2022, 10, 1061667. [Google Scholar] [CrossRef]
- Jönsson, L.J.; Alriksson, B.; Nilvebrant, N. Bioconversion of lignocellulose: Inhibitors and detoxification. Biotechnol. Biofuels 2013, 6, 16. [Google Scholar] [CrossRef]
- Li, Q.; Feng, P.; Tang, H.; Lu, F.; Mou, B.; Zhao, L.; Li, N.; Yang, Y.; Fu, C.; Long, W.; et al. Genome-wide identification of resistance genes and cellular analysis of key gene knockout strain under 5-hydroxymethylfurfural stress in Saccharomyces cerevisiae. BMC Microbiol. 2023, 23, 382. [Google Scholar] [CrossRef]
- Cassells, B.; Karhumaa, K.; Sànchez, I.; Nogué, V.; Lidén, G. Hybrid SSF/SHF Processing of SO2 Pretreated Wheat Straw-Tuning Co-fermentation by Yeast Inoculum Size and Hydrolysis Time. Appl. Biochem. Biotechnol. 2017, 181, 536–547. [Google Scholar] [CrossRef]
- Allen, S.A.; Clark, W.; McCaffery, J.M.; Cai, Z.; Lanctot, A.; Slininger, P.J.; Liu, Z.L.; Gorsich, S.W. Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol. Biofuels 2010, 3, 2. [Google Scholar] [CrossRef]
- Li, Y.; Sun, L.; Cao, D.; Cao, X.; Sun, S. One-step conversion of corn stalk to glucose and furfural in molten salt hydrate/organic solvent biphasic system. Bioresour. Technol. 2023, 386, 129520. [Google Scholar] [CrossRef]
- Wang, L.; Wu, D.; Tang, P.; Yuan, Q. Effect of organic acids found in cottonseed hull hydrolysate on the xylitol fermentation by Candida tropicalis. Bioprocess Biosyst. Eng. 2013, 36, 1053–1061. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, J.; Yuan, J.; Jiang, X.; Jiang, L.; Zhao, G.; Huang, D.; Liu, B. Omics-based analyses revealed metabolic responses of Clostridium acetobutylicum to lignocellulose-derived inhibitors furfural, formic acid and phenol stress for butanol fermentation. Biotechnol. Biofuels 2019, 12, 101. [Google Scholar] [CrossRef]
- Pan, X.; Liu, H.; Liu, J.; Wang, C.; Wen, J. Omics-based approaches reveal phospholipids remodeling of Rhizopus oryzae responding to furfural stress for fumaric acid-production from xylose. Bioresour. Technol. 2016, 222, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cheng, G.; Joshua, C.; He, Z.; Sun, X.; Li, R.; Liu, L.; Yuan, Q. Furfural tolerance and detoxification mechanism in Candida tropicalis. Biotechnol. Biofuels 2016, 9, 250. [Google Scholar] [CrossRef]
- Huang, S.; Xue, T.; Wang, Z.; Ma, Y.; He, X.; Hong, J.; Zou, S.; Song, H.; Zhang, M. Furfural-tolerant Zymomonas mobilis derived from error-prone PCR-based whole genome shuffling and their tolerant mechanism. Appl. Microbiol. Biotechnol. 2018, 102, 3337–3347. [Google Scholar] [CrossRef]
- Wang, L.; Qi, A.; Liu, J.; Shen, Y.; Wang, J. Comparative metabolic analysis of the adaptive Candida tropicalis to furfural stress response. Chem. Eng. Sci. 2023, 267, 118348. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, R.; Li, Z.; Dai, D.; Li, C.; Zhou, X. A novel pathway construction in Candida tropicalis for direct xylitol conversion from corncob xylan. Bioresour. Technol. 2013, 128, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Mateo, S.; Puentes, J.G.; Moya, A.J.; Sánchez, S. Ethanol and xylitol production by fermentation of acid hydrolysate from olive pruning with Candida tropicalis NBRC 0618. Bioresour. Technol. 2015, 190, 1–6. [Google Scholar] [CrossRef]
- Tizazu, B.Z.; Roy, K.; Moholkar, V.S. Ultrasonic enhancement of xylitol production from sugarcane bagasse using immobilized Candida tropicalis MTCC 184. Bioresour. Technol. 2018, 268, 247–258. [Google Scholar] [CrossRef]
- Wang, S.; He, Z.; Yuan, Q. Xylose enhances furfural tolerance in Candida tropicalis by improving NADH recycle. Chem. Eng. Sci. 2017, 158, 37–40. [Google Scholar] [CrossRef]
- He, Y.; Song, Z.; Dong, X.; Zheng, Q.; Peng, X.; Jia, X. Candida tropicalis prompted effectively simultaneous removal of carbon, nitrogen and phosphorus in activated sludge reactor: Microbial community succession and functional characteristics. Bioresour. Technol. 2022, 348, 126820. [Google Scholar] [CrossRef]
- Wang, S.; Li, H.; Fan, X.; Zhang, J.; Tang, P.; Yuan, Q. Metabolic responses in Candida tropicalis to complex inhibitors during xylitol bioconversion. Fungal Genet. Biol. 2015, 82, 1–8. [Google Scholar] [CrossRef]
- Wang, J.; Peng, J.; Fan, H.; Xiu, X.; Xue, L.; Wang, L.; Su, J.; Yang, X.; Wang, R. Development of mazF-based markerless genome editing system and metabolic pathway engineering in Candida tropicalis for producing long-chain dicarboxylic acids. J. Ind. Microbiol. Biotechnol. 2018, 45, 971–981. [Google Scholar] [CrossRef] [PubMed]
- Erian, A.M.; Sauer, M. Utilizing yeasts for the conversion of renewable feedstocks to sugar alcohols—A review. Bioresour. Technol. 2022, 346, 126296. [Google Scholar] [CrossRef]
- Narisetty, V.; Okibe, M.C.; Amulya, K.; Jokodola, E.O.; Coulon, F.; Tyagi, V.K.; Lens, P.N.L.; Parameswaran, B.; Kumar, V. Technological advancements in valorization of second generation (2G) feedstocks for bio-based succinic acid production. Bioresour. Technol. 2022, 360, 127513. [Google Scholar] [CrossRef]
- Wang, L.; Wu, D.; Tang, P.; Fan, X.; Yuan, Q. Xylitol production from corncob hydrolysate using polyurethane foam with immobilized Candida tropicalis. Carbohydr. Polym. 2012, 90, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Jiang, B.; Zhang, T.; Chen, J. Combined mutagenesis and metabolic regulation to enhance D-arabitol production from Candida parapsilosis. J. Ind. Microbiol. Biotechnol. 2020, 47, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Toledano, M.B.; Delaunay-Moisan, A.; Outten, C.E.; Igbaria, A. Functions and cellular compartmentation of the thioredoxin and glutathione pathways in yeast. Antioxid. Redox Signal. 2013, 18, 1699–1711. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Huang, X.; Zhou, Y.; Li, J.; Liu, F.; Li, X.; Hu, X.; Wang, J.; Guo, L.; Liu, R.; et al. Cytosol Peroxiredoxin and Cell Surface Catalase Differentially Respond to H2O2 Stress in Aspergillus nidulans. Antioxidants 2023, 12, 1333. [Google Scholar] [CrossRef]
- Hatem, E.; Berthonaud, V.; Dardalhon, M.; Lagniel, G.; Baudouin-Cornu, P.; Huang, M.; Labarre, J.; Chédin, S. Glutathione is essential to preserve nuclear function and cell survival under oxidative stress. Free Radic. Biol. Med. 2014, 67, 103–114. [Google Scholar] [CrossRef]
- Kildegaard, K.R.; Hallström, B.M.; Blicher, T.H.; Sonnenschein, N.; Jensen, N.B.; Sherstyk, S.; Harrison, S.J.; Maury, J.; Herrgård, M.J.; Juncker, A.S.; et al. Evolution reveals a glutathione-dependent mechanism of 3-hydroxypropionic acid tolerance. Metab. Eng. 2014, 26, 57–66. [Google Scholar] [CrossRef]
- Ma, M.; Liu, L.Z. Quantitative transcription dynamic analysis reveals candidate genes and key regulators for ethanol tolerance in Saccharomyces cerevisiae. BMC Microbiol. 2010, 10, 169. [Google Scholar] [CrossRef]
- Goldberg, A.L. Protein degradation and protection against misfolded or damaged proteins. Nature 2003, 426, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, H.; Ha, S.; Ju, D.; Xie, Y. Proteasomal degradation of Rpn4 in Saccharomyces cerevisiae is critical for cell viability under stressed conditions. Genetics 2010, 184, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Liu, Z.L. Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genom. 2010, 11, 660. [Google Scholar] [CrossRef]
- Almario, M.P.; Reyes, L.H.; Kao, K.C. Evolutionary engineering of Saccharomyces cerevisiae for enhanced tolerance to hydrolysates of lignocellulosic biomass. Biotechnol. Biofuels 2013, 110, 2616–2623. [Google Scholar] [CrossRef]
- Luo, Y.; Qin, G.; Zhang, J.; Liang, Y.; Song, Y.; Zhao, M.; Tsuge, T.; Aoyama, T.; Liu, J.; Gu, H.; et al. D-myo-inositol-3-phosphate affects phosphatidylinositol-mediated endomembrane function in Arabidopsis and is essential for auxin-regulated embryogenesis. Plant Cell. 2011, 23, 1352–1372. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Sahraeian, S.M.E.; Mohiyuddin, M.; Sebra, R.; Tilgner, H.; Afshar, P.T.; Au, K.F.; Bani Asadi, N.; Gerstein, M.B.; Wong, W.H.; Snyder, M.P.; et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Commun. 2017, 8, 59. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Ni, Z.; Jiao, B.; Hu, Y.; Sun, Z.; Wu, D.; Yuan, Q.; Han, Y.; Wang, L. Comprehensive Transcriptomic Analysis of the Isolated Candida tropicalis with Enhanced Tolerance of Furfural Inhibitor. Int. J. Mol. Sci. 2025, 26, 2999. https://doi.org/10.3390/ijms26072999
Liu J, Ni Z, Jiao B, Hu Y, Sun Z, Wu D, Yuan Q, Han Y, Wang L. Comprehensive Transcriptomic Analysis of the Isolated Candida tropicalis with Enhanced Tolerance of Furfural Inhibitor. International Journal of Molecular Sciences. 2025; 26(7):2999. https://doi.org/10.3390/ijms26072999
Chicago/Turabian StyleLiu, Jianguang, Zifu Ni, Bingyu Jiao, Yuansen Hu, Zhongke Sun, Dapeng Wu, Qipeng Yuan, Yuhuan Han, and Le Wang. 2025. "Comprehensive Transcriptomic Analysis of the Isolated Candida tropicalis with Enhanced Tolerance of Furfural Inhibitor" International Journal of Molecular Sciences 26, no. 7: 2999. https://doi.org/10.3390/ijms26072999
APA StyleLiu, J., Ni, Z., Jiao, B., Hu, Y., Sun, Z., Wu, D., Yuan, Q., Han, Y., & Wang, L. (2025). Comprehensive Transcriptomic Analysis of the Isolated Candida tropicalis with Enhanced Tolerance of Furfural Inhibitor. International Journal of Molecular Sciences, 26(7), 2999. https://doi.org/10.3390/ijms26072999





