Genome-Wide Analysis and Expression Profiles of AhCOLs Family in Peanut (Arachis hypogaea L.)
Abstract
:1. Introduction
2. Results
2.1. Physical and Chemical Properties and Chromosomal Localization of the AhCOL Gene Family
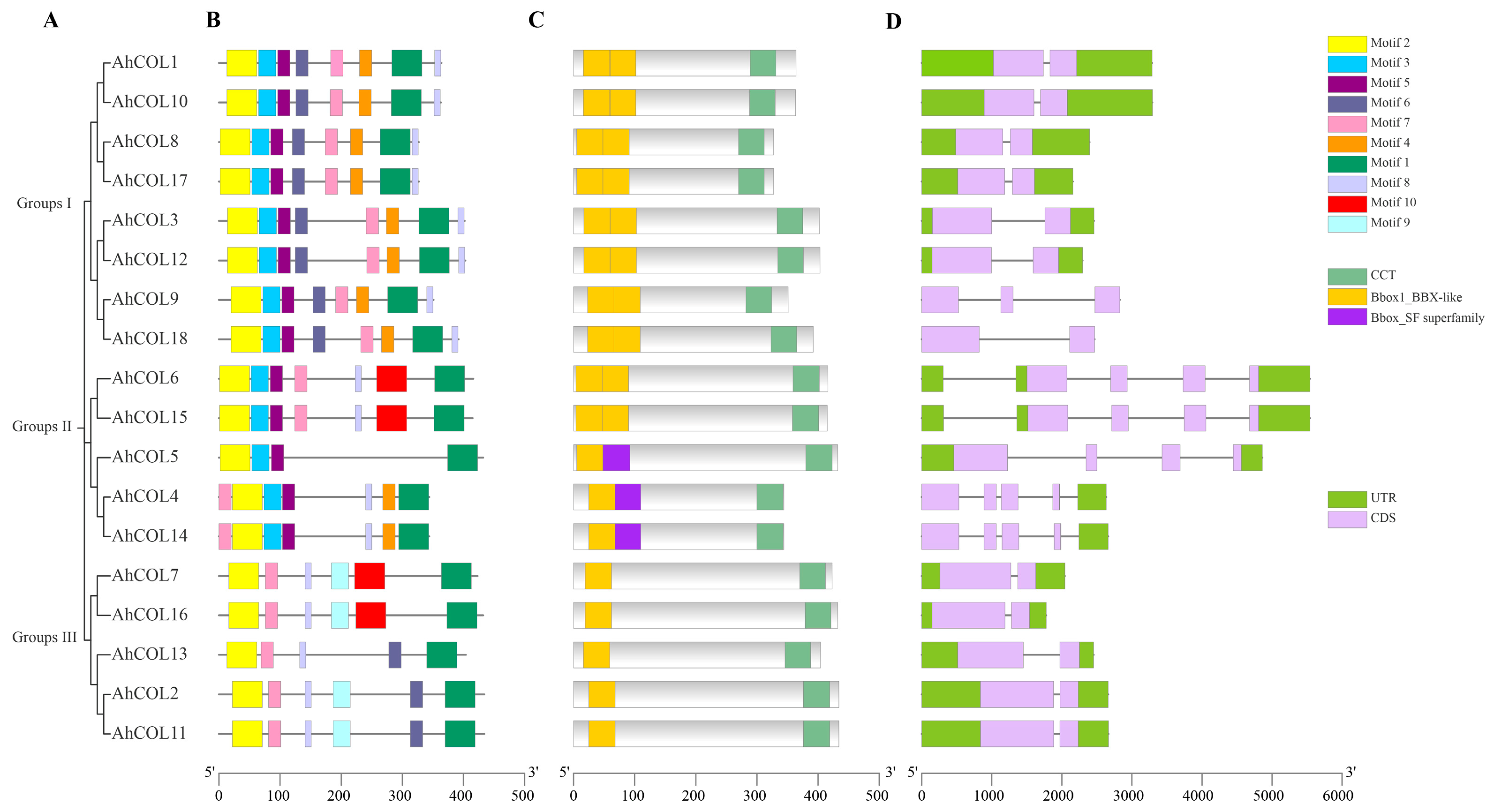
2.2. Phylogenetic Analysis of AhCOL Gene Family in Peanut
2.3. Genetic Structure Analysis of AhCOL Genes in Peanut
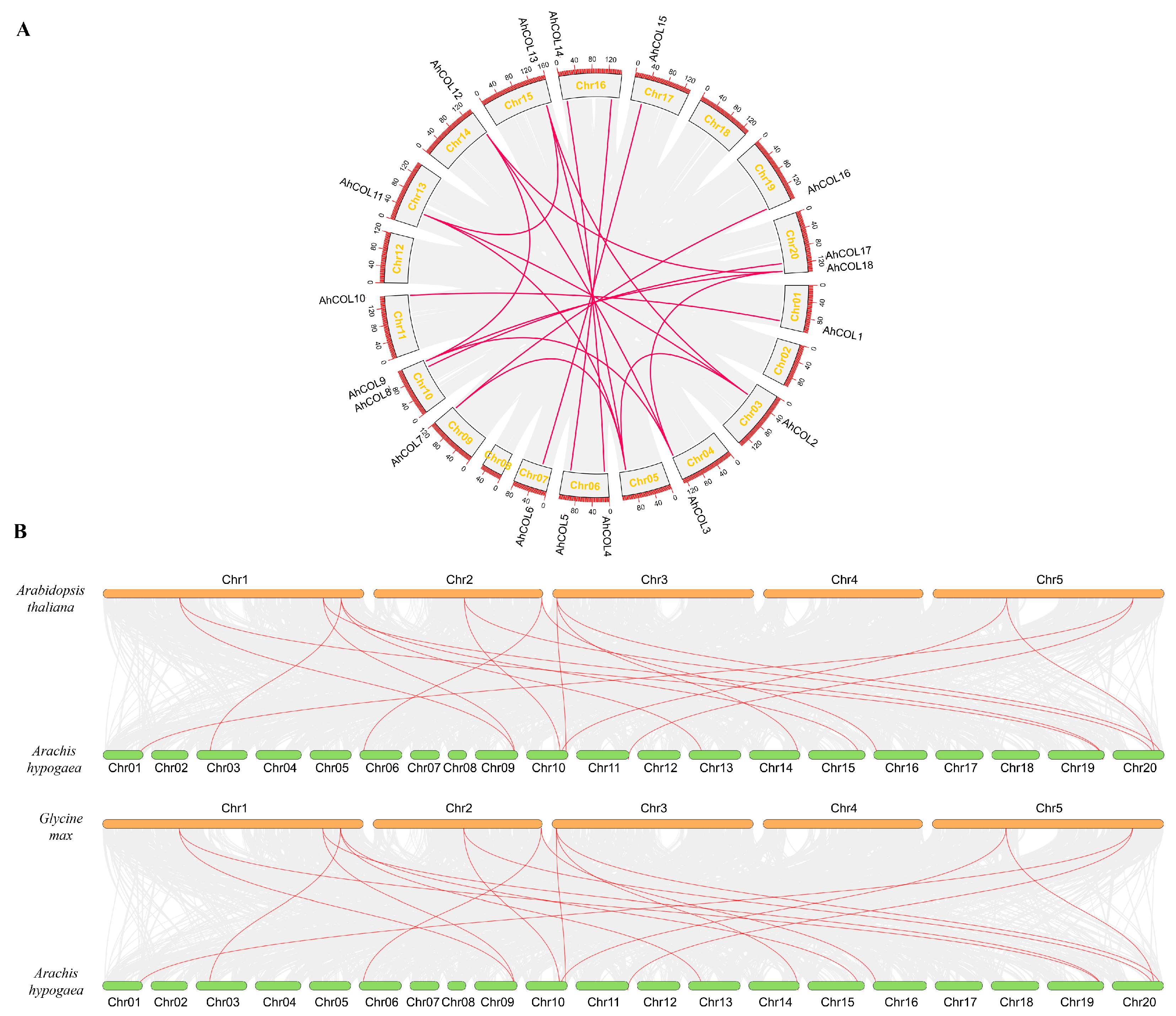
2.4. Collinearity Analysis of AhCOL Gene Family in Peanut
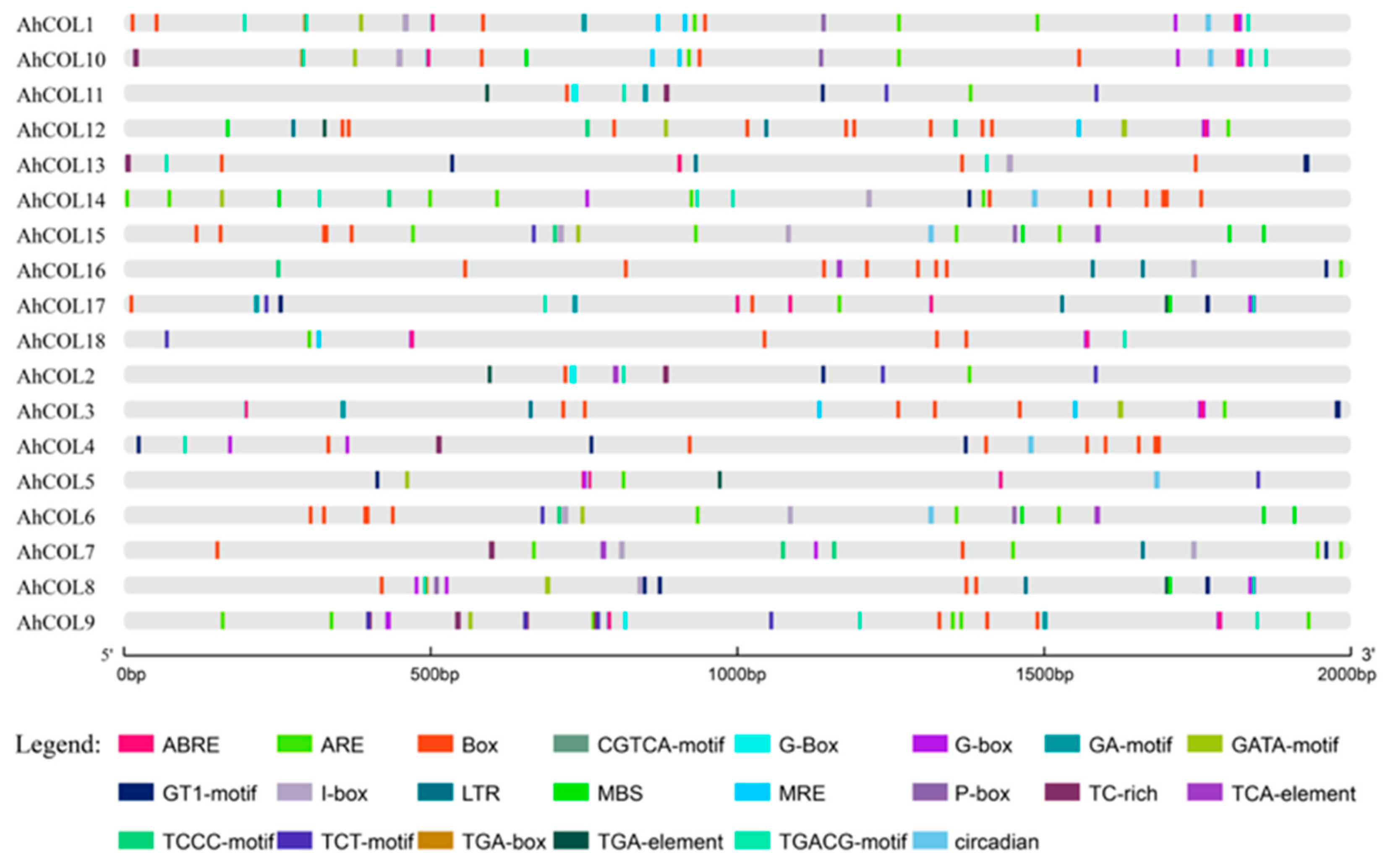
2.5. Cis-Element Analysis of AhCOL Gene Promoters in Peanut
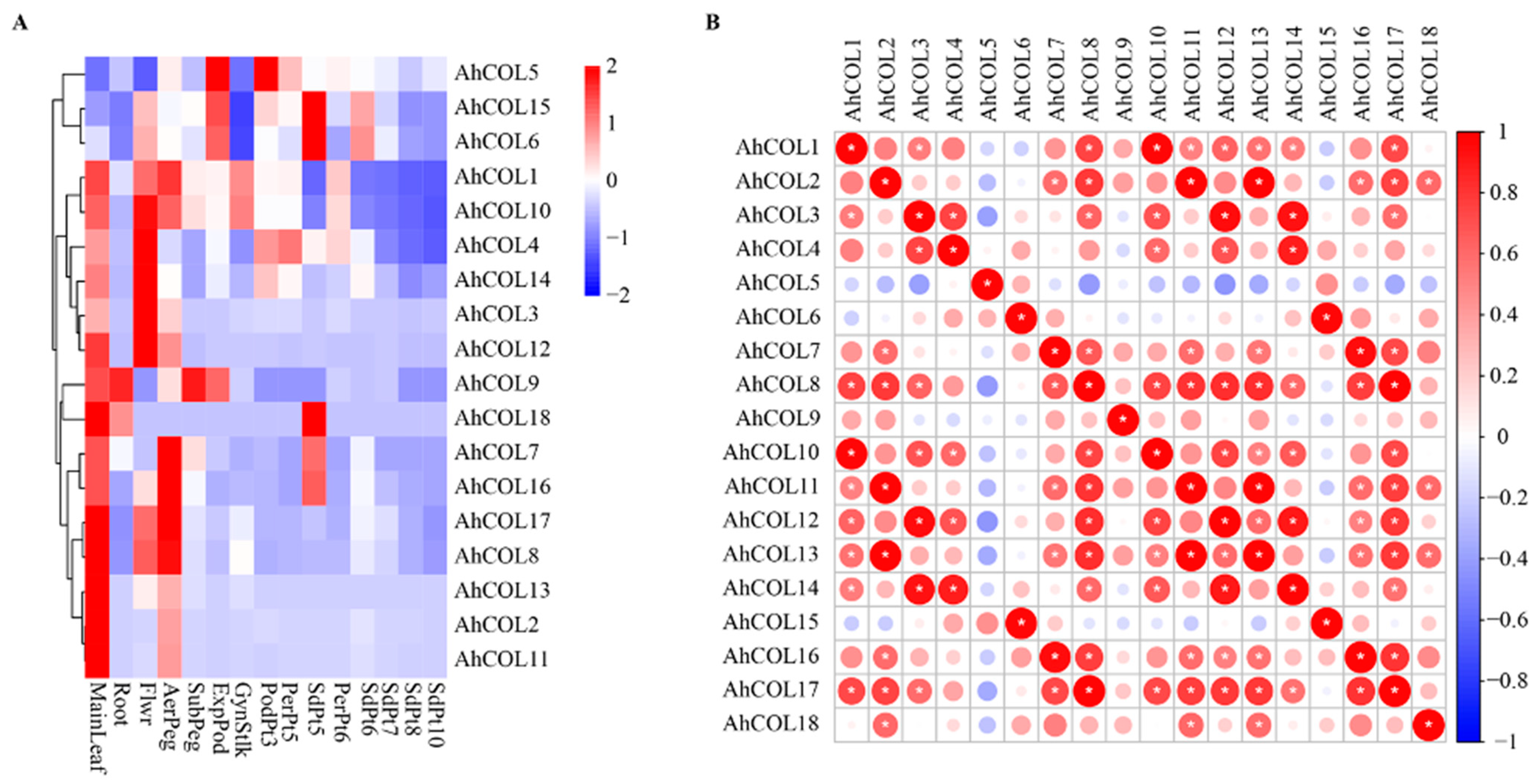
2.6. Expression of AhCOL Genes in Different Tissues of Peanut
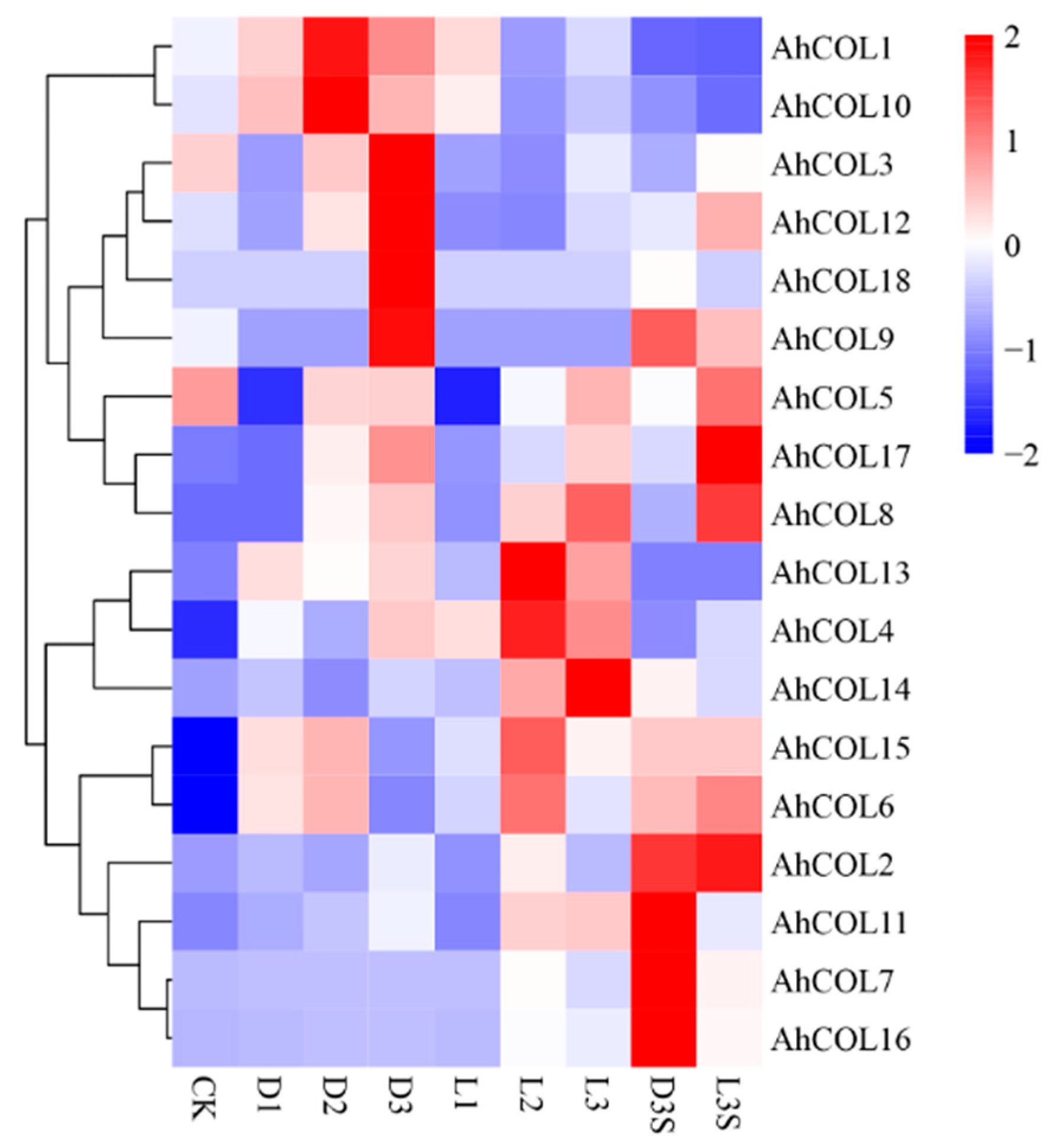
2.7. Expression of Peanut AhCOL Gene Family Members Under Light and Mechanical Pressure
2.8. Subcellular Localization of AhCOL Proteins in Peanut
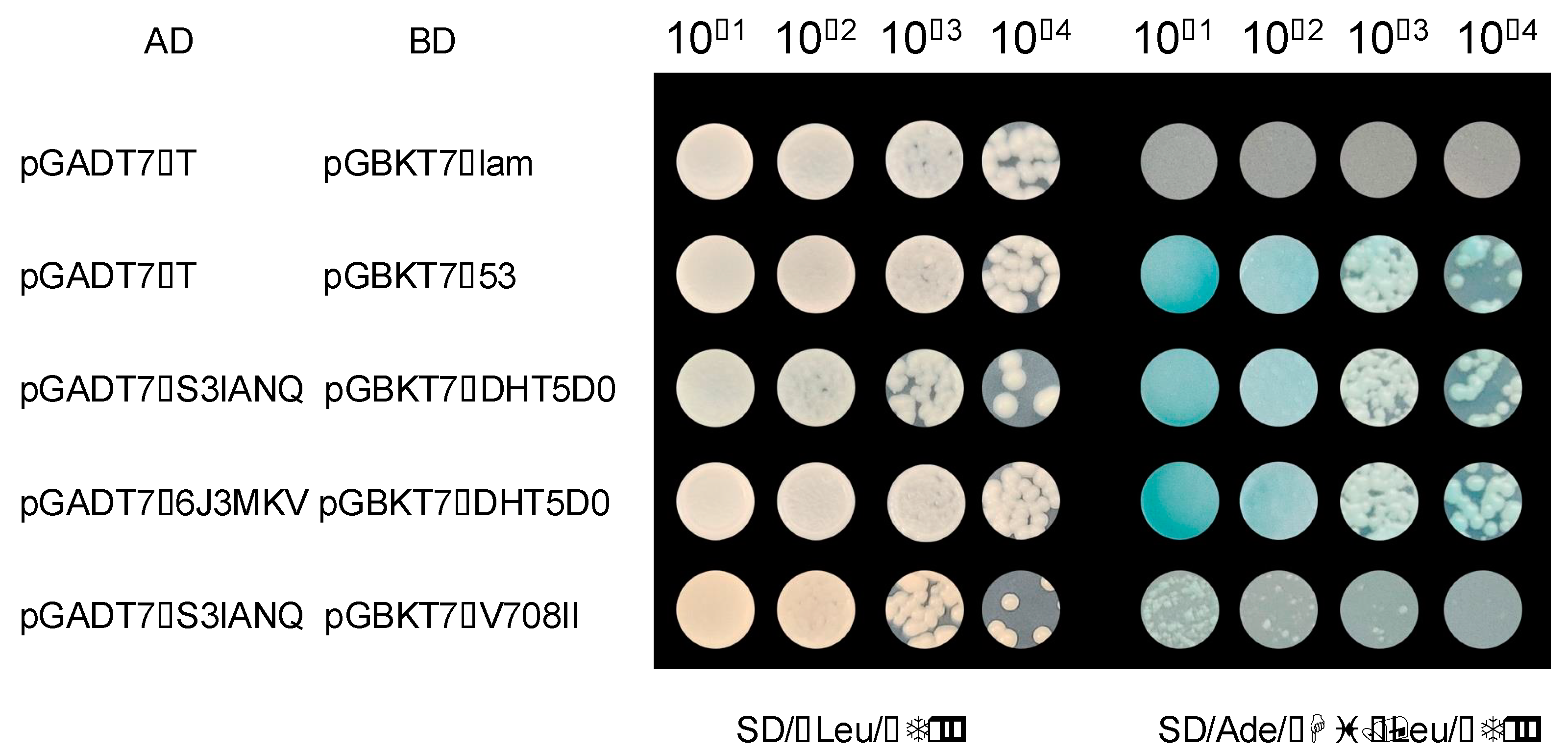
2.9. Peanut AhCOL Protein Interaction
3. Discussion
4. Materials and Methods
4.1. Experimental Materials
4.2. Identification of AhCOL Gene Family Members in Arachis hypogaea
4.3. Molecular Characterization and Chromosomal Location of AhCOL Genes
4.4. Phylogenetic and Genetic Structure Analysis of AhCOL Genes
4.5. Predictive Analysis of Promoter Elements of AhCOL Gene in Peanut
4.6. RNA-Seq and Bioinformatics Analysis
4.7. Subcellular Localization
4.8. Yeast Two-Hybrid Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, H.; Luo, L.; Li, Y.; Li, H.; Zhang, X.; Zhang, K.; Zhu, S.; Li, X.; Li, Y.; Wan, Y.; et al. Fine mapping of qAHPS07 and functional studies of AhRUVBL2 controlling pod size in peanut (Arachis hypogaea L.). Plant Biotechnol. J. 2023, 21, 1785–1798. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Simpson, C.; Singh, A.K.; Simpson, C.E. Biosystematic and Genetic Resources. In The Groundnut Crop, A Scientific Basis for Improvement; Smartt, J., Ed.; Chapman and Hall: London, UK, 1994; pp. 96–138. [Google Scholar]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Kaba, J.S.; Kumaga, F.K.; Ofori, K. Effect of flower production and time of flowering on pod yield of peanut (Arachis hypogaea L.) genotypes. IOSR J. Agric. Vet. Sci. 2014, 7, 44–49. [Google Scholar] [CrossRef]
- Nigam, S.N.; Dwivedi, S.L.; Ramraj, V.M.; Chandra, S. Combining Ability of Response to Photoperiod in Peanut. Crop Sci. 1997, 37, 1159–1162. [Google Scholar] [CrossRef]
- Chopra, R.; Simpson, C.E.; Hillhouse, A.; Payton, P.; Sharma, J.; Burow, M.D. SNP genotyping reveals major QTLs for plant architectural traits between A-genome peanut wild species. Mol. Genet. Genom. 2018, 293, 1477–1491. [Google Scholar] [CrossRef]
- Li, W.; Liao, Y.; Huang, L.; Lu, Q.; Li, S.; Chen, X.; Jin, J.; Wang, R. Genome-wide associate analysis of flowering traits and identification of candidate genes in peanut. Acta Agron. Sin. 2024, 1–20. [Google Scholar]
- Zhong, C.; Li, Z.; Cheng, Y.; Zhang, H.; Liu, Y.; Wang, X.; Jiang, C.; Zhao, X.; Zhao, S.; Wang, J.; et al. Comparative Genomic and Expression Analysis Insight into Evolutionary Characteristics of PEBP Genes in Cultivated Peanuts and Their Roles in Floral Induction. Int. J. Mol. Sci. 2022, 23, 12429. [Google Scholar] [CrossRef]
- Liu, Y.; Luo, C.; Guo, Y.; Liang, R.; Yu, H.; Chen, S.; Mo, X.; Yang, X.; He, X. Isolation and Functional Characterization of Two CONSTANS-like 16 (MiCOL16) Genes from Mango. Int. J. Mol. Sci. 2022, 23, 3075. [Google Scholar] [CrossRef]
- Wickland, D.P.; Hanzawa, Y. The Flowering Locus T/Terminal Flower 1 Gene Family: Functional Evolution and Molecular Mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef]
- Okamuro, J.K.; Szeto, W.; Lotys-Prass, C.; Jofuku, K.D. Photo and hormonal control of meristem identity in the Arabidopsis flower mutants apetala2 and apetala1. Plant Cell 1997, 9, 37–47. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Blazquez, M.A.; Green, R.; Nilsson, O.; Sussman, M.R.; Weigel, D. Gibberellins promote flowering of arabidopsis by activating the LEAFY promoter. Plant Cell 1998, 10, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Ma, J.; Han, Y.; Chen, X.; Fu, Y.-F. Cloning and Expression Analysis of the Soybean CO-Like Gene GmCOL9. Plant Mol. Biol. Report. 2010, 29, 352–359. [Google Scholar] [CrossRef]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.H. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef]
- Robson, F.; Costa, M.M.; Hepworth, S.R.; Vizir, I.; Pineiro, M.; Reeves, P.H.; Putterill, J.; Coupland, G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001, 28, 619–631. [Google Scholar] [CrossRef]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2484. [Google Scholar] [CrossRef]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A Developmental Transcriptome Map for Allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef]
- Lagercrantz, U.; Axelsson, T. Rapid evolution of the family of CONSTANS LIKE genes in plants. Mol. Biol. Evol. 2000, 17, 1499–1507. [Google Scholar] [CrossRef]
- Griffiths, S.; Dunford, R.P.; Coupland, G.; Laurie, D.A. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 2003, 131, 1855–1867. [Google Scholar] [CrossRef]
- Khatun, K.; Debnath, S.; Robin, A.H.K.; Wai, A.H.; Nath, U.K.; Lee, D.J.; Kim, C.K.; Chung, M.Y. Genome-wide identification, genomic organization, and expression profiling of the CONSTANS-like (COL) gene family in petunia under multiple stresses. BMC Genom. 2021, 22, 727. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, G.; Shao, C.; Gao, K.; Ma, N.; Rao, J.; Miao, X. Genome-wide exploration of the CONSTANS-like (COL) gene family and its potential role in regulating plant flowering time in foxtail millet (Setaria italica). Sci. Rep. 2024, 14, 24518. [Google Scholar] [CrossRef]
- Zhao, Y.; Xu, J.; Xu, X.; Liu, H.; Chang, Q.; Xu, L.; Liang, Z. Genome-Wide Identification of CONSTANS-like (COL) Gene Family and the Potential Function of ApCOL08 Under Salt Stress in Andrographis paniculata. Int. J. Mol. Sci. 2025, 26, 724. [Google Scholar] [CrossRef]
- Ledger, S.; Strayer, C.; Ashton, F.; Kay, S.A.; Putterill, J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 2001, 26, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Graeff, M.; Straub, D.; Eguen, T.; Dolde, U.; Rodrigues, V.; Brandt, R.; Wenkel, S. MicroProtein-Mediated Recruitment of CONSTANS into a TOPLESS Trimeric Complex Represses Flowering in Arabidopsis. PLoS Genet. 2016, 12, e1005959. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Z.; Li, H.; Zhao, X.; Liu, X.; Ortiz, M.; Lin, C.; Liu, B. CONSTANS-LIKE 7 regulates branching and shade avoidance response in Arabidopsis. J. Exp. Bot. 2013, 64, 1017–1024. [Google Scholar] [CrossRef]
- Takase, T.; Kakikubo, Y.; Nakasone, A.; Nishiyama, Y.; Yasuhara, M.; Tokioka-Ono, Y.; Kiyosue, T. Characterization and transgenic study of CONSTANS-LIKE8 (COL8) gene in Arabidopsis thaliana: Expression of 35S:COL8 delays flowering under long-day conditions. Plant Biotechnol. 2011, 28, 439–446. [Google Scholar] [CrossRef]
- Tsuji, H.; Taoka, K.; Shimamoto, K. Regulation of flowering in rice: Two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 2011, 14, 45–52. [Google Scholar] [CrossRef]
- Wu, W.; Zheng, X.M.; Chen, D.; Zhang, Y.; Ma, W.; Zhang, H.; Sun, L.; Yang, Z.; Zhao, C.; Zhan, X.; et al. OsCOL16, encoding a CONSTANS-like protein, represses flowering by up-regulating Ghd7 expression in rice. Plant Sci. 2017, 260, 60–69. [Google Scholar] [CrossRef]
- Gonzalez-Schain, N.D.; Diaz-Mendoza, M.; Zurczak, M.; Suarez-Lopez, P. Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J. 2012, 70, 678–690. [Google Scholar] [CrossRef]
- Jeong, D.-H.; Sung, S.-K.; An, G. Molecular cloning and characterization of constans-like cDNA clones of the fuji apple. J. Plant Biol. 1999, 42, 23–31. [Google Scholar] [CrossRef]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Serrano, G.; Herrera-Palau, R.; Romero, J.M.; Serrano, A.; Coupland, G.; Valverde, F. Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr. Biol. 2009, 19, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yang, R.; Shen, S.; Zhao, J. Molecular mechanism of flowering time regulation in Brassica rapa: Similarities and differences with Arabidopsis. Hortic. Plant J. 2024, 10, 615–628. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Large-scale plant protein subcellular location prediction. J. Cell Biochem. 2007, 100, 665–678. [Google Scholar] [CrossRef]
- Chou, K.C. Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 2005, 21, 10–19. [Google Scholar] [CrossRef]
- Shen, H.B.; Chou, K.C. Ensemble classifier for protein fold pattern recognition. Bioinformatics 2006, 22, 1717–1722. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Cui, Y.; Bian, J.; Lv, Y.; Li, J.; Deng, X.W.; Liu, X. Analysis of the Transcriptional Dynamics of Regulatory Genes During Peanut Pod Development Caused by Darkness and Mechanical Stress. Front. Plant Sci. 2022, 13, 904162. [Google Scholar] [CrossRef]
- Pattee, H.E.; Johns, E.B.; Singleton, J.A.; Sanders, T.H. Composition changes of peanut fruit parts during maturation. Peanut Sci. 1974, 1, 57–62. [Google Scholar] [CrossRef]
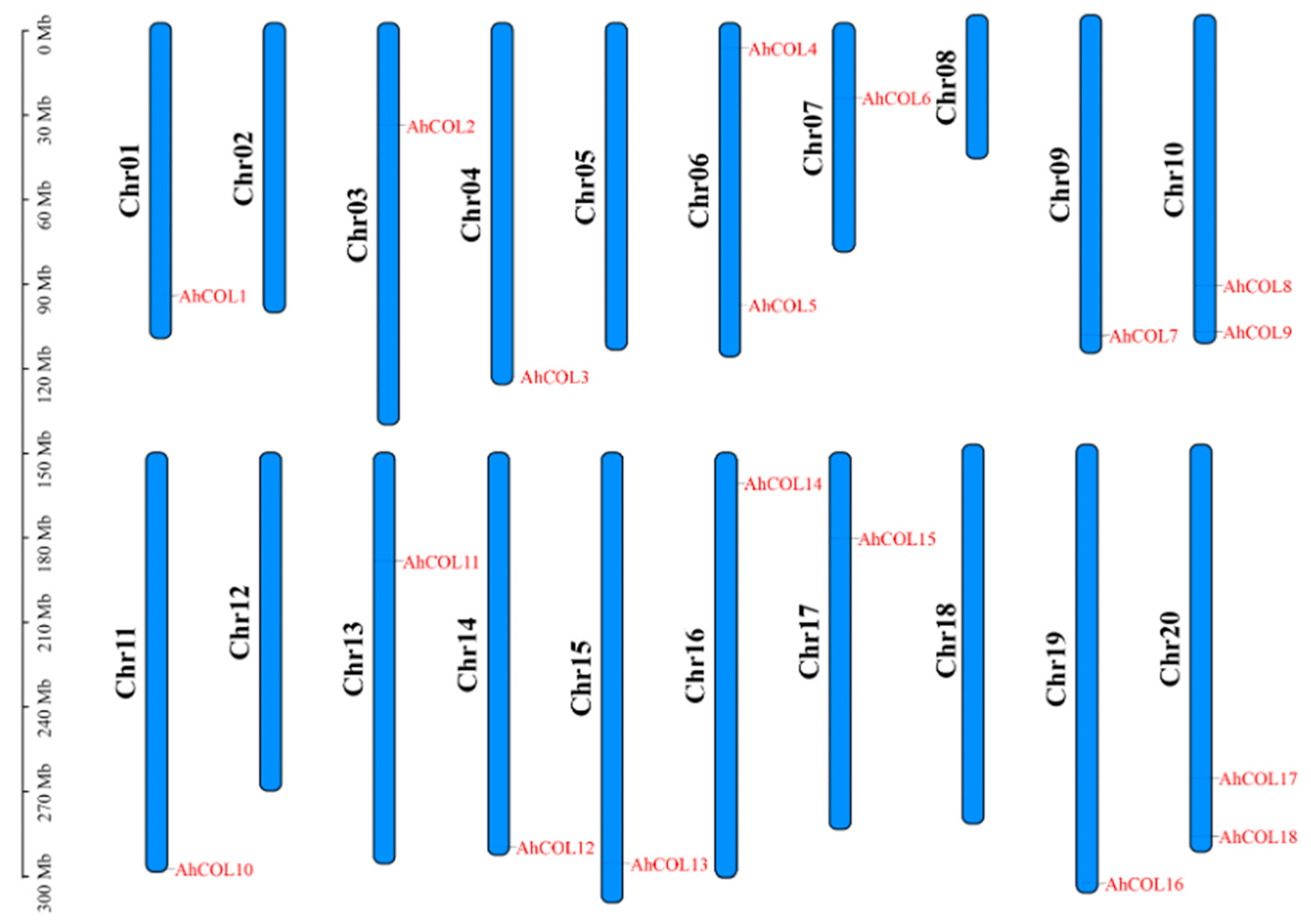


| AhCOL Member | Gene ID (arahy.Tifrunner. gnm2.ann1.) | Chromosome Location | Strand | Number of Amino Acids | Molecular Weight/Da | PI | Subcellular Location |
|---|---|---|---|---|---|---|---|
| AhCOL1 | arahy.6J3MKV.1 | Chr01: 96,565,988_96,569,275 | + | 364 | 39,606.40 | 6.59 | Nucleus. |
| AhCOL2 | arahy.8MV22J.1 | Chr03: 36,453,808_36,456,469 | + | 434 | 48,577.11 | 5.42 | Nucleus. |
| AhCOL3 | arahy.S3IANQ.1 | Chr04: 125,410,273_125,412,732 | − | 402 | 43,764.84 | 5.41 | Nucleus. |
| AhCOL4 | arahy.7A5VUD.1 | Chr06: 8,891,338_8,893,972 | + | 344 | 38,530.81 | 6.17 | Nucleus. |
| AhCOL5 | arahy.P8XRWE.1 | Chr06: 100,018,431_100,023,293 | + | 432 | 47,768.07 | 5.94 | Nucleus. |
| AhCOL6 | arahy.NK4UTA.1 | Chr07: 26,461,465_26,467,006 | + | 416 | 45,150.21 | 5.14 | Nucleus. |
| AhCOL7 | arahy.WVH2H8.1 | Chr09: 113,590,832_113,592,875 | − | 423 | 47,372.99 | 6.24 | Nucleus. |
| AhCOL8 | arahy.ZS9VWH.1 | Chr10: 96,097,643_96,100,039 | − | 327 | 35,690.76 | 6.51 | Nucleus. |
| AhCOL9 | arahy.LV381L.1 | Chr10: 112,354,889_112,357,717 | + | 351 | 38,906.28 | 5.34 | Nucleus. |
| AhCOL10 | arahy.GMWG2V.1 | Chr11: 147,626,175_147,629,467 | − | 363 | 39,479.34 | 6.96 | Nucleus. |
| AhCOL11 | arahy.ZUL46H.1 | Chr13: 38,639,582_38,642,247 | + | 434 | 48,523.97 | 5.48 | Nucleus. |
| AhCOL12 | arahy.669CET.1 | Chr14: 139,785,864_139,788,159 | − | 403 | 44,104.19 | 5.51 | Nucleus. |
| AhCOL13 | arahy.WEZ503.1 | Chr15: 145,833,862_145,836,316 | − | 404 | 46,144.44 | 5.48 | Nucleus. |
| AhCOL14 | arahy.N2A79V.1 | Chr16: 10,817,157_10,819,819 | − | 344 | 38,510.83 | 6.17 | Nucleus. |
| AhCOL15 | arahy.TFJ3CW.1 | Chr17: 30,349,549_30,355,089 | − | 415 | 45,053.10 | 5.14 | Nucleus. |
| AhCOL16 | arahy.AY8I6Y.1 | Chr19: 155,464,256_155,466,032 | + | 432 | 48,501.95 | 6.04 | Nucleus. |
| AhCOL17 | arahy.B9QMIW.1 | Chr20: 118,304,787_118,306,945 | − | 327 | 35,721.79 | 6.51 | Nucleus. |
| AhCOL18 | arahy.E88P3Q.1 | Chr20: 138,909,248_138,911,716 | + | 392 | 43,383.72 | 4.82 | Nucleus. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Liu, X.; Liu, C.; Liu, X. Genome-Wide Analysis and Expression Profiles of AhCOLs Family in Peanut (Arachis hypogaea L.). Int. J. Mol. Sci. 2025, 26, 3404. https://doi.org/10.3390/ijms26073404
Wang W, Liu X, Liu C, Liu X. Genome-Wide Analysis and Expression Profiles of AhCOLs Family in Peanut (Arachis hypogaea L.). International Journal of Molecular Sciences. 2025; 26(7):3404. https://doi.org/10.3390/ijms26073404
Chicago/Turabian StyleWang, Wei, Xiaoyu Liu, Che Liu, and Xiaoqin Liu. 2025. "Genome-Wide Analysis and Expression Profiles of AhCOLs Family in Peanut (Arachis hypogaea L.)" International Journal of Molecular Sciences 26, no. 7: 3404. https://doi.org/10.3390/ijms26073404
APA StyleWang, W., Liu, X., Liu, C., & Liu, X. (2025). Genome-Wide Analysis and Expression Profiles of AhCOLs Family in Peanut (Arachis hypogaea L.). International Journal of Molecular Sciences, 26(7), 3404. https://doi.org/10.3390/ijms26073404





