Monoclonality and Low Genetic Diversity in Vanilla shenzhenica: Highlighting Urgent Need for Genetic Preservation of China’s Only Endangered Vanilla
Abstract
:1. Introduction
2. Results
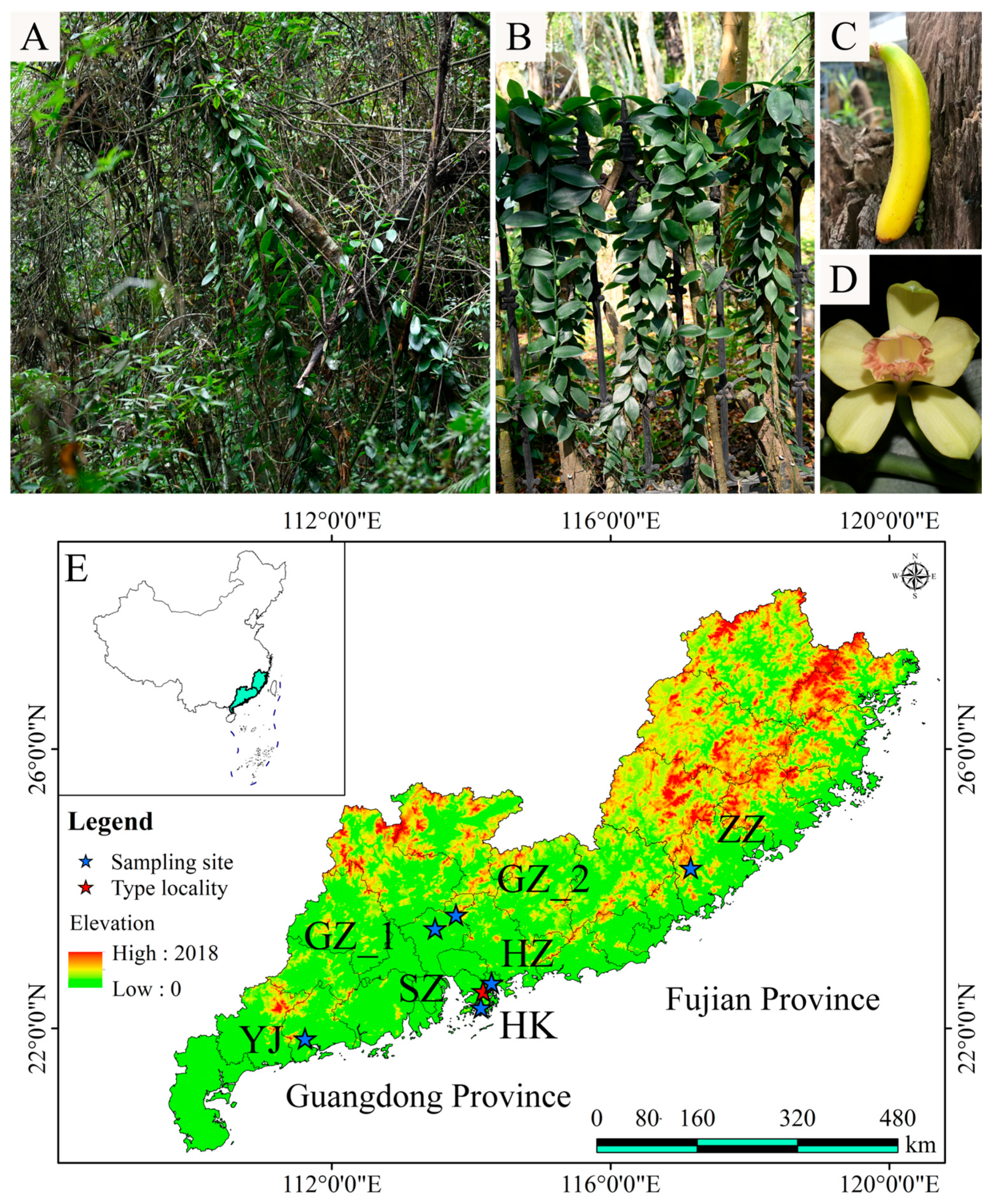
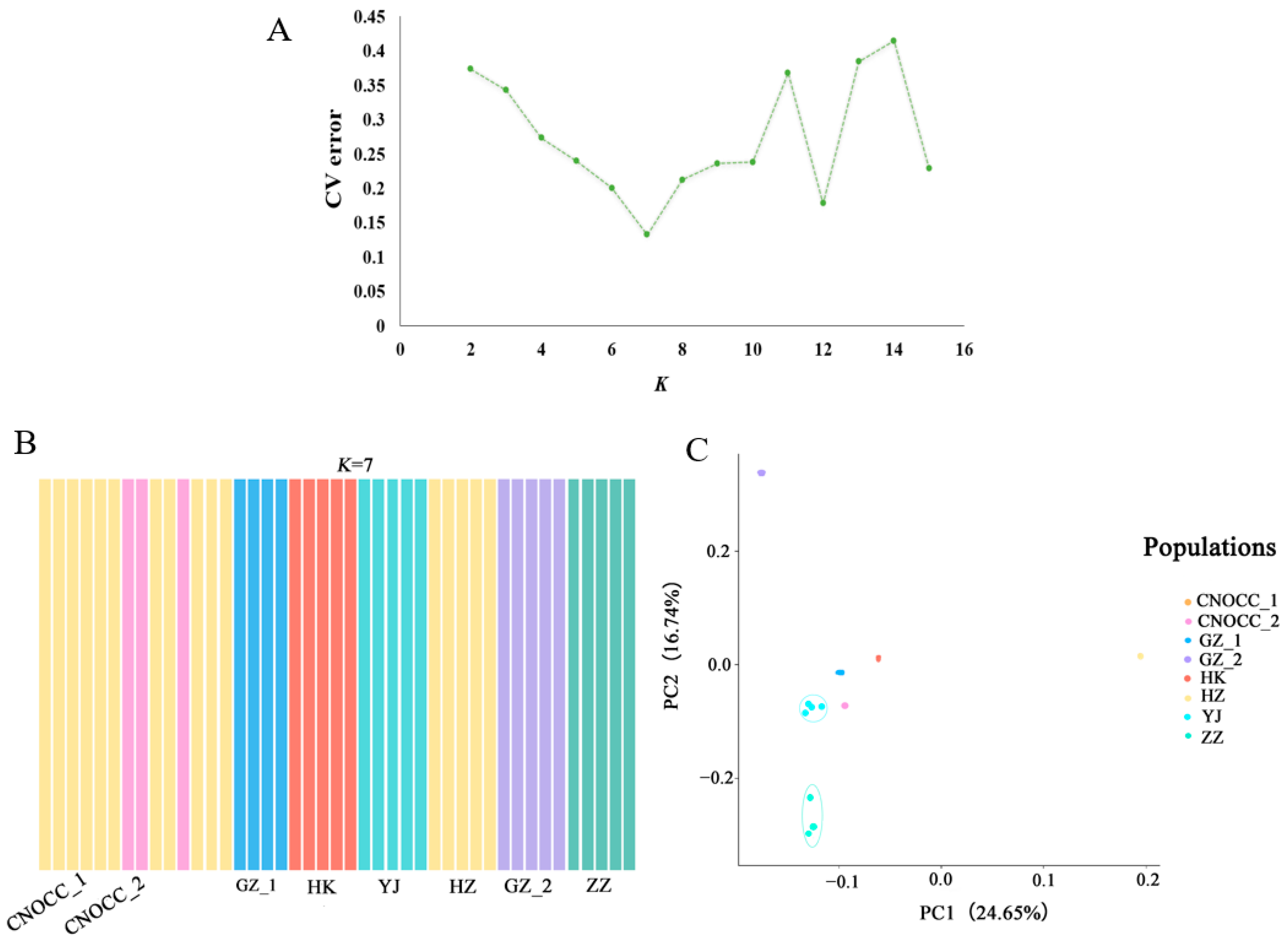
2.1. Sequencing Data, Phylogeny, and Genetic Structure
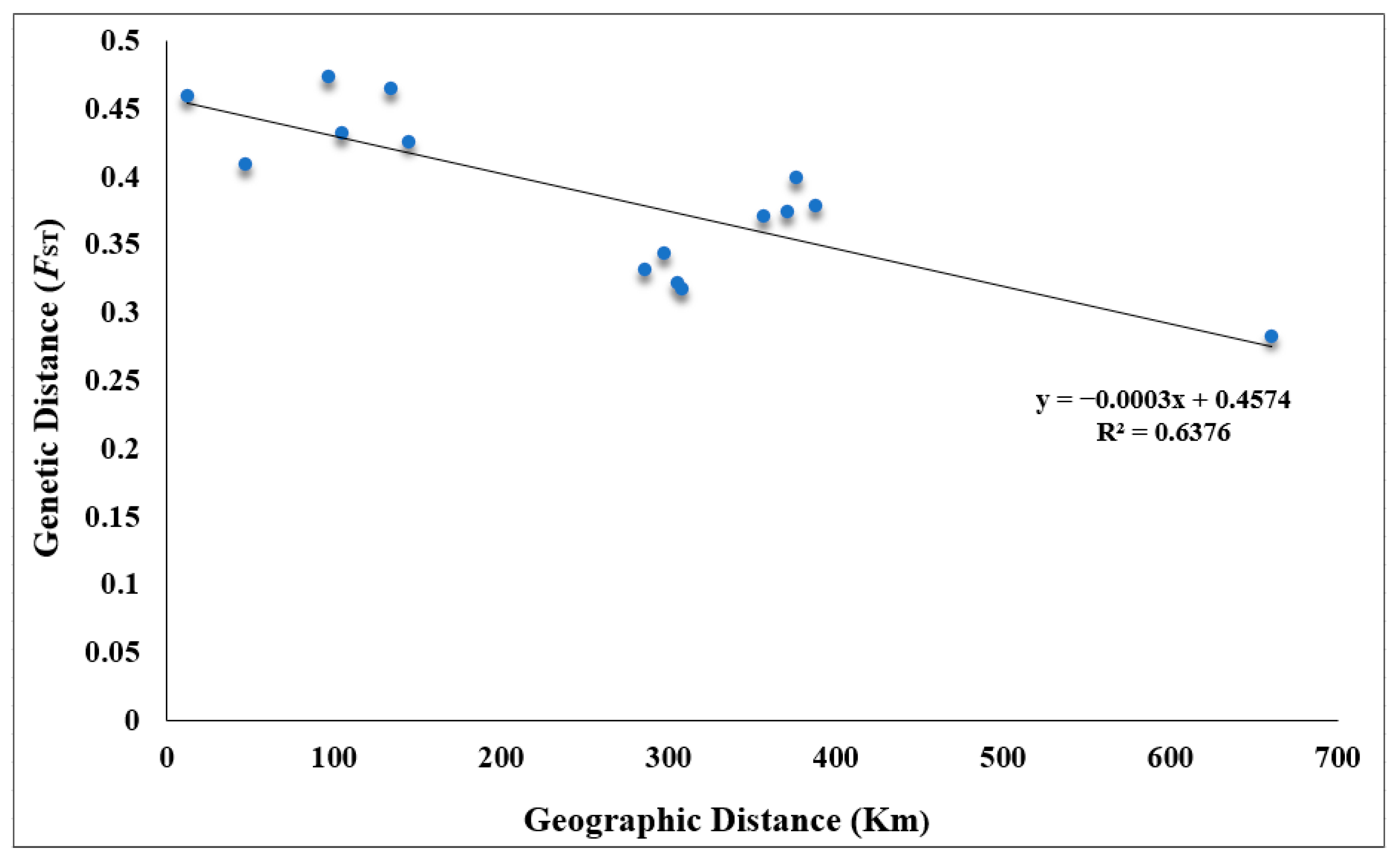
2.2. Genetic Diversity
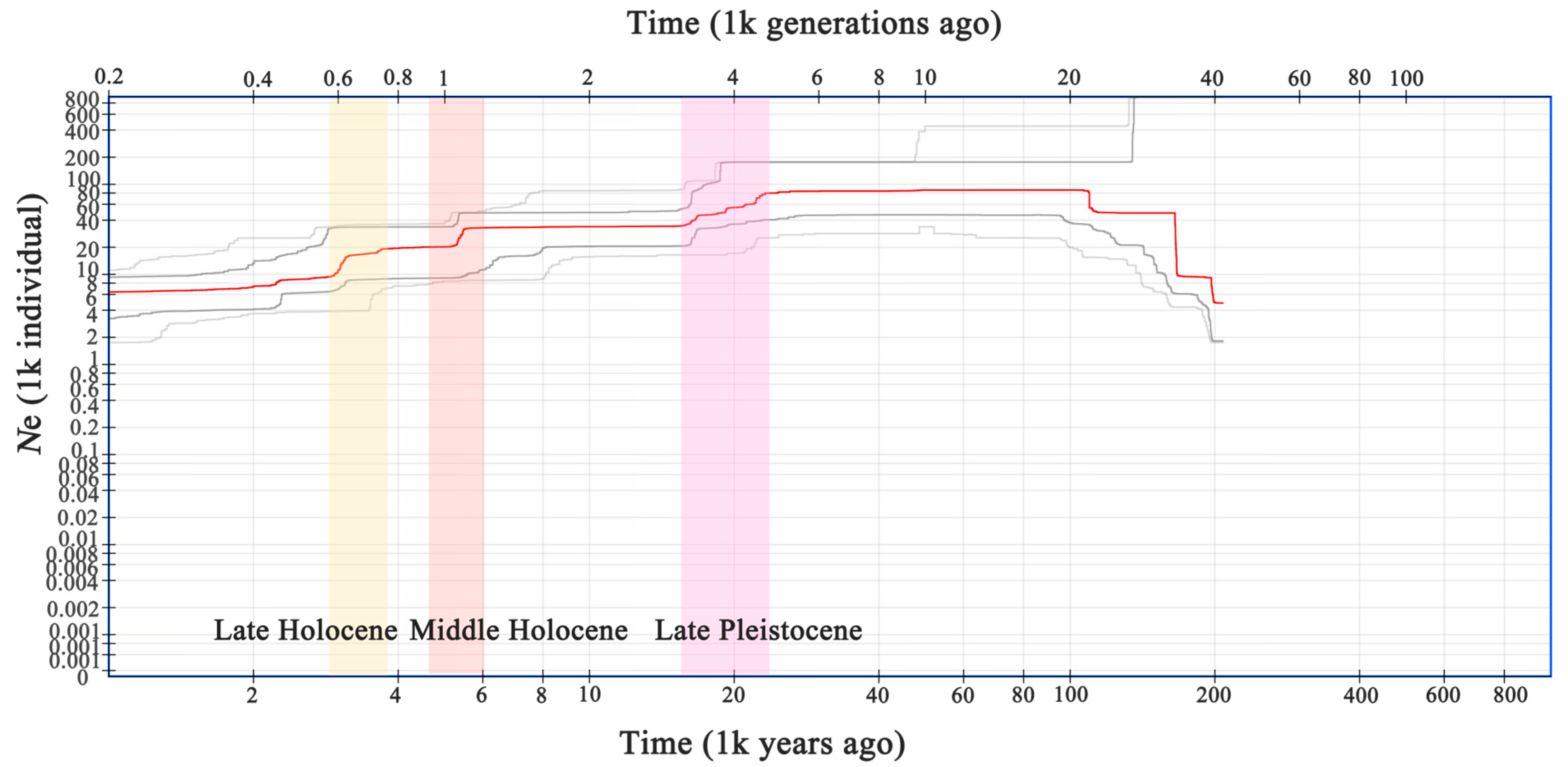
2.3. Historical Population Dynamics
3. Discussion
3.1. Clonal Propagation and Ultimately Monoclonal Populations of V. shenzhenica
3.2. Genetic Diversity of V. shenzhenica
3.3. Conservation Implications
4. Materials and Methods
4.1. Taxon Sampling, Sequencing, SNP Calling, and Chloroplast Genome Assembly
4.2. Phylogenetic, Genetic Diversity, and Population Structure Analyses
4.3. Demographic History
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bona, A.; Kulesza, U.; Jadwiszczak, K.A. Clonal diversity, gene flow and seed production in endangered populations of Betula humilis Schrk. Tree Genet. Genomes 2019, 15, 50. [Google Scholar] [CrossRef]
- Stueffer, J.F.; De Kroon, H.; During, H.J. Exploitation of environmental heterogeneity by spatial division of labor in a clonal plant. Funct. Ecol. 1996, 10, 328–334. [Google Scholar] [CrossRef]
- Caraco, T.; Kelly, C.K. On the adaptive value of physiological integration in clonal plants. Ecology 1991, 72, 81–93. [Google Scholar] [CrossRef]
- Silander, J.A. Microevolution and clone structure in Spartina patens. Science 1979, 203, 658–660. [Google Scholar] [CrossRef]
- Honnay, O.; Bossuyt, B. Prolonged clonal growth: Escape route or route to extinction? Oikos 2005, 108, 427–432. [Google Scholar] [CrossRef]
- Barrett, S.C. Influences of clonality on plant sexual reproduction. Proc. Natl. Acad. Sci. USA 2015, 112, 8859–8866. [Google Scholar] [CrossRef]
- Meloni, M.; Reid, A.; Caujapé-Castells, J.; Marrero, A.; Fernández-Palacios, J.M.; Mesa-Coelo, R.A.; Conti, E. Effects of clonality on the genetic variability of rare, insular species: The case of Ruta microcarpa from the Canary Islands. Ecol. Evol. 2013, 3, 1569–1579. [Google Scholar] [CrossRef]
- Eriksson, O. Seedling dynamics and life histories in clonal plants. Oikos 1989, 55, 231–238. [Google Scholar] [CrossRef]
- Warburton, C.L.; James, E.A.; Fripp, Y.J.; Trueman, S.J.; Wallace, H.M. Clonality and sexual reproductive failure in remnant populations of Santalum lanceolatum (Santalaceae). Biol. Conserv. 2000, 96, 45–54. [Google Scholar] [CrossRef]
- Bory, S.; Grisoni, M.; Duval, M.F.; Besse, P. Biodiversity and preservation of Vanilla: Present state of knowledge. Genet. Resour. Crop Evol. 2008, 55, 551–571. [Google Scholar] [CrossRef]
- Liu, Z.J.; Chen, S.C.; Ru, Z.Z. Vanilla shenzhenica Z.J. Liu & S.C. Chen, the first new species of Orchidaceae found in Shenzhen, South China. Acta Phytotaxon. Sin. 2007, 45, 301–303. [Google Scholar] [CrossRef]
- National Forestry and Grassland Administration. List of National Key Protected Wild Plants in China (Revised Edition); NFGA: Beijing, China, 2021. [Google Scholar]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Stoeckel, S.; Grange, J.; Fernández-Manjarres, J.F.; Bilger, I.; Frascaria-Lacost, N.; Mariette, S. Heterozygote excess in a self-incompatible and partially clonal forest tree species—Prunus avium L. Mol. Ecol. 2006, 15, 2109–2118. [Google Scholar] [CrossRef]
- Wehenkel, C.; Hernández-Díaz, J.C.; Hernández-Velasco, J.; Simental-Rodríguez, S.L.; Porth, I.; Goessen, R.; González-Elizondo, M.S.; Fladung, M.; Groppe, K.; Jaramillo-Correa, J.P.; et al. Chapter 9-Mexican Populus tremuloides Michx: Adaptation to Climate Change Under Extreme Heterozygote Excess. In The Poplar Genome; Porth, I., Klápště, J., McKown, A., Eds.; Springer International Publishing: Cham, Switzerland, 2024; pp. 183–190. [Google Scholar] [CrossRef]
- Reynes, L.; Thibaut, T.; Mauger, S.; Blanfuné, A.; Holon, F.; Cruaud, C.; Couloux, A.; Valero, M.; Aurelle, D. Genomic signatures of clonality in the deep water kelp Laminaria rodriguezii. Mol. Ecol. 2021, 30, 1806–1822. [Google Scholar] [CrossRef]
- Hewitt, A.; Rymer, P.; Holford, P.; Morris, E.C.; Renshaw, A. Evidence for clonality, breeding system, genetic diversity and genetic structure in large and small populations of Melaleuca deanei (Myrtaceae). Aust. J. Bot. 2019, 67, 36–45. [Google Scholar] [CrossRef]
- Bory, S.; Brown, S.; Duval, M.-F.; Besse, P. Chapter 2—Evolutionary Processes and Diversification in the Genus Vanilla. In Vanilla; Odoux, E., Grisoni, M., Eds.; CRC Press: Boca Raton, FL, USA, 2010; pp. 15–28. [Google Scholar] [CrossRef]
- Soto Arenas, M.A. Vainilla: Los Retos de un Cultivo Basado en una Especie Amenazada Con una Historia de Vida Compleja; Congreso Internacional de Productores de Vainilla: Papantla, Veracruz, Mexico, 2006. [Google Scholar]
- Stoeckel, S.; Masson, J.P. The exact distributions of FIS under partial asexuality in small finite populations with mutation. PLoS ONE 2014, 9, e85228. [Google Scholar] [CrossRef]
- Lin, C.H.; Miriti, M.N.; Goodell, K. Demographic consequences of greater clonal than sexual reproduction in Dicentra canadensis. Ecol. Evol. 2016, 6, 3871–3883. [Google Scholar] [CrossRef]
- Vallejo-Marín, M.; Dorken, M.E.; Barrett, S.C. The ecological and evolutionary consequences of clonality for plant mating. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 193–213. [Google Scholar] [CrossRef]
- Hu, A.Q.; Gale, S.W.; Kumar, P.; Saunders, R.M.; Sun, M.; Fischer, G.A. Preponderance of clonality triggers loss of sex in Bulbophyllum bicolor, an obligately outcrossing epiphytic orchid. Mol. Ecol. 2017, 26, 3358–3372. [Google Scholar] [CrossRef]
- Dorken, M.E.; Eckert, C.G. Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). J. Ecol. 2001, 89, 339–350. [Google Scholar] [CrossRef]
- Eckert, C.G. Chapter 15—The loss of sex in clonal plants. In Ecology and Evolutionary Biology of Clonal Plants; Stuefer, J.F., Erschbamer, B., Huber, H., Suzuki, J.-I., Eds.; Springer International Publishing: Cham, Switzerland, 2024; pp. 279–298. [Google Scholar] [CrossRef]
- Wiberg, R.A.W.; Scobie, A.R.; A’Hara, S.W.; Ennos, R.A.; Cottrell, J.E. The genetic consequences of long-term habitat fragmen-tation on a self-incompatible clonal plant, Linnaea borealis L. Biol. Conserv. 2016, 201, 405–413. [Google Scholar] [CrossRef]
- Zobel, M.; Moora, M.; Herben, T. Clonal mobility and its implications for spatio-temporal patterns of plant communities: What do we need to know next? Oikos 2010, 119, 802–806. [Google Scholar] [CrossRef]
- Buckley, H.L.; Freckleton, R.P. Understanding the role of species dynamics in abundance–occupancy relationships. J. Ecol. 2010, 98, 645–658. [Google Scholar] [CrossRef]
- Xiao, J.H.; Ding, X.; Li, L.; Ma, H.; Ci, X.Q.; van der Merwe, M.; Conran, J.G.; Li, J. Miocene diversification of a golden-thread nanmu tree species (Phoebe zhennan, Lauraceae) around the Sichuan Basin shaped by the East Asian monsoon. Ecol. Evol. 2020, 10, 10543–10557. [Google Scholar] [CrossRef]
- Cai, C.N.; Hou, Q.X.; Ci, X.Q.; Xiao, J.H.; Zhang, C.Y.; Li, J. Genetic Diversity of Horsfieldia hainanensis: An Endangered Species with Extremely Small Populations. J. Trop. Subtrop. Bot. 2021, 29, 547–555. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, X.; Sun, G.; Li, F.; Gong, X. Application of RAD sequencing for evaluating the genetic diversity of domesticated Panax notoginseng (Araliaceae). PLoS ONE 2016, 11, e0166419. [Google Scholar] [CrossRef]
- Xu, Y.; Zang, R. Theoretical and practical research on conservation of Wild Plants with Extremely Small Populations in China. Biodivers. Sci. 2022, 30, 84–105. [Google Scholar] [CrossRef]
- Ellegren, H.; Galtier, N. Determinants of genetic diversity. Nat. Rev. Genet. 2016, 17, 422–433. [Google Scholar] [CrossRef]
- Cheon, C.P.; Chung, M.Y.; Chung, S.G.; Chung, M.G. Allozyme variation of a small subshrub Ardisia japonica (Myrsinaceae) in north eastern Asia. Silvae Genet. 2002, 51, 1–6. [Google Scholar]
- Kollmann, J.; Steinger, T.; Roy, B.A. Evidence of sexuality in European Rubus (Rosaceae) species based on AFLP and allozyme analysis. Am. J. Bot. 2000, 87, 1592–1598. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-González, H.; Raggio, M.; Labrín-Sotomayor, N.Y.; Ferrer, M.M.; Peña-Ramírez, Y.J. Genetic variability of Tabebuia rosea (Bignoniaceae) from plantations and remnant populations in the Mayan Forest. Forests 2023, 14, 2006. [Google Scholar] [CrossRef]
- Furlan, E.; Stoklosa, J.; Griffiths, J.; Gust, N.; Ellis, R.; Huggins, R.M.; Weeks, A.R. Small population size and extremely low levels of genetic diversity in island populations of the platypus, Ornithorhynchus anatinus. Ecol. Evol. 2012, 2, 844–857. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Deng, Y.; Zhang, H.; Yu, C.H.; Chen, C.X. Holocene environment changes in the tropical and subtropical areas of the South China and the relation to human activities. Quat. Sci. 2004, 24, 387–393. (In Chinese) [Google Scholar]
- Ma, H.; Liu, Y.; Liu, D.; Sun, W.; Liu, X.; Wan, Y.; Zhang, X.; Zhang, R.; Yun, Q.; Wang, J.; et al. Chromosome-level genome assembly and population genetic analysis of a critically endangered rhododendron provide insights into its conservation. Plant J. 2021, 107, 1533–1545. [Google Scholar] [CrossRef]
- Abeli, T.; Dalrymple, S.; Godefroid, S.; Mondoni, A.; Müller, J.V.; Rossi, G.; Orsenigo, S. Ex situ collections and their potential for the restoration of extinct plants. Conserv. Biol. 2020, 34, 303–313. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, M.; Wen, X.; Li, N.; Ren, H. Strengthen ex situ conservation of plants and promote protection and utilization of plant resources. Bull. Chin. Acad. Sci. 2021, 36, 417–424. [Google Scholar] [CrossRef]
- Wang, Z.; Xie, L.; Prather, C.M.; Guo, H.; Han, G.; Ma, C. What drives the shift between sexual and clonal reproduction of Caragana stenophylla along a climatic aridity gradient? BMC Plant Biol. 2018, 18, 91. [Google Scholar] [CrossRef]
- Hollister, J.D.; Greiner, S.; Wang, W.; Wang, J.; Zhang, Y.; Wong, G.K.S.; Wright, S.I.; Johnson, M.T. Recurrent loss of sex is associated with accumulation of deleterious mutations in Oenothera. Mol. Biol. Evol. 2015, 32, 896–905. [Google Scholar] [CrossRef]
- de Oliveira, R.T.; da Silva Oliveira, J.P.; Macedo, A.F. Vanilla beyond Vanilla planifolia and Vanilla × tahitensis: Taxonomy and historical notes, reproductive biology, and metabolites. Plants 2022, 11, 3311. [Google Scholar] [CrossRef]
- Chen, L.J.; Rao, W.H. The occurrence of Vanilla shenzhenica in Huizhou of Guangdong. J. South China Agric. Univ. 2009, 30, 115–116. (In Chinese) [Google Scholar]
- Barretto, G.; Cribb, P.J.; Gale, S.W. The Wild Orchids of Hong Kong; Natural History Publications (Borneo): Sabah, Malaysia, 2011. [Google Scholar]
- Ma, L.; Chen, X.Y.; Su, X.X.; Lan, S.R.; Chen, S.P. Four new records species of Orchidaceae from Fujian Province. Chin. J. Trop. Crops 2020, 41, 1779–1782. (In Chinese) [Google Scholar] [CrossRef]
- Averyanov, L.V.; Nguyen, V.C.; Truong, B.V.; Nguyen, K.S.; Nguyen, C.H.; Maisak, T.V.; Doan, N.T.; Nguyen, T.H.; Pham, V.T.; Dat, P.T.T.; et al. New orchids in the flora of Vietnam VI (Orchidaceae, tribes Arethuseae, Cymbidieae, Diurideae, Epidendreae, Vandeae, and Vanilleae). Phytotaxa 2023, 597, 87–110. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. Imeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Jian, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von-Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De-Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. A layered grammar of graphics. J. Comput. Graph. Stat. 2010, 19, 3–28. [Google Scholar] [CrossRef]
- R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org (accessed on 29 February 2024).
- Dray, S.; Dufour, A.-B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Liu, X.; Fu, Y.X. Exploring population size changes using SNP frequency spectra. Nat. Genet. 2015, 47, 555–559. [Google Scholar] [CrossRef]
- Gutenkunst, R.N.; Hernandez, R.D.; Williamson, S.H.; Bustamante, C.D. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009, 5, e1000695. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, D.; Wariss, H.M.; Zhang, R.; Tao, L.; Milne, R.I.; Sun, W. Demographic history and identification of threats revealed by population genomic analysis provide insights into conservation for an endangered maple. Mol. Ecol. 2022, 31, 767–779. [Google Scholar] [CrossRef]
- Hasing, T.; Tang, H.; Brym, M.; Khazi, F.; Huang, T.; Chambers, A.H. A phased Vanilla planifolia genome enables genetic improvement of flavour and production. Nat. Food 2020, 1, 811–819. [Google Scholar] [CrossRef]

| Population | Ho | He | π | FIS |
|---|---|---|---|---|
| GZ_1 | 0.198 | 0.106 | 0.123 | −0.131 |
| GZ_2 | 0.215 | 0.113 | 0.127 | −0.158 |
| HK | 0.205 | 0.109 | 0.121 | −0.150 |
| HZ | 0.184 | 0.098 | 0.109 | −0.134 |
| YJ | 0.210 | 0.165 | 0.185 | −0.040 |
| ZZ | 0.172 | 0.142 | 0.159 | −0.016 |
| CNOCC_1 | 0.177 | 0.094 | 0.099 | −0.148 |
| CNOCC_2 | 0.219 | 0.114 | 0.136 | −0.137 |
| Species | 0.214 | 0.135 | 0.148 | −0.138 |
| Population | GZ_1 | GZ_2 | HK | HZ | YJ | ZZ | CNOCC_1 | CNOCC_2 |
|---|---|---|---|---|---|---|---|---|
| GZ_1 | 0 | 0.430 | 0.437 | 0.467 | 0.327 | 0.380 | 0.455 | 0.471 |
| GZ_2 | 0 | 0.420 | 0.449 | 0.316 | 0.372 | 0.445 | 0.452 | |
| HK | 0 | 0.424 | 0.329 | 0.375 | 0.418 | 0.454 | ||
| HZ | 0 | 0.334 | 0.384 | 0.009 | 0.478 | |||
| YJ | 0 | 0.282 | 0.344 | 0.331 | ||||
| ZZ | 0 | 0.388 | 0.375 | |||||
| CNOCC_1 | 0 | 0.470 | ||||||
| CNOCC_2 | 0 |
| Population | Location | Altitude (m) | Sample Size | Voucher |
|---|---|---|---|---|
| GZ_1 | Conghua, Guangzhou, Guangdong | 460 | 4 | JBC15371~JBC15374 |
| GZ_2 | Conghua, Guangzhou, Guangdong | 356 | 5 | JG12731~JG12735 |
| HK | Hong Kong | 75 | 5 | JG12671~JG12672 |
| HZ | Huiyang, Huizhou, Guangdong | 181 | 5 | JG12721~JG12725 |
| YJ | Yangxi, Yangjiang, Guangdong | 327 | 5 | JG12691~JG12695 |
| ZZ | Nanjing, Zhangzhou, Fujian | 413 | 5 | JG12741~JG12745 |
| CNOCC_1 | Living collections at CNOCC, potentially collected from Huizhou | - | 11 | C11~C13 D11~D13 D23, D24 D31~D3 |
| CNOCC_2 | Living collections at CNOCC, potentially collected from Shenzhen | - | 3 | D21, D22, D25 |
| Total | 43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, L.; Hu, A.-Q.; Wang, M.-N.; Cheng, Z.; Chi, K.-B.; Long, C.-L.; Liu, J.-G. Monoclonality and Low Genetic Diversity in Vanilla shenzhenica: Highlighting Urgent Need for Genetic Preservation of China’s Only Endangered Vanilla. Int. J. Mol. Sci. 2025, 26, 3451. https://doi.org/10.3390/ijms26073451
Xiao L, Hu A-Q, Wang M-N, Cheng Z, Chi K-B, Long C-L, Liu J-G. Monoclonality and Low Genetic Diversity in Vanilla shenzhenica: Highlighting Urgent Need for Genetic Preservation of China’s Only Endangered Vanilla. International Journal of Molecular Sciences. 2025; 26(7):3451. https://doi.org/10.3390/ijms26073451
Chicago/Turabian StyleXiao, Li, Ai-Qun Hu, Mei-Na Wang, Zhuo Cheng, Kuan-Bo Chi, Chun-Lin Long, and Jin-Gang Liu. 2025. "Monoclonality and Low Genetic Diversity in Vanilla shenzhenica: Highlighting Urgent Need for Genetic Preservation of China’s Only Endangered Vanilla" International Journal of Molecular Sciences 26, no. 7: 3451. https://doi.org/10.3390/ijms26073451
APA StyleXiao, L., Hu, A.-Q., Wang, M.-N., Cheng, Z., Chi, K.-B., Long, C.-L., & Liu, J.-G. (2025). Monoclonality and Low Genetic Diversity in Vanilla shenzhenica: Highlighting Urgent Need for Genetic Preservation of China’s Only Endangered Vanilla. International Journal of Molecular Sciences, 26(7), 3451. https://doi.org/10.3390/ijms26073451







