The Role of Bacteria-Derived Hydrogen Sulfide in Multiple Axes of Disease
Abstract
1. Introduction
1.1. Endogenous H2S
1.2. Bacteria-Derived H2S
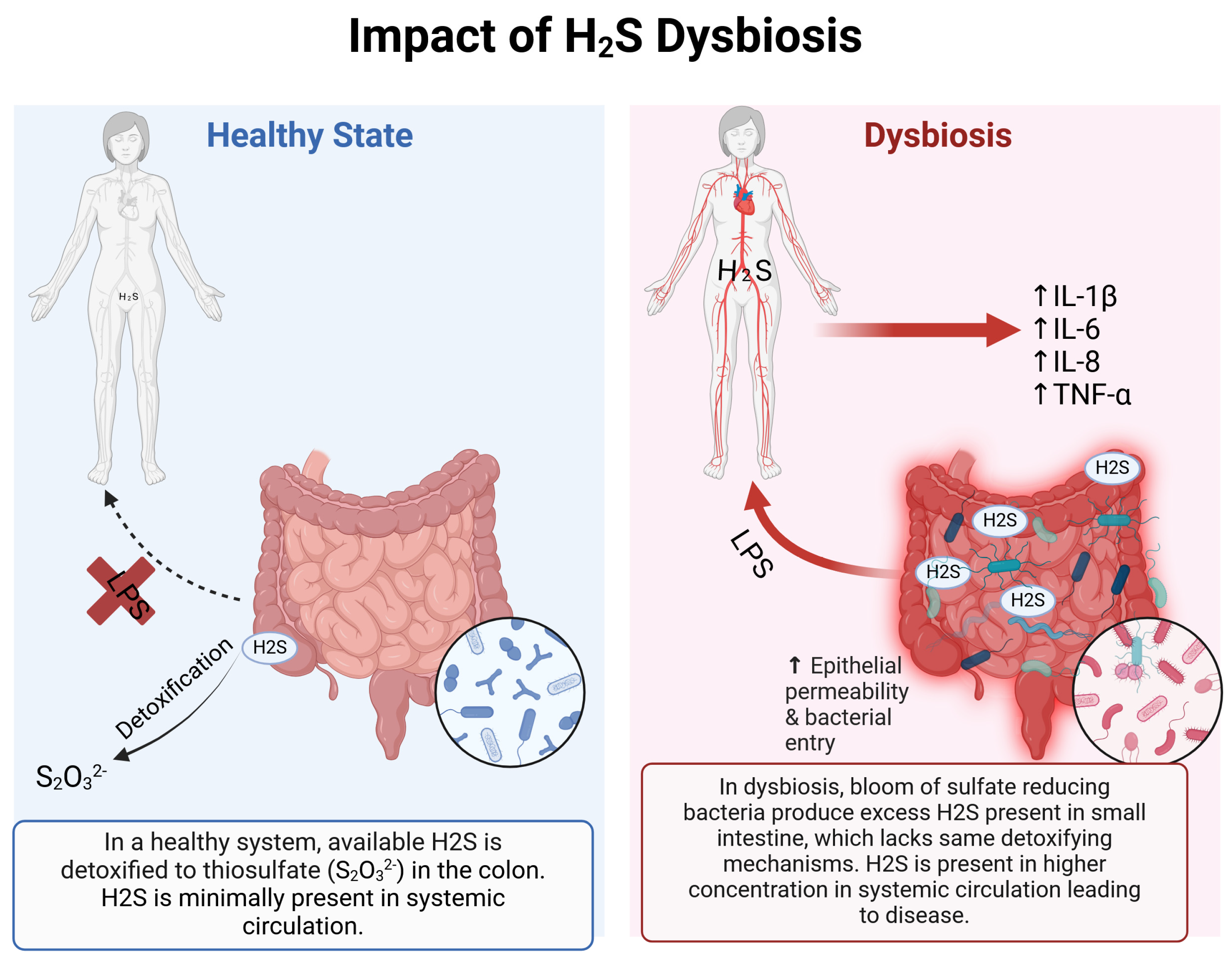
1.3. Detoxification of H2S
1.4. H2S and Disease
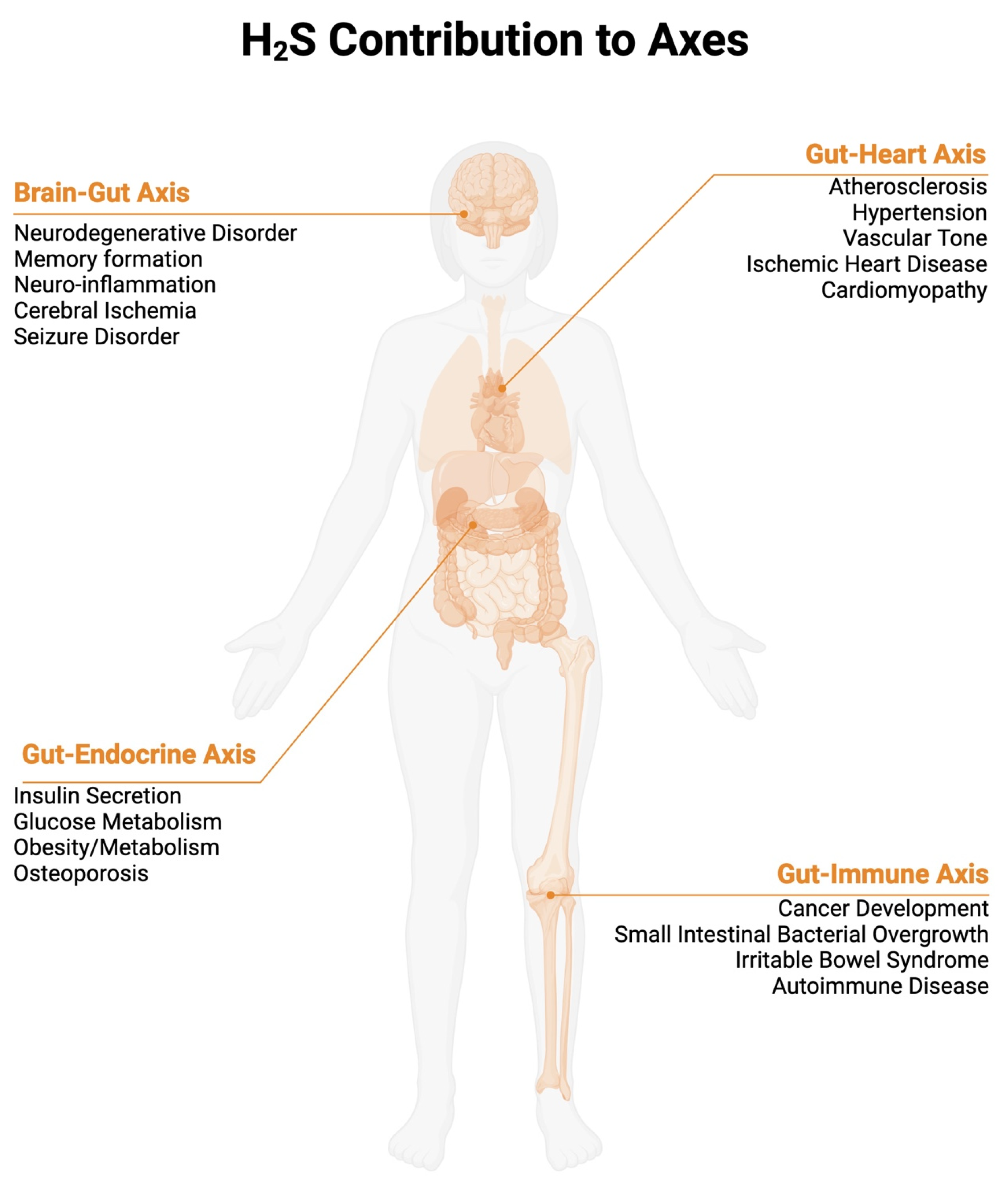
2. Effects of Hydrogen Sulfide in Different Axes
2.1. Gut–Immune Axis/Inflammatory Bowel Disease
- Colorectal Cancer development
- Irritable Bowel Syndrome/SIBO
- Ulcer healing
2.2. Brain–Gut Axis
- Parkinson’s Disease
- Alzheimer’s disease
- Ischemic stroke
2.3. Gut–Heart Axis
- Hypertension
- Atherosclerosis
2.4. Gut–Endocrine Axis
- Disturbances in energy metabolism
3. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Figliuolo, V.R.; dos Santos, L.M.; Abalo, A.; Nanini, H.; Santos, A.; Brittes, N.M.; Bernardazzi, C.; de Souza, H.S.P.; Vieira, L.Q.; Coutinho-Silva, R.; et al. Sulfate-reducing bacteria stimulate gut immune responses and contribute to inflammation in experimental colitis. Life Sci. 2017, 189, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota-gut-brain axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Mahony, S.M. The microbiome-gut-brain axis: From bowel to behavior. Neurogastroenterol. Motil. 2011, 23, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Morais, L.H.; Schreiber, H.L.t.; Mazmanian, S.K. The gut microbiota-brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef]
- Yan, M.; Man, S.; Sun, B.; Ma, L.; Guo, L.; Huang, L.; Gao, W. Gut liver brain axis in diseases: The implications for therapeutic interventions. Signal Transduct. Target. Ther. 2023, 8, 443. [Google Scholar] [CrossRef]
- Matsiras, D.; Bezati, S.; Ventoulis, I.; Verras, C.; Parissis, J.; Polyzogopoulou, E. Gut Failure: A Review of the Pathophysiology and Therapeutic Potentials in the Gut–Heart Axis. J. Clin. Med. 2023, 12, 2567. [Google Scholar] [CrossRef]
- Mendiola, P.J.; Naik, J.S.; Gonzalez Bosc, L.V.; Gardiner, A.S.; Birg, A.; Kanagy, N.L. Hydrogen Sulfide Actions in the Vasculature. Compr. Physiol. 2021, 11, 2467–2488. [Google Scholar] [CrossRef]
- Donertas Ayaz, B.; Zubcevic, J. Gut microbiota and neuroinflammation in pathogenesis of hypertension: A potential role for hydrogen sulfide. Pharmacol. Res. 2020, 153, 104677. [Google Scholar] [CrossRef]
- Wallace, J.L.; Vong, L.; McKnight, W.; Dicay, M.; Martin, G.R. Endogenous and Exogenous Hydrogen Sulfide Promotes Resolution of Colitis in Rats. Gastroenterology 2009, 137, 569–578.e561. [Google Scholar] [CrossRef]
- Kanagy, N.L.; Szabo, C.; Papapetropoulos, A. Vascular biology of hydrogen sulfide. Am. J. Physiol. Cell Physiol. 2017, 312, C537–C549. [Google Scholar] [CrossRef]
- Zhang, J.X.; Pegoli, W.; Clemens, M.G. Endothelin-1 induces direct constriction of hepatic sinusoids. Am. J. Physiol. Gastrointest. Liver Physiol. 1994, 266, G624–G632. [Google Scholar] [CrossRef] [PubMed]
- Randi, E.B.; Casili, G.; Jacquemai, S.; Szabo, C. Selenium-Binding Protein 1 (SELENBP1) Supports Hydrogen Sulfide Biosynthesis and Adipogenesis. Antioxidants 2021, 10, 361. [Google Scholar] [CrossRef]
- Kartha, R.V.; Zhou, J.; Hovde, L.B.; Cheung, B.W.Y.; Schröder, H. Enhanced detection of hydrogen sulfide generated in cell culture using an agar trap method. Anal. Biochem. 2012, 423, 102–108. [Google Scholar] [CrossRef]
- Shen, X.; Kolluru, G.K.; Yuan, S.; Kevil, C. Measurement of H2S in vivo and in vitro by the monobromobimane method. Methods Enzymol. 2015, 554, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Huang, D.; Sun, Z.; Chen, X. Effects of intestinal Desulfovibrio bacteria on host health and its potential regulatory strategies: A review. Microbiol. Res. 2024, 284, 127725. [Google Scholar] [CrossRef]
- Wallace, J.L.; Motta, J.P.; Buret, A.G. Hydrogen sulfide: An agent of stability at the microbiome-mucosa interface. Am. J. Physiol.-Gastrointest. Liver Physiol. 2018, 314, G143–G149. [Google Scholar] [CrossRef]
- Kushkevych, I.; Cejnar, J.; Treml, J.; Dordević, D.; Kollar, P.; Vítězová, M. Recent Advances in Metabolic Pathways of Sulfate Reduction in Intestinal Bacteria. Cells 2020, 9, 698. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; Stams, A.J.M. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef]
- Pfennig, N.; Widdel, F.; Trüper, H.G. The Dissimilatory Sulfate-Reducing Bacteria. In The Prokaryotes: A Handbook on Habitats, Isolation, and Identification of Bacteria; Starr, M.P., Stolp, H., Trüper, H.G., Balows, A., Schlegel, H.G., Eds.; Springer: Berlin/Heidelberg, Germany, 1981; pp. 926–940. [Google Scholar]
- Dordević, D.; Jančíková, S.; Vítězová, M.; Kushkevych, I. Hydrogen sulfide toxicity in the gut environment: Meta-analysis of sulfate-reducing and lactic acid bacteria in inflammatory processes. J. Adv. Res. 2021, 27, 55–69. [Google Scholar] [CrossRef]
- Voordouw, G.; Niviere, V.; Ferris, F.G.; Fedorak, P.M.; Westlake, D.W.S. Distribution of Hydrogenase Genes in Desulfovibrio spp. and Their Use in Identification of Species from the Oil Field Environment. Appl. Environ. Microbiol. 1990, 56, 3748–3754. [Google Scholar] [CrossRef]
- Kushkevych, I.; Dordević, D.; Vítězová, M. Toxicity of hydrogen sulfide toward sulfate-reducing bacteria Desulfovibrio piger Vib-7. Arch. Microbiol. 2019, 201, 389–397. [Google Scholar] [CrossRef]
- Basic, A.; Blomqvist, S.; Carlen, A.; Dahlen, G. Estimation of bacterial hydrogen sulfide production in vitro. J. Oral Microbiol. 2015, 7, 28166. [Google Scholar] [CrossRef]
- Claesson, R.; Edlund, M.B.; Persson, S.; Carlsson, J. Production of volatile sulfur compounds by various Fusobacterium species. Oral Microbiol. Immunol. 1990, 5, 137–142. [Google Scholar] [CrossRef] [PubMed]
- László, M.G.A. Hydrogen Sulphide SIBO: Pathological Condition or Physiological Adaptation A Narrative Review. May 2023. Available online: https://s3.amazonaws.com/kajabi-storefronts-production/file-uploads/sites/114257/themes/2153533682/downloads/dbb0ee-55ca-e636-063a-3328b61e1f_Thesis_Hydrogen_Sulfide_SIBO_Final.pdf (accessed on 1 December 2024).
- Levitt, M.D.; Furne, J.; Springfield, J.; Suarez, F.; DeMaster, E. Detoxification of hydrogen sulfide and methanethiol in the cecal mucosa. J. Clin. Investig. 1999, 104, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- Suarez, F.; Furne, J.; Springfield, J.; Levitt, M. Production and elimination of sulfur-containing gases in the rat colon. Am. J. Physiol. Gastrointest. Liver Physiol. 1998, 274, G727–G733. [Google Scholar] [CrossRef]
- Strickland, J.; Cummings, A.; Spinnato, J.A., III; Liccione, J.J.; Foureman, G.L. Toxicological Review of Hydrogen Sulfide; United States Environmental Protection Agency: Washington, DC, USA, 2003.
- Peter, E.A.; Shen, X.; Shah, S.H.; Pardue, S.; Glawe, J.D.; Zhang, W.W.; Reddy, P.; Akkus, N.I.; Varma, J.; Kevil, C.G. Plasma free H2S levels are elevated in patients with cardiovascular disease. J. Am. Heart Assoc. 2013, 2, e000387. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, M.; Chow, E.J.; Lin, H.C. Eradication of small intestinal bacterial overgrowth reduces symptoms of irritable bowel syndrome. Am. J. Gastroenterol. 2000, 95, 3503–3506. [Google Scholar] [CrossRef]
- Nakamura, N.; Lin, H.C.; McSweeney, C.S.; Mackie, R.I.; Gaskins, H.R. Mechanisms of Microbial Hydrogen Disposal in the Human Colon and Implications for Health and Disease. Annu. Rev. Food Sci. Technol. 2010, 1, 363–395. [Google Scholar] [CrossRef]
- Nicholls, P.; Marshall, D.C.; Cooper, C.E.; Wilson, M.T. Sulfide inhibition of and metabolism by cytochrome c oxidase. Biochem. Soc. Trans. 2013, 41, 1312–1316. [Google Scholar] [CrossRef]
- Singh, S.; Lin, H. Hydrogen Sulfide in Physiology and Diseases of the Digestive Tract. Microorganisms 2015, 3, 866–889. [Google Scholar] [CrossRef]
- Buret, A.G.; Allain, T.; Motta, J.-P.; Wallace, J.L. Effects of Hydrogen Sulfide on the Microbiome: From Toxicity to Therapy. Antioxid. Redox Signal. 2021, 36, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Dordević, D.; Kollar, P.; Vítězová, M.; Drago, L. Hydrogen Sulfide as a Toxic Product in the Small–Large Intestine Axis and its Role in IBD Development. J. Clin. Med. 2019, 8, 1054. [Google Scholar] [CrossRef] [PubMed]
- Deplancke, B.; Gaskins, H.R. Hydrogen sulfide induces serum-independent cell cycle entry in nontransformed rat intestinal epithelial cells. FASEB J. 2003, 17, 1310–1312. [Google Scholar] [CrossRef]
- Singh, S.B.; Carroll-Portillo, A.; Lin, H.C. Desulfovibrio in the Gut: The Enemy within? Microorganisms 2023, 11, 1772. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Slevin, E.; Koyama, S.; Harrison, K.; Wan, Y.; Klaunig, J.E.; Wu, C.; Shetty, A.K.; Meng, F. Dysbiosis in gastrointestinal pathophysiology: Role of the gut microbiome in Gulf War Illness. J. Cell. Mol. Med. 2023, 27, 891–905. [Google Scholar] [CrossRef]
- Di Vincenzo, F.; Del Gaudio, A.; Petito, V.; Lopetuso, L.R.; Scaldaferri, F. Gut microbiota, intestinal permeability, and systemic inflammation: A narrative review. Intern. Emerg. Med. 2023, 19, 275–293. [Google Scholar] [CrossRef]
- Alhasson, F.; Das, S.; Seth, R.; Dattaroy, D.; Chandrashekaran, V.; Ryan, C.N.; Chan, L.S.; Testerman, T.; Burch, J.; Hofseth, L.J.; et al. Altered gut microbiome in a mouse model of Gulf War Illness causes neuroinflammation and intestinal injury via leaky gut and TLR4 activation. PLoS ONE 2017, 12, e0172914. [Google Scholar] [CrossRef]
- Mimoun, S.; Andriamihaja, M.; Chaumontet, C.; Atanasiu, C.; Benamouzig, R.; Blouin, J.M.; Tomé, D.; Bouillaud, F.; Blachier, F. Detoxification of H2S by differentiated colonic epithelial cells: Implication of the sulfide oxidizing unit and of the cell respiratory capacity. Antioxid. Redox Signal. 2012, 17, 1–10. [Google Scholar] [CrossRef]
- Wallace, J.L.; Blackler, R.W.; Chan, M.V.; Da Silva, G.J.; Elsheikh, W.; Flannigan, K.L.; Gamaniek, I.; Manko, A.; Wang, L.; Motta, J.-P.; et al. Anti-Inflammatory and Cytoprotective Actions of Hydrogen Sulfide: Translation to Therapeutics. Antioxid. Redox Signal. 2014, 22, 398–410. [Google Scholar] [CrossRef]
- Wallace, J.L.; Dicay, M.; McKnight, W.; Martin, G.R. Hydrogen sulfide enhances ulcer healing in rats. FASEB J. 2007, 21, 4070–4076. [Google Scholar] [CrossRef] [PubMed]
- Mottawea, W.; Chiang, C.K.; Mühlbauer, M.; Starr, A.E.; Butcher, J.; Abujamel, T.; Deeke, S.A.; Brandel, A.; Zhou, H.; Shokralla, S.; et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn’s disease. Nat. Commun. 2016, 7, 13419. [Google Scholar] [CrossRef]
- Chassard, C.; Dapoigny, M.; Scott, K.P.; Crouzet, L.; Del’Homme, C.; Marquet, P.; Martin, J.C.; Pickering, G.; Ardid, D.; Eschalier, A.; et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment. Pharmacol. Ther. 2012, 35, 828–838. [Google Scholar] [CrossRef]
- Parajuli, S.P.; Choi, S.; Lee, J.; Kim, Y.D.; Park, C.G.; Kim, M.Y.; Kim, H.I.; Yeum, C.H.; Jun, J.Y. The Inhibitory Effects of Hydrogen Sulfide on Pacemaker Activity of Interstitial Cells of Cajal from Mouse Small Intestine. Korean J. Physiol. Pharmacol. 2010, 14, 83–89. [Google Scholar] [CrossRef][Green Version]
- Attene-Ramos, M.S.; Nava, G.M.; Muellner, M.G.; Wagner, E.D.; Plewa, M.J.; Gaskins, H.R. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ. Mol. Mutagen. 2010, 51, 304–314. [Google Scholar] [CrossRef]
- Motta, J.-P.; Flannigan, K.L.; Agbor, T.A.; Beatty, J.K.; Blackler, R.W.; Workentine, M.L.; Da Silva, G.J.; Wang, R.; Buret, A.G.; Wallace, J.L. Hydrogen Sulfide Protects from Colitis and Restores Intestinal Microbiota Biofilm and Mucus Production. Inflamm. Bowel Dis. 2015, 21, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Drucker, N.A.; Jensen, A.R.; Ferkowicz, M.; Markel, T.A. Hydrogen sulfide provides intestinal protection during a murine model of experimental necrotizing enterocolitis. J. Pediatr. Surg. 2018, 53, 1692–1698. [Google Scholar] [CrossRef]
- Elrod, J.W.; Calvert, J.W.; Morrison, J.; Doeller, J.E.; Kraus, D.W.; Tao, L.; Jiao, X.; Scalia, R.; Kiss, L.; Szabo, C.; et al. Hydrogen sulfide attenuates myocardial ischemia-reperfusion injury by preservation of mitochondrial function. Proc. Natl. Acad. Sci. USA 2007, 104, 15560–15565. [Google Scholar] [CrossRef] [PubMed]
- Head, K.A.; Jurenka, J.S. Inflammatory bowel disease part I: Ulcerative colitis—Pathophysiology and conventional and alternative treatment options. Altern. Med. Rev. 2003, 8, 247–283. [Google Scholar]
- Press, A.G.; Hauptmann, I.A.; Hauptmann, L.; Fuchs, B.; Fuchs, M.; Ewe, K.; Ramadori, G. Gastrointestinal pH profiles in patients with inflammatory bowel disease. Aliment. Pharmacol. Ther. 1998, 12, 673–678. [Google Scholar] [CrossRef]
- Thibault, R.; Blachier, F.; Darcy-Vrillon, B.; de Coppet, P.; Bourreille, A.; Segain, J.-P. Butyrate utilization by the colonic mucosa in inflammatory bowel diseases: A transport deficiency. Inflamm. Bowel Dis. 2010, 16, 684–695. [Google Scholar] [CrossRef] [PubMed]
- Hond, E.D.; Hiele, M.; Evenepoel, P.; Peeters, M.; Ghoos, Y.; Rutgeerts, P. In vivo butyrate metabolism and colonic permeability in extensive ulcerative colitis. Gastroenterology 1998, 115, 584–590. [Google Scholar] [CrossRef]
- Taglialegna, A. Fat, Desulfovibrio and cancer. Nat. Rev. Microbiol. 2024, 22, 388. [Google Scholar] [CrossRef]
- Módis, K.; Bos, E.M.; Calzia, E.; van Goor, H.; Coletta, C.; Papapetropoulos, A.; Hellmich, M.R.; Radermacher, P.; Bouillaud, F.; Szabo, C. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part II. Pathophysiological and therapeutic aspects. Br. J. Pharmacol. 2014, 171, 2123–2146. [Google Scholar] [CrossRef] [PubMed]
- Szabo, C.; Ransy, C.; Módis, K.; Andriamihaja, M.; Murghes, B.; Coletta, C.; Olah, G.; Yanagi, K.; Bouillaud, F. Regulation of mitochondrial bioenergetic function by hydrogen sulfide. Part I. Biochemical and physiological mechanisms. Br. J. Pharmacol. 2014, 171, 2099–2122. [Google Scholar] [CrossRef] [PubMed]
- Kolluru, G.K.; Shen, X.; Bir, S.C.; Kevil, C.G. Hydrogen sulfide chemical biology: Pathophysiological roles and detection. Nitric Oxide 2013, 35, 5–20. [Google Scholar] [CrossRef]
- Wang, R. Physiological implications of hydrogen sulfide: A whiff exploration that blossomed. Physiol. Rev. 2012, 92, 791–896. [Google Scholar] [CrossRef]
- Rivas-Domínguez, A.; Pastor, N.; Martínez-López, L.; Colón-Pérez, J.; Bermúdez, B.; Orta, M.L. The Role of DNA Damage Response in Dysbiosis-Induced Colorectal Cancer. Cells 2021, 10, 1934. [Google Scholar] [CrossRef]
- Dahmus, J.D.; Kotler, D.L.; Kastenberg, D.M.; Kistler, C.A. The gut microbiome and colorectal cancer: A review of bacterial pathogenesis. J. Gastrointest. Oncol. 2018, 9, 769–777. [Google Scholar] [CrossRef]
- Panza, E.; De Cicco, P.; Armogida, C.; Scognamiglio, G.; Gigantino, V.; Botti, G.; Germano, D.; Napolitano, M.; Papapetropoulos, A.; Bucci, M.; et al. Role of the cystathionine γ lyase/hydrogen sulfide pathway in human melanoma progression. Pigment. Cell Melanoma Res. 2015, 28, 61–72. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, L.; Schmid, J.; Ebert, M.; Soucek, P.; Kunicka, T.; Liska, V.; Bruha, J.; Neary, P.; Dezeeuw, N.; Tommasino, M.; et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1381–1390. [Google Scholar] [CrossRef]
- Zepeda-Rivera, M.; Minot, S.S.; Bouzek, H.; Wu, H.; Blanco-Miguez, A.; Manghi, P.; Jones, D.S.; LaCourse, K.D.; Wu, Y.; McMahon, E.F.; et al. A distinct Fusobacterium nucleatum clade dominates the colorectal cancer niche. Nature 2024, 628, 424–432. [Google Scholar] [CrossRef]
- Dukowicz, A.C.; Lacy, B.E.; Levine, G.M. Small intestinal bacterial overgrowth: A comprehensive review. Gastroenterol. Hepatol. 2007, 3, 112–122. [Google Scholar]
- Takakura, W.; Pimentel, M. Small Intestinal Bacterial Overgrowth and Irritable Bowel Syndrome—An Update. Front. Psychiatry 2020, 11, 664. [Google Scholar] [CrossRef]
- Rezaie, A.; Buresi, M.; Lembo, A.; Lin, H.; McCallum, R.; Rao, S.; Schmulson, M.; Valdovinos, M.; Zakko, S.; Pimentel, M. Hydrogen and Methane-Based Breath Testing in Gastrointestinal Disorders: The North American Consensus. Am. J. Gastroenterol. 2017, 112, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Ritz, N.L.; Lin, D.M.; Wilson, M.R.; Barton, L.L.; Lin, H.C. Sulfate-reducing bacteria slow intestinal transit in a bismuth-reversible fashion in mice. Neurogastroenterol. Motil. Off. J. Eur. Gastrointest. Motil. Soc. 2017, 29, e12907. [Google Scholar] [CrossRef]
- Banik, G.D.; De, A.; Som, S.; Jana, S.; Daschakraborty, S.B.; Chaudhuri, S.; Pradhan, M. Hydrogen sulphide in exhaled breath: A potential biomarker for small intestinal bacterial overgrowth in IBS. J. Breath Res. 2016, 10, 026010. [Google Scholar] [CrossRef]
- Lin, H.C. Small Intestinal Bacterial Overgrowth. JAMA 2004, 292, 852–858. [Google Scholar] [CrossRef]
- Chen, B.; Kim, J.J.-W.; Zhang, Y.; Du, L.; Dai, N. Prevalence and predictors of small intestinal bacterial overgrowth in irritable bowel syndrome: A systematic review and meta-analysis. J. Gastroenterol. 2018, 53, 807–818. [Google Scholar] [CrossRef]
- Coffman, C.N.; Varga, M.G.; Alcock, J.; Carrol-Portillo, A.; Singh, S.B.; Xue, X.; Lin, H.C. Norepinephrine induces growth of Desulfovibrio vulgaris in an iron dependent manner. Anaerobe 2022, 75, 102582. [Google Scholar] [CrossRef]
- Coffman, C.N.; Carroll-Portillo, A.; Alcock, J.; Singh, S.B.; Rumsey, K.; Braun, C.A.; Xue, B.; Lin, H.C. Magnesium Oxide Reduces Anxiety-like Behavior in Mice by Inhibiting Sulfate-Reducing Bacteria. Microorganisms 2024, 12, 1429. [Google Scholar] [CrossRef]
- Deplancke, B.; Hristova, K.R.; Oakley, H.A.; McCracken, V.J.; Aminov, R.; Mackie, R.I.; Gaskins, H.R. Molecular Ecological Analysis of the Succession and Diversity of Sulfate-Reducing Bacteria in the Mouse Gastrointestinal Tract. Appl. Environ. Microbiol. 2000, 66, 2166–2174. [Google Scholar] [PubMed]
- Bollinger, R.R.; Barbas, A.S.; Bush, E.L.; Lin, S.S.; Parker, W. Biofilms in the normal human large bowel: Fact rather than fiction. Gut 2007, 56, 1481–1482. [Google Scholar] [CrossRef][Green Version]
- Ijssennagger, N.; Belzer, C.; Hooiveld, G.J.; Dekker, J.; Van Mil, S.W.C.; Müller, M.; Kleerebezem, M.; Van Der Meer, R.; Klaenhammer, T.R. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc. Natl. Acad. Sci. USA 2015, 112, 10038–10043. [Google Scholar] [CrossRef]
- Rowan, F.E.; Docherty, N.G.; Coffey, J.C.; O’Connell, P.R. Sulphate-reducing bacteria and hydrogen sulphide in the aetiology of ulcerative colitis. Br. J. Surg. 2009, 96, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Blachier, F.; Beaumont, M.; Kim, E. Cysteine-derived hydrogen sulfide and gut health: A matter of endogenous or bacterial origin. Curr. Opin. Clin. Nutr. Metab. Care 2019, 22, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Wiest, R.; Garcia-Tsao, G. Bacterial translocation (BT) in cirrhosis. Hepatology 2005, 41, 422–433. [Google Scholar] [CrossRef]
- Runyon, B.A.; Squier, S.; Borzio, M. Translocation of gut bacteria in rats with cirrhosis to mesenteric lymph nodes partially explains the pathogenesis of spontaneous bacterial peritonitis. J. Hepatol. 1994, 21, 792–796. [Google Scholar] [CrossRef]
- Singh, S.; Coffman, C.; Varga, M.; Carroll-Portillo, A.; Braun, C.; Lin, H. Intestinal Alkaline Phosphatase Prevents Sulfate Reducing Bacteria-Induced Increased Tight Junction Permeability by Inhibiting Snail Pathway. Front. Cell. Infect. Microbiol. 2022, 12, 882498. [Google Scholar] [CrossRef]
- Singh, S.B.; Braun, C.A.; Carroll-Portillo, A.; Coffman, C.N.; Lin, H.C. Sulfate-Reducing Bacteria Induce Pro-Inflammatory TNF-alpha and iNOS via PI3K/Akt Pathway in a TLR 2-Dependent Manner. Microorganisms 2024, 12, 1833. [Google Scholar] [CrossRef]
- Birg, A.; Hu, S.; Lin, H.C. Reevaluating our understanding of lactulose breath tests by incorporating hydrogen sulfide measurements. JGH Open 2019, 3, 228–233. [Google Scholar] [CrossRef]
- Joshua, Z.G.; Britta, N.; Anna, E.W.; Ryan, B.; Allison, S. Hydrogen sulfide small intestinal bacterial overgrowth case registry. medRxiv 2023. [Google Scholar] [CrossRef]
- Singer-Englar, T.; Rezaie, A.; Gupta, K.; Pichetshote, N.; Sedighi, R.; Lin, E.; Chua, K.S.; Pimentel, M. Competitive hydrogen gas utilization by methane-and hydrogen sulfide-producing microorganisms and associated symptoms: Results of a novel 4-gas breath test machine. Gastroenterology 2018, 154, S47. [Google Scholar]
- Martin-Viñas, J.J.; Quigley, E.M.M. Immune response in irritable bowel syndrome: A systematic review of systemic and mucosal inflammatory mediators. J. Dig. Dis. 2016, 17, 572–581. [Google Scholar] [CrossRef]
- Mitselou, A.; Grammeniatis, V.; Varouktsi, A.; Papadatos, S.S.; Katsanos, K.; Galani, V. Proinflammatory cytokines in irritable bowel syndrome: A comparison with inflammatory bowel disease. Intest. Res. 2020, 18, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Rizos, E.; Pyleris, E.; Pimentel, M.; Triantafyllou, K.; Giamarellos-Bourboulis, E.J. Small Intestine Bacterial Overgrowth Can Form an Indigenous Proinflammatory Environment in the Duodenum: A Prospective Study. Microorganisms 2022, 10, 960. [Google Scholar] [CrossRef]
- Burns, G.L.; Talley, N.J.; Keely, S. Immune responses in the irritable bowel syndromes: Time to consider the small intestine. BMC Med. 2022, 20, 115. [Google Scholar] [CrossRef]
- Wallace, J.L.; Syer, S.; Denou, E.; De Palma, G.; Vong, L.; McKnight, W.; Jury, J.; Bolla, M.; Bercik, P.; Collins, S.M.; et al. Proton pump inhibitors exacerbate NSAID-induced small intestinal injury by inducing dysbiosis. Gastroenterology 2011, 141, 1314–1322.e5. [Google Scholar] [CrossRef]
- Del Piano, M.; Pagliarulo, M.; Tari, R.; Carmagnola, S.; Balzarini, M.; Lorenzini, P.; Pane, M. Correlation Between Chronic Treatment with Proton Pump Inhibitors and Bacterial Overgrowth in the Stomach: Any Possible Beneficial Role for Selected Lactobacilli? J. Clin. Gastroenterol. 2014, 48, S40–S46. [Google Scholar]
- Lo, W.K.; Chan, W.W. Proton pump inhibitor use and the risk of small intestinal bacterial overgrowth: A meta-analysis. Clin. Gastroenterol. Hepatol. 2013, 11, 483–490. [Google Scholar] [CrossRef]
- Abe, K.; Kimura, H. The Possible Role of Hydrogen sulfide as an Endogenous Neuromodulator. J. Neurosci. 1996, 16, 1066–1071. [Google Scholar] [PubMed]
- Kimura, H.; Shibuya, N.; Kimura, Y. Hydrogen Sulfide Is a Signaling Molecule and a Cytoprotectant. Antioxid. Redox Signal. 2012, 17, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H. Hydrogen sulfide induces cyclic AMP and modulates the NMDA receptor. Biochem. Biophys. Res. Commun. 2000, 267, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Nagai, Y.; Tsugane, M.; Oka, J.-I.; Kimura, H. Hydrogen sulfide induces calcium waves in astrocytes. FASEB J. 2004, 18, 557–559. [Google Scholar] [CrossRef]
- Kimura, Y.; Kimura, H. Hydrogen sulfide protects neurons from oxidative stress. FASEB J. 2004, 18, 1165–1167. [Google Scholar] [CrossRef]
- Chidambaram, S.B.; Rathipriya, A.G.; Mahalakshmi, A.M.; Sharma, S.; Hediyal, T.A.; Ray, B.; Sunanda, T.; Rungratanawanich, W.; Kashyap, R.S.; Qoronfleh, M.W.; et al. The Influence of Gut Dysbiosis in the Pathogenesis and Management of Ischemic Stroke. Cells 2022, 11, 1239. [Google Scholar] [CrossRef]
- Scott, K.P.; Jean-Michel, A.; Midtvedt, T.; van Hemert, S. Manipulating the gut microbiota to maintain health and treat disease. Microb. Ecol. Health Dis. 2015, 26, 25877. [Google Scholar] [CrossRef]
- Matsunami, M.; Kirishi, S.; Okui, T.; Kawabata, A. Hydrogen sulfide-induced colonic mucosal cytoprotection involves T-type calcium channel-dependent neuronal excitation in rats. J. Physiol. Pharmacol. 2012, 63, 61–68. [Google Scholar]
- Braniste, V.; Al-Asmakh, M.; Kowal, C.; Anuar, F.; Abbaspour, A.; Toth, M.; Korecka, A.; Bakocevic, N.; Ng, L.G.; Kundu, P.; et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 2014, 6, 263ra158. [Google Scholar] [CrossRef]
- Erny, D.; Hrabě de Angelis, A.L.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef]
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016, 167, 1469–1480.e1412. [Google Scholar] [CrossRef]
- Bollinger, J.L.; Collins, K.E.; Patel, R.; Wellman, C.L. Behavioral stress alters corticolimbic microglia in a sex- and brain region-specific manner. PLoS ONE 2017, 12, e0187631. [Google Scholar] [CrossRef] [PubMed]
- Murros, K.E. Hydrogen Sulfide Produced by Gut Bacteria May Induce Parkinson’s Disease. Cells 2022, 11, 978. [Google Scholar] [CrossRef]
- Hertel, J.; Harms, A.C.; Heinken, A.; Baldini, F.; Thinnes, C.C.; Glaab, E.; Vasco, D.A.; Pietzner, M.; Stewart, I.D.; Wareham, N.J.; et al. Integrated Analyses of Microbiome and Longitudinal Metabolome Data Reveal Microbial-Host Interactions on Sulfur Metabolism in Parkinson’s Disease. Cell Rep. 2019, 29, 1767–1777.e1768. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.F.; Lu, M.; Tiong, C.X.; Dawe, G.S.; Hu, G.; Bian, J.S. Neuroprotective effects of hydrogen sulfide on Parkinson’s disease rat models. Aging Cell 2010, 9, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Wallen, Z.D.; Appah, M.; Dean, M.N.; Sesler, C.L.; Factor, S.A.; Molho, E.; Zabetian, C.P.; Standaert, D.G.; Payami, H. Characterizing dysbiosis of gut microbiome in PD: Evidence for overabundance of opportunistic pathogens. npj Park. Dis. 2020, 6, 11. [Google Scholar] [CrossRef]
- Romano, S.; Savva, G.M.; Bedarf, J.R.; Charles, I.G.; Hildebrand, F.; Narbad, A. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. npj Park. Dis. 2021, 7, 27. [Google Scholar] [CrossRef]
- Thapa, M.; Kumari, A.; Chin, C.Y.; Choby, J.E.; Jin, F.; Bogati, B.; Chopyk, D.M.; Koduri, N.; Pahnke, A.; Elrod, E.J.; et al. Translocation of gut commensal bacteria to the brain. bioRxiv 2023. [Google Scholar] [CrossRef]
- Ryman, S.G.; Shaff, N.; Dodd, A.; Nitschke, S.; Wertz, C.; Julio, K.; Suarez Cedeno, G.; Deligtisch, A.; Erhardt, E.; Lin, H.; et al. Reduced and Delayed Cerebrovascular Reactivity in Patients with Parkinson’s Disease. Mov. Disord. 2023, 38, 1262–1272. [Google Scholar] [CrossRef]
- Rayhan, R.U.; Stevens, B.W.; Raksit, M.P.; Ripple, J.A.; Timbol, C.R.; Adewuyi, O.; VanMeter, J.W.; Baraniuk, J.N. Exercise challenge in Gulf War Illness reveals two subgroups with altered brain structure and function. PLoS ONE 2013, 8, e63903. [Google Scholar] [CrossRef]
- Hasegawa, S.; Goto, S.; Tsuji, H.; Okuno, T.; Asahara, T.; Nomoto, K.; Shibata, A.; Fujisawa, Y.; Minato, T.; Okamoto, A.; et al. Intestinal Dysbiosis and Lowered Serum Lipopolysaccharide-Binding Protein in Parkinson’s Disease. PLoS ONE 2015, 10, e0142164. [Google Scholar] [CrossRef]
- Wang, Q.; Luo, Y.; Ray Chaudhuri, K.; Reynolds, R.; Tan, E.-K.; Pettersson, S. The role of gut dysbiosis in Parkinson’s disease: Mechanistic insights and therapeutic options. Brain 2021, 144, 2571–2593. [Google Scholar] [CrossRef]
- Hoyles, L.; Snelling, T.; Umlai, U.-K.; Nicholson, J.K.; Carding, S.R.; Glen, R.C.; McArthur, S. Microbiome–host systems interactions: Protective effects of propionate upon the blood–brain barrier. Microbiome 2018, 6, 55. [Google Scholar] [CrossRef]
- Matt, S.M.; Allen, J.M.; Lawson, M.A.; Mailing, L.J.; Woods, J.A.; Johnson, R.W. Butyrate and Dietary Soluble Fiber Improve Neuroinflammation Associated with Aging in Mice. Front. Immunol. 2018, 9, 1832. [Google Scholar] [CrossRef]
- Touyz, R.M.; Camargo, L.L. Microglia, the Missing Link in the Brain-Gut-Hypertension Axis. Circ. Res. 2019, 124, 671–673. [Google Scholar] [CrossRef]
- Hu, L.-F.; Lu, M.; Wong, P.T.H.; Bian, J.-S. Hydrogen sulfide: Neurophysiology and neuropathology. Antioxid. Redox Signal. 2011, 15, 405–419. [Google Scholar]
- Schreier, S.M.; Muellner, M.K.; Steinkellner, H.; Hermann, M.; Esterbauer, H.; Exner, M.; Gmeiner, B.M.; Kapiotis, S.; Laggner, H. Hydrogen sulfide scavenges the cytotoxic lipid oxidation product 4-HNE. Neurotox. Res. 2010, 17, 249–256. [Google Scholar] [CrossRef]
- Xuan, A.; Long, D.; Li, J.; Ji, W.; Zhang, M.; Hong, L.; Liu, J. Hydrogen sulfide attenuates spatial memory impairment and hippocampal neuroinflammation in beta-amyloid rat model of Alzheimer’s disease. J. Neuroinflamm. 2012, 9, 202. [Google Scholar] [CrossRef]
- Das, T.K.; Ganesh, B.P. Interlink between the gut microbiota and inflammation in the context of oxidative stress in Alzheimer’s disease progression. Gut Microbes 2023, 15, 2206504. [Google Scholar] [CrossRef]
- Loffredo, L.; Ettorre, E.; Zicari, A.M.; Inghilleri, M.; Nocella, C.; Perri, L.; Spalice, A.; Fossati, C.; De Lucia, M.C.; Pigozzi, F.; et al. Oxidative Stress and Gut-Derived Lipopolysaccharides in Neurodegenerative Disease: Role of NOX2. Oxidative Med. Cell. Longev. 2020, 2020, 8630275. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.T.H.; Qu, K.; Chimon, G.N.; Seah, A.B.H.; Chang, H.M.; Wong, M.C.; Ng, Y.K.; Rumpel, H.; Halliwell, B.; Chen, C.P.L.H. High Plasma Cyst(e)ine Level May Indicate Poor Clinical Outcome in Patients with Acute Stroke: Possible Involvement of Hydrogen Sulfide. J. Neuropathol. Exp. Neurol. 2006, 65, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Qu, K.; Chen, C.P.L.H.; Halliwell, B.; Moore, P.K.; Wong, P.T.H. Hydrogen Sulfide Is a Mediator of Cerebral Ischemic Damage. Stroke 2006, 37, 889–893. [Google Scholar] [CrossRef]
- Warenycia, M.W.; Goodwin, L.R.; Benishin, C.G.; Reiffenstein, R.J.; Francom, D.M.; Taylor, J.D.; Dieken, F.P. Acute hydrogen sulfide poisoning: Demonstration of selective uptake of sulfide by the brainstem by measurement of brain sulfide levels. Biochem. Pharmacol. 1989, 38, 973–981. [Google Scholar] [CrossRef]
- Bui, T.V.A.; Hwangbo, H.; Lai, Y.; Hong, S.B.; Choi, Y.-J.; Park, H.-J.; Ban, K. The Gut-Heart Axis: Updated Review for The Roles of Microbiome in Cardiovascular Health. Korean Circ. J. 2023, 53, 499–518. [Google Scholar] [PubMed]
- Birg, A.; Lin, H.C.; Kanagy, N. Portal Venous Flow Is Increased by Jejunal but Not Colonic Hydrogen Sulfide in a Nitric Oxide-Dependent Fashion in Rats. Dig. Dis. Sci. 2020, 66, 2661–2668. [Google Scholar] [CrossRef]
- Dongó, E.; Kiss, L. The Potential Role of Hydrogen Sulfide in the Regulation of Cerebrovascular Tone. Biomolecules 2020, 10, 1685. [Google Scholar] [CrossRef]
- Whelton, P.K.; Carey, R.M.; Aronow, W.S.; Casey, D.E., Jr.; Collins, K.J.; Dennison Himmelfarb, C.; DePalma, S.M.; Gidding, S.; Jamerson, K.A.; Jones, D.W.; et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 2018, 71, e13–e115. [Google Scholar] [CrossRef]
- Dan, X.; Mushi, Z.; Baili, W.; Han, L.; Enqi, W.; Huanhu, Z.; Shuchun, L. Differential Analysis of Hypertension-Associated Intestinal Microbiota. Int. J. Med. Sci. 2019, 16, 872–881. [Google Scholar] [CrossRef]
- Finke, N.; Vandieken, V.; Jørgensen, B.B. Acetate, lactate, propionate, and isobutyrate as electron donors for iron and sulfate reduction in Arctic marine sediments, Svalbard. FEMS Microbiol. Ecol. 2007, 59, 10–22. [Google Scholar] [CrossRef]
- Moore, J.W.; Babidge, W.; Millard, S.; Roediger, W.E. Effect of sulphide on short chain acyl-CoA metabolism in rat colonocytes. Gut 1997, 41, 77. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Babidge, W.; Millard, S.; Roediger, W. Sulfides impair short chain fatty acid beta-oxidation at acyl-CoA dehydrogenase level in colonocytes: Implications for ulcerative colitis. Mol. Cell. Biochem. 1998, 181, 117–124. [Google Scholar] [CrossRef]
- Hsu, C.-N.; Tain, Y.-L. Preventing Developmental Origins of Cardiovascular Disease: Hydrogen Sulfide as a Potential Target? Antioxidants 2021, 10, 247. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Pattillo, C.B.; Pardue, S.; Bir, S.C.; Wang, R.; Kevil, C.G. Measurement of plasma hydrogen sulfide in vivo and in vitro. Free Radic. Biol. Med. 2011, 50, 1021–1031. [Google Scholar] [CrossRef]
- Gui, D.-D.; Luo, W.; Yan, B.-J.; Ren, Z.; Tang, Z.-H.; Liu, L.-S.; Zhang, J.-F.; Jiang, Z.-S. Effects of gut microbiota on atherosclerosis through hydrogen sulfide. Eur. J. Pharmacol. 2021, 896, 173916. [Google Scholar] [CrossRef]
- Kruth, H.S. Macrophage foam cells and atherosclerosis. Front. Biosci. 2001, 6, D429–D455. [Google Scholar] [CrossRef] [PubMed]
- Jie, Z.; Xia, H.; Zhong, S.-L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef]
- Gribble, F.M.; Reimann, F. Signalling in the gut endocrine axis. Physiol. Behav. 2017, 176, 183–188. [Google Scholar] [CrossRef]
- Yang, S.; Cao, J.; Sun, C.; Yuan, L. The Regulation Role of the Gut-Islets Axis in Diabetes. Diabetes Metab. Syndr. Obes. 2024, 17, 1415–1423. [Google Scholar] [CrossRef]
- Gutierrez-Aguilar, R.; Woods, S.C. Nutrition and L and K-enteroendocrine cells. Curr. Opin. Endocrinol. Diabetes Obes. 2011, 18, 35–41. [Google Scholar] [CrossRef]
- Grasset, E.; Puel, A.; Charpentier, J.; Collet, X.; Christensen, J.E.; Tercé, F.; Burcelin, R. A Specific Gut Microbiota Dysbiosis of Type 2 Diabetic Mice Induces GLP-1 Resistance through an Enteric NO-Dependent and Gut-Brain Axis Mechanism. Cell Metab. 2017, 25, 1075–1090.e1075. [Google Scholar] [CrossRef]
- Yamane, S.; Inagaki, N. Regulation of glucagon-like peptide-1 sensitivity by gut microbiota dysbiosis. J. Diabetes Investig. 2018, 9, 262–264. [Google Scholar] [CrossRef] [PubMed]
- Pichette, J.; Fynn-Sackey, N.; Gagnon, J. Hydrogen Sulfide and Sulfate Prebiotic Stimulates the Secretion of GLP-1 and Improves Glycemia in Male Mice. Endocrinology 2017, 158, 3416–3425. [Google Scholar] [CrossRef]
- Qi, Q.; Zhang, H.; Jin, Z.; Wang, C.; Xia, M.; Chen, B.; Lv, B.; Peres Diaz, L.; Li, X.; Feng, R.; et al. Hydrogen sulfide produced by the gut microbiota impairs host metabolism via reducing GLP-1 levels in male mice. Nat. Metab. 2024, 6, 1601–1615. [Google Scholar] [CrossRef] [PubMed]
- Bełtowski, J.; Wójcicka, G.; Jamroz-Wiśniewska, A. Hydrogen sulfide in the regulation of insulin secretion and insulin sensitivity: Implications for the pathogenesis and treatment of diabetes mellitus. Biochem. Pharmacol. 2018, 149, 60–76. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Yang, W.; Jia, X.; Yang, G.; Duridanova, D.; Cao, K.; Wang, R. Pancreatic islet overproduction of H2S and suppressed insulin release in Zucker diabetic rats. Lab. Investig. 2009, 89, 59–67. [Google Scholar] [CrossRef]
- Xue, R.; Hao, D.D.; Sun, J.P.; Li, W.W.; Zhao, M.M.; Li, X.H.; Chen, Y.; Zhu, J.H.; Ding, Y.J.; Liu, J.; et al. Hydrogen Sulfide Treatment Promotes Glucose Uptake by Increasing Insulin Receptor Sensitivity and Ameliorates Kidney Lesions in Type 2 Diabetes. Antioxid. Redox Signal. 2013, 19, 5–23. [Google Scholar] [CrossRef]
- Okamoto, M.; Yamaoka, M.; Takei, M.; Ando, T.; Taniguchi, S.; Ishii, I.; Tohya, K.; Ishizaki, T.; Niki, I.; Kimura, T. Endogenous hydrogen sulfide protects pancreatic beta-cells from a high-fat diet-induced glucotoxicity and prevents the development of type 2 diabetes. Biochem. Biophys. Res. Commun. 2013, 442, 227–233. [Google Scholar] [CrossRef]
- Carter, R.N.; Gibbins, M.T.G.; Barrios-Llerena, M.E.; Wilkie, S.E.; Freddolino, P.L.; Libiad, M.; Vitvitsky, V.; Emerson, B.; Le Bihan, T.; Brice, M.; et al. The hepatic compensatory response to elevated systemic sulfide promotes diabetes. Cell Rep. 2021, 37, 109958. [Google Scholar] [CrossRef]
- Perez, N.B.; Dorsen, C.; Squires, A. Dysbiosis of the Gut Microbiome: A Concept Analysis. J. Holist. Nurs. 2020, 38, 223–232. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Birg, A.; Lin, H.C. The Role of Bacteria-Derived Hydrogen Sulfide in Multiple Axes of Disease. Int. J. Mol. Sci. 2025, 26, 3340. https://doi.org/10.3390/ijms26073340
Birg A, Lin HC. The Role of Bacteria-Derived Hydrogen Sulfide in Multiple Axes of Disease. International Journal of Molecular Sciences. 2025; 26(7):3340. https://doi.org/10.3390/ijms26073340
Chicago/Turabian StyleBirg, Aleksandr, and Henry C. Lin. 2025. "The Role of Bacteria-Derived Hydrogen Sulfide in Multiple Axes of Disease" International Journal of Molecular Sciences 26, no. 7: 3340. https://doi.org/10.3390/ijms26073340
APA StyleBirg, A., & Lin, H. C. (2025). The Role of Bacteria-Derived Hydrogen Sulfide in Multiple Axes of Disease. International Journal of Molecular Sciences, 26(7), 3340. https://doi.org/10.3390/ijms26073340





