Latest News from the “Guardian”: p53 Directly Activates Asymmetric Stem Cell Division Regulators
Abstract
1. The “Guardian of the Genome” and More: Newly Emerged p53 Functions
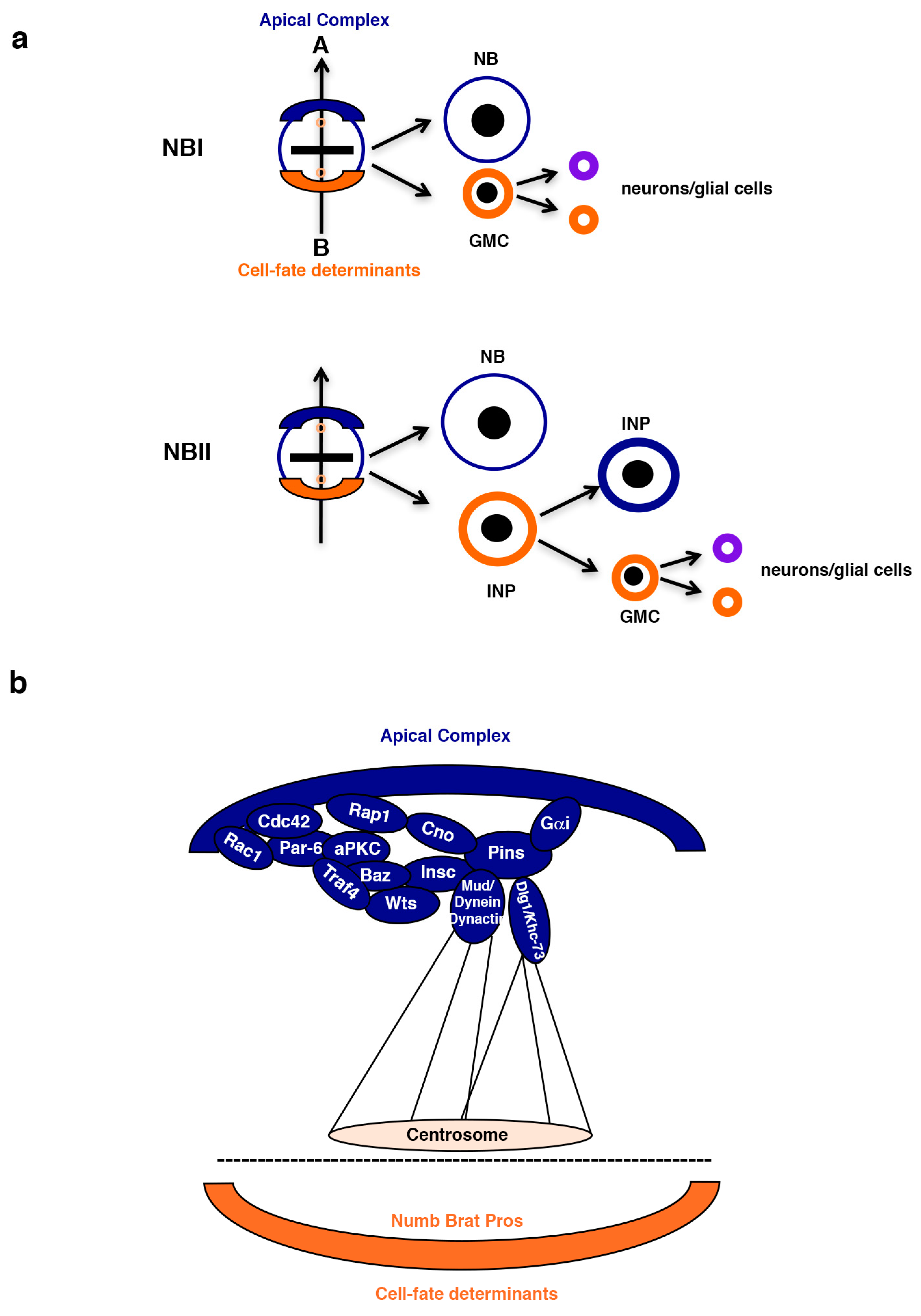
2. Asymmetric Stem Cell Division
2.1. The Cell-Fate Determinants Numb and Brat
2.2. ASCD Failures Can Cause Tumor-like Overgrowth
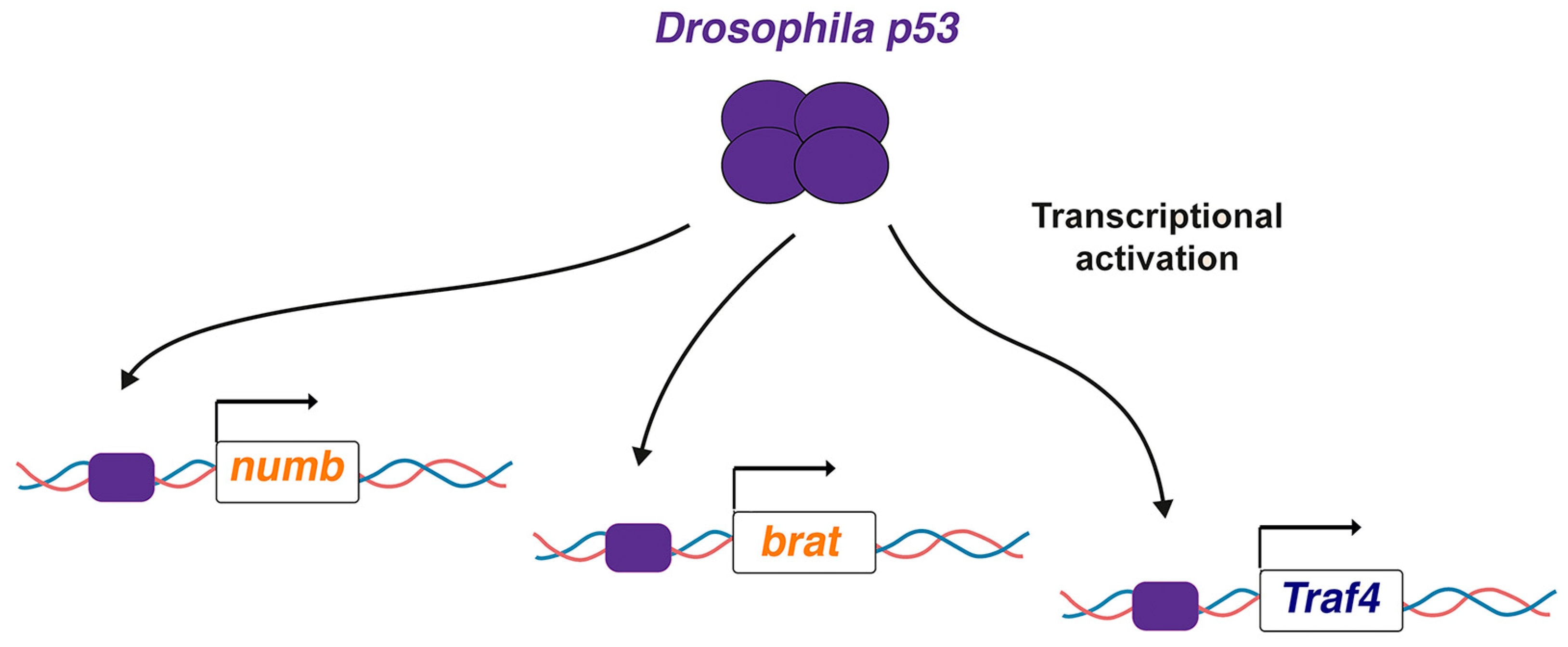
3. Drosophila p53 Directly Activates Key ASCD Regulators: Why So Mild Phenotypes?
4. Future Directions
Funding
Conflicts of Interest
References
- Kress, M.; May, E.; Cassingena, R.; May, P. Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J. Virol. 1979, 31, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.P.; Crawford, L.V. T antigen is bound to a host protein in SV40-transformed cells. Nature 1979, 278, 261–263. [Google Scholar] [CrossRef]
- Linzer, D.I.; Levine, A.J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 1979, 17, 43–52. [Google Scholar] [CrossRef]
- Lane, D.P. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, T.; Su, W.; Dou, Z.; Zhao, D.; Jin, X.; Lei, H.; Wang, J.; Xie, X.; Cheng, B.; et al. Mutant p53 in cancer: From molecular mechanism to therapeutic modulation. Cell Death Dis. 2022, 13, 974. [Google Scholar] [CrossRef]
- Bouaoun, L.; Sonkin, D.; Ardin, M.; Hollstein, M.; Byrnes, G.; Zavadil, J.; Olivier, M. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum. Mutat. 2016, 37, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lozano, G. p53: Multiple Facets of a Rubik’s Cube. Annu. Rev. Cancer Biol. 2017, 1, 185–201. [Google Scholar] [CrossRef]
- Boutelle, A.M.; Attardi, L.D. p53 and Tumor Suppression: It Takes a Network. Trends Cell Biol. 2021, 31, 298–310. [Google Scholar] [CrossRef]
- Pant, V.; Lozano, G. Limiting the power of p53 through the ubiquitin proteasome pathway. Genes. Dev. 2014, 28, 1739–1751. [Google Scholar] [CrossRef]
- Hernández Borrero, L.J.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Cicalese, A.; Bonizzi, G.; Pasi, C.E.; Faretta, M.; Ronzoni, S.; Giulini, B.; Brisken, C.; Minucci, S.; Di Fiore, P.P.; Pelicci, P.G. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 2009, 138, 1083–1095. [Google Scholar] [CrossRef]
- Vousden, K.H.; Prives, C. Blinded by the Light: The Growing Complexity of p53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef]
- Tyner, S.D.; Venkatachalam, S.; Choi, J.; Jones, S.; Ghebranious, N.; Igelmann, H.; Lu, X.; Soron, G.; Cooper, B.; Brayton, C.; et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 2002, 415, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Van Nostrand, J.L.; Brady, C.A.; Jung, H.; Fuentes, D.R.; Kozak, M.M.; Johnson, T.M.; Lin, C.Y.; Lin, C.J.; Swiderski, D.L.; Vogel, H.; et al. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature 2014, 514, 228–232. [Google Scholar] [CrossRef] [PubMed]
- McGowan, K.A.; Li, J.Z.; Park, C.Y.; Beaudry, V.; Tabor, H.K.; Sabnis, A.J.; Zhang, W.; Fuchs, H.; de Angelis, M.H.; Myers, R.M.; et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 2008, 40, 963–970. [Google Scholar] [CrossRef]
- Levine, A.J.; Berger, S.L. The interplay between epigenetic changes and the p53 protein in stem cells. Genes. Dev. 2017, 31, 1195–1201. [Google Scholar] [CrossRef] [PubMed]
- Cooks, T.; Harris, C.C.; Oren, M. Caught in the cross fire: p53 in inflammation. Carcinogenesis 2014, 35, 1680–1690. [Google Scholar] [CrossRef]
- Jain, A.K.; Barton, M.C. p53: Emerging roles in stem cells, development and beyond. Development 2018, 145. [Google Scholar] [CrossRef]
- Aylon, Y.; Oren, M. The Paradox of p53: What, How, and Why? Cold Spring Harb. Perspect. Med. 2016, 6. [Google Scholar] [CrossRef]
- Ingaramo, M.C.; Sánchez, J.A.; Dekanty, A. Regulation and function of p53: A perspective from Drosophila studies. Mech. Dev. 2018, 154, 82–90. [Google Scholar] [CrossRef]
- Pappas, K.; Xu, J.; Zairis, S.; Resnick-Silverman, L.; Abate, F.; Steinbach, N.; Ozturk, S.; Saal, L.H.; Su, T.; Cheung, P.; et al. p53 Maintains Baseline Expression of Multiple Tumor Suppressor Genes. Mol. Cancer Res. 2017, 15, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Spike, B.T.; Wahl, G.M. p53, Stem Cells, and Reprogramming: Tumor Suppression beyond Guarding the Genome. Genes. Cancer 2011, 2, 404–419. [Google Scholar] [CrossRef] [PubMed]
- Ouadah, Y.; Rojas, E.R.; Riordan, D.P.; Capostagno, S.; Kuo, C.S.; Krasnow, M.A. Rare Pulmonary Neuroendocrine Cells Are Stem Cells Regulated by Rb, p53, and Notch. Cell 2019, 179, 403–416.e423. [Google Scholar] [CrossRef] [PubMed]
- Knoblich, J.A. Mechanisms of asymmetric stem cell division. Cell 2008, 132, 583–597. [Google Scholar] [CrossRef]
- Homem, C.C.; Knoblich, J.A. Drosophila neuroblasts: A model for stem cell biology. Development 2012, 139, 4297–4310. [Google Scholar] [CrossRef]
- Petronczki, M.; Knoblich, J.A. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 2001, 3, 43–49. [Google Scholar]
- Schober, M.; Schaefer, M.; Knoblich, J.A. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 1999, 402, 548–551. [Google Scholar] [CrossRef]
- Wodarz, A.; Ramrath, A.; Grimm, A.; Knust, E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 2000, 150, 1361–1374. [Google Scholar]
- Wodarz, A.; Ramrath, A.; Kuchinke, U.; Knust, E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 1999, 402, 544–547. [Google Scholar]
- de Torres-Jurado, A.; Manzanero-Ortiz, S.; Carmena, A. Glial-secreted Netrins regulate Robo1/Rac1-Cdc42 signaling threshold levels during Drosophila asymmetric neural stem/progenitor cell division. Curr. Biol. 2022, 32, 2174–2188.e2173. [Google Scholar] [CrossRef]
- Carmena, A.; Makarova, A.; Speicher, S. The Rap1-Rgl-Ral signaling network regulates neuroblast cortical polarity and spindle orientation. J. Cell Biol. 2011, 195, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Atwood, S.X.; Chabu, C.; Penkert, R.R.; Doe, C.Q.; Prehoda, K.E. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J. Cell Sci. 2007, 120, 3200–3206. [Google Scholar] [CrossRef]
- Carmena, A.; Speicher, S.; Baylies, M. The PDZ protein Canoe/AF-6 links Ras-MAPK, Notch and Wingless/Wnt signaling pathways by directly interacting with Ras, Notch and Dishevelled. PLoS ONE 2006, 1, e66. [Google Scholar]
- Parmentier, M.L.; Woods, D.; Greig, S.; Phan, P.G.; Radovic, A.; Bryant, P.; O’Kane, C.J. Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J. Neurosci. 2000, 20, RC84. [Google Scholar] [CrossRef]
- Yu, F.; Cai, Y.; Kaushik, R.; Yang, X.; Chia, W. Distinct roles of Galphai and Gbeta13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J. Cell Biol. 2003, 162, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.K.; Neumuller, R.A.; Novatchkova, M.; Du, Q.; Knoblich, J.A. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell 2006, 10, 731–742. [Google Scholar] [CrossRef]
- Izumi, Y.; Ohta, N.; Hisata, K.; Raabe, T.; Matsuzaki, F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 2006, 8, 586–593. [Google Scholar] [CrossRef]
- Siller, K.H.; Cabernard, C.; Doe, C.Q. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 2006, 8, 594–600. [Google Scholar] [CrossRef]
- Johnston, C.A.; Hirono, K.; Prehoda, K.E.; Doe, C.Q. Identification of an Aurora-A/PinsLINKER/Dlg spindle orientation pathway using induced cell polarity in S2 cells. Cell 2009, 138, 1150–1163. [Google Scholar] [CrossRef]
- Keder, A.; Rives-Quinto, N.; Aerne, B.L.; Franco, M.; Tapon, N.; Carmena, A. The hippo pathway core cassette regulates asymmetric cell division. Curr. Biol. 2015, 25, 2739–2750. [Google Scholar] [CrossRef]
- Speicher, S.; Fischer, A.; Knoblich, J.; Carmena, A. The PDZ protein Canoe regulates the asymmetric division of Drosophila neuroblasts and muscle progenitors. Curr. Biol. 2008, 18, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Bello, B.; Reichert, H.; Hirth, F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 2006, 133, 2639–2648. [Google Scholar] [CrossRef]
- Betschinger, J.; Mechtler, K.; Knoblich, J.A. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 2006, 124, 1241–1253. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.K.; Rolland, V.; Betschinger, J.; Kinsey, K.A.; Emery, G.; Knoblich, J.A. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 2008, 14, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Wilkinson, B.D.; Siegrist, S.E.; Wharton, R.P.; Doe, C.Q. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell 2006, 10, 441–449. [Google Scholar] [CrossRef]
- Knoblich, J.A.; Jan, L.Y.; Jan, Y.N. Asymmetric segregation of Numb and Prospero during cell division. Nature 1995, 377, 624–627. [Google Scholar] [CrossRef]
- Uemura, T.; Shepherd, S.; Ackerman, L.; Jan, L.Y.; Jan, Y.N. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 1989, 58, 349–360. [Google Scholar] [CrossRef]
- Rhyu, M.S.; Jan, L.Y.; Jan, Y.N. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 1994, 76, 477–491. [Google Scholar] [CrossRef]
- Smith, C.A.; Lau, K.M.; Rahmani, Z.; Dho, S.E.; Brothers, G.; She, Y.M.; Berry, D.M.; Bonneil, E.; Thibault, P.; Schweisguth, F.; et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. Embo J. 2007, 26, 468–480. [Google Scholar]
- Sun, Y.; Goderie, S.K.; Temple, S. Asymmetric distribution of EGFR receptor during mitosis generates diverse CNS progenitor cells. Neuron 2005, 45, 873–886. [Google Scholar] [CrossRef]
- Wakamatsu, Y.; Maynard, T.M.; Jones, S.U.; Weston, J.A. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron 1999, 23, 71–81. [Google Scholar]
- Shen, Q.; Zhong, W.; Jan, Y.N.; Temple, S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development 2002, 129, 4843–4853. [Google Scholar]
- Zhong, W.; Jiang, M.M.; Schonemann, M.D.; Meneses, J.J.; Pedersen, R.A.; Jan, L.Y.; Jan, Y.N. Mouse numb is an essential gene involved in cortical neurogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 6844–6849. [Google Scholar] [CrossRef] [PubMed]
- Verdi, J.M.; Bashirullah, A.; Goldhawk, D.E.; Kubu, C.J.; Jamali, M.; Meakin, S.O.; Lipshitz, H.D. Distinct human NUMB isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc. Natl. Acad. Sci. USA 1999, 96, 10472–10476. [Google Scholar] [CrossRef] [PubMed]
- Dho, S.E.; French, M.B.; Woods, S.A.; McGlade, C.J. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J. Biol. Chem. 1999, 274, 33097–33104. [Google Scholar] [CrossRef]
- Toriya, M.; Tokunaga, A.; Sawamoto, K.; Nakao, K.; Okano, H. Distinct functions of human numb isoforms revealed by misexpression in the neural stem cell lineage in the Drosophila larval brain. Dev. Neurosci. 2006, 28, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Verdi, J.M.; Schmandt, R.; Bashirullah, A.; Jacob, S.; Salvino, R.; Craig, C.G.; Program, A.E.; Lipshitz, H.D.; McGlade, C.J. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr. Biol. 1996, 6, 1134–1145. [Google Scholar] [CrossRef]
- Yan, B. Numb--from flies to humans. Brain Dev. 2010, 32, 293–298. [Google Scholar] [CrossRef]
- Guo, M.; Jan, L.Y.; Jan, Y.N. Control of daughter cell fates during asymmetric division: Interaction of Numb and Notch. Neuron 1996, 17, 27–41. [Google Scholar] [CrossRef]
- Spana, E.P.; Doe, C.Q. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 1996, 17, 21–26. [Google Scholar] [CrossRef]
- McGill, M.A.; McGlade, C.J. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 2003, 278, 23196–23203. [Google Scholar] [CrossRef] [PubMed]
- Pece, S.; Serresi, M.; Santolini, E.; Capra, M.; Hulleman, E.; Galimberti, V.; Zurrida, S.; Maisonneuve, P.; Viale, G.; Di Fiore, P.P. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 2004, 167, 215–221. [Google Scholar] [CrossRef]
- Pece, S.; Confalonieri, S.; R Romano, P.; Di Fiore, P.P. NUMB-ing down cancer by more than just a NOTCH. Biochim. Biophys. Acta 2011, 1815, 26–43. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Campos, S.M.; García-Heredia, J.M. The Multitasker Protein: A Look at the Multiple Capabilities of NUMB. Cells 2023, 12, 333. [Google Scholar] [CrossRef]
- Colaluca, I.N.; Tosoni, D.; Nuciforo, P.; Senic-Matuglia, F.; Galimberti, V.; Viale, G.; Pece, S.; Di Fiore, P.P. NUMB controls p53 tumour suppressor activity. Nature 2008, 451, 76–80. [Google Scholar] [CrossRef]
- Tosoni, D.; Zecchini, S.; Coazzoli, M.; Colaluca, I.; Mazzarol, G.; Rubio, A.; Caccia, M.; Villa, E.; Zilian, O.; Di Fiore, P.P.; et al. The Numb/p53 circuitry couples replicative self-renewal and tumor suppression in mammary epithelial cells. J. Cell Biol. 2015, 211, 845–862. [Google Scholar] [CrossRef] [PubMed]
- Faraldo, M.M.; Glukhova, M.A. Regulating the regulator: Numb acts upstream of p53 to control mammary stem and progenitor cell. J. Cell Biol. 2015, 211, 737–739. [Google Scholar] [CrossRef]
- Reymond, A.; Meroni, G.; Fantozzi, A.; Merla, G.; Cairo, S.; Luzi, L.; Riganelli, D.; Zanaria, E.; Messali, S.; Cainarca, S.; et al. The tripartite motif family identifies cell compartments. EMBO J. 2001, 20, 2140–2151. [Google Scholar] [CrossRef]
- Henry, J.; Mather, I.H.; McDermott, M.F.; Pontarotti, P. B30.2-like domain proteins: Update and new insights into a rapidly expanding family of proteins. Mol. Biol. Evol. 1998, 15, 1696–1705. [Google Scholar] [CrossRef]
- Valletti, A.; Marzano, F.; Pesole, G.; Sbisà, E.; Tullo, A. Targeting Chemoresistant Tumors: Could TRIM Proteins-p53 Axis Be a Possible Answer? Int. J. Mol. Sci. 2019, 20, 1776. [Google Scholar] [CrossRef]
- Elabd, S.; Meroni, G.; Blattner, C. TRIMming p53’s anticancer activity. Oncogene 2016, 35, 5577–5584. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Wang, X.L.; Ly, P.; Belyi, V.; Xu-Monette, Z.Y.; Young, K.H.; Hu, W.; Feng, Z. E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ. 2014, 21, 1792–1804. [Google Scholar] [CrossRef]
- Wang, H.; Somers, G.W.; Bashirullah, A.; Heberlein, U.; Yu, F.; Chia, W. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes. Dev. 2006, 20, 3453–3463. [Google Scholar] [CrossRef] [PubMed]
- Caussinus, E.; Gonzalez, C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 2005, 37, 1125–1129. [Google Scholar] [CrossRef]
- Gateff, E.; Schneiderman, H.A. Neoplasms in mutant and cultured wild-tupe tissues of Drosophila. Natl. Cancer Inst. Monogr. 1969, 31, 365–397. [Google Scholar] [PubMed]
- Gateff, E. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science 1978, 200, 1448–1459. [Google Scholar]
- Gateff, E. Tumor suppressor and overgrowth suppressor genes of Drosophila melanogaster: Developmental aspects. Int. J. Dev. Biol. 1994, 38, 565–590. [Google Scholar] [PubMed]
- Albertson, R.; Doe, C.Q. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat. Cell Biol. 2003, 5, 166–170. [Google Scholar] [CrossRef]
- Ohshiro, T.; Yagami, T.; Zhang, C.; Matsuzaki, F. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature 2000, 408, 593–596. [Google Scholar] [CrossRef]
- Peng, C.Y.; Manning, L.; Albertson, R.; Doe, C.Q. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature 2000, 408, 596–600. [Google Scholar] [CrossRef]
- Carmena, A. The Case of the Scribble Polarity Module in Asymmetric Neuroblast Division in Development and Tumorigenesis. Int. J. Mol. Sci. 2020, 21, 2865. [Google Scholar] [CrossRef] [PubMed]
- Rives-Quinto, N.; Franco, M.; de Torres-Jurado, A.; Carmena, A. Synergism between canoe and scribble mutations causes tumor-like overgrowth via Ras activation in neural stem cells and epithelia. Development 2017, 144, 2570–2583. [Google Scholar] [CrossRef] [PubMed]
- Carmena, A. Compromising asymmetric stem cell division in Drosophila central brain: Revisiting the connections with tumorigenesis. Fly 2018, 12, 71–80. [Google Scholar] [CrossRef]
- Manzanero-Ortiz, S.; de Torres-Jurado, A.; Hernández-Rojas, R.; Carmena, A. Pilot RNAi Screen in Drosophila Neural Stem Cell Lineages to Identify Novel Tumor Suppressor Genes Involved in Asymmetric Cell Division. Int. J. Mol. Sci. 2021, 22, 1332. [Google Scholar] [CrossRef] [PubMed]
- Manzanero-Ortiz, S.; Franco, M.; Laxmeesha, M.; Carmena, A. p53 tumor suppressor directly activates conserved asymmetric stem cell division regulators. iScience 2024, 27, 111118. [Google Scholar] [CrossRef]
- Cai, Y.; Chia, W.; Yang, X. A family of snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. EMBO J. 2001, 20, 1704–1714. [Google Scholar] [CrossRef]
- Fischer, M. Conservation and divergence of the p53 gene regulatory network between mice and humans. Oncogene 2019, 38, 4095–4109. [Google Scholar] [CrossRef]
- Wang, H.; Cai, Y.; Chia, W.; Yang, X. Drosophila homologs of mammalian TNF/TNFR-related molecules regulate segregation of Miranda/Prospero in neuroblasts. EMBO J. 2006, 25, 5783–5793. [Google Scholar] [CrossRef]
- Jin, S.; Martinek, S.; Joo, W.S.; Wortman, J.R.; Mirkovic, N.; Sali, A.; Yandell, M.D.; Pavletich, N.P.; Young, M.W.; Levine, A.J. Identification and characterization of a p53 homologue in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2000, 97, 7301–7306. [Google Scholar] [CrossRef]
- Ollmann, M.; Young, L.M.; Di Como, C.J.; Karim, F.; Belvin, M.; Robertson, S.; Whittaker, K.; Demsky, M.; Fisher, W.W.; Buchman, A.; et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 2000, 101, 91–101. [Google Scholar] [CrossRef]
- Brodsky, M.H.; Weinert, B.T.; Tsang, G.; Rong, Y.S.; McGinnis, N.M.; Golic, K.G.; Rio, D.C.; Rubin, G.M. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell Biol. 2004, 24, 1219–1231. [Google Scholar] [CrossRef] [PubMed]
- Sogame, N.; Kim, M.; Abrams, J.M. Drosophila p53 preserves genomic stability by regulating cell death. Proc. Natl. Acad. Sci. USA 2003, 100, 4696–4701. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Freije, W.A.; Guptan, P.; Banerjee, U. Metabolic control of G1-S transition: Cyclin E degradation by p53-induced activation of the ubiquitin-proteasome system. J. Cell Biol. 2010, 188, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Yagishita, N.; Sasaki, T.; Nakazawa, M.; Kato, Y.; Yamadera, T.; Bae, E.; Toriyama, S.; Ikeda, R.; Zhang, L.; et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ’Synoviolin’. EMBO J. 2007, 26, 113–122. [Google Scholar] [CrossRef]
- Chakraborty, R.; Li, Y.; Zhou, L.; Golic, K.G. Corp Regulates P53 in Drosophila melanogaster via a Negative Feedback Loop. PLoS Genet. 2015, 11, e1005400. [Google Scholar] [CrossRef]
- Chen, G.; Kong, J.; Tucker-Burden, C.; Anand, M.; Rong, Y.; Rahman, F.; Moreno, C.S.; Van Meir, E.G.; Hadjipanayis, C.G.; Brat, D.J. Human Brat ortholog TRIM3 is a tumor suppressor that regulates asymmetric cell division in glioblastoma. Cancer Res. 2014, 74, 4536–4548. [Google Scholar] [CrossRef]
- de la Cova, C.; Senoo-Matsuda, N.; Ziosi, M.; Wu, D.C.; Bellosta, P.; Quinzii, C.M.; Johnston, L.A. Supercompetitor status of Drosophila Myc cells requires p53 as a fitness sensor to reprogram metabolism and promote viability. Cell Metab. 2014, 19, 470–483. [Google Scholar] [CrossRef]
- Barrio, L.; Dekanty, A.; Milán, M. MicroRNA-mediated regulation of Dp53 in the Drosophila fat body contributes to metabolic adaptation to nutrient deprivation. Cell Rep. 2014, 8, 528–541. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carmena, A. Latest News from the “Guardian”: p53 Directly Activates Asymmetric Stem Cell Division Regulators. Int. J. Mol. Sci. 2025, 26, 3171. https://doi.org/10.3390/ijms26073171
Carmena A. Latest News from the “Guardian”: p53 Directly Activates Asymmetric Stem Cell Division Regulators. International Journal of Molecular Sciences. 2025; 26(7):3171. https://doi.org/10.3390/ijms26073171
Chicago/Turabian StyleCarmena, Ana. 2025. "Latest News from the “Guardian”: p53 Directly Activates Asymmetric Stem Cell Division Regulators" International Journal of Molecular Sciences 26, no. 7: 3171. https://doi.org/10.3390/ijms26073171
APA StyleCarmena, A. (2025). Latest News from the “Guardian”: p53 Directly Activates Asymmetric Stem Cell Division Regulators. International Journal of Molecular Sciences, 26(7), 3171. https://doi.org/10.3390/ijms26073171





