A Comparative Study of a Potent CNS-Permeable RARβ-Modulator, Ellorarxine, in Neurons, Glia and Microglia Cells In Vitro
Abstract
:1. Introduction
2. Results
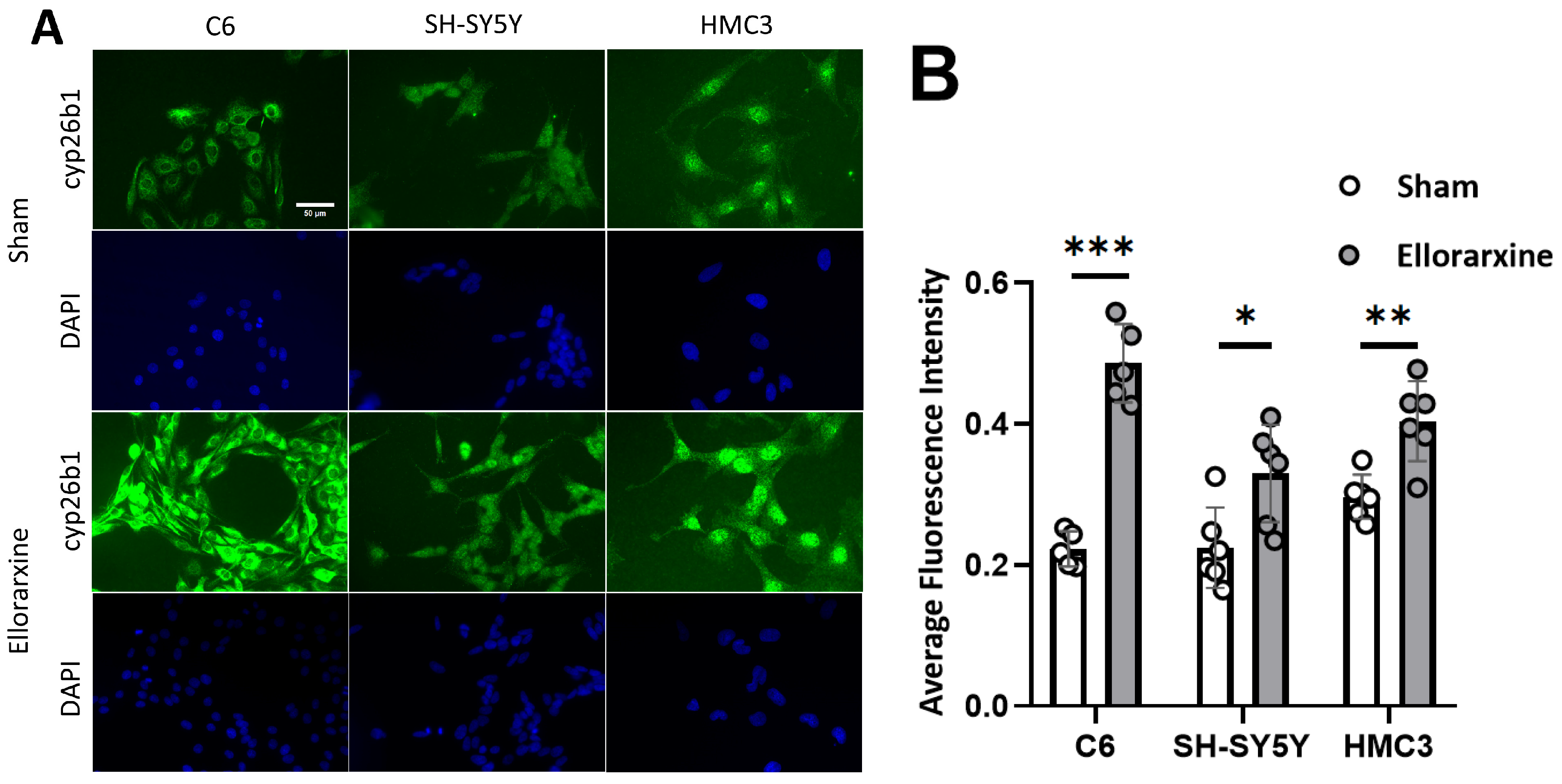
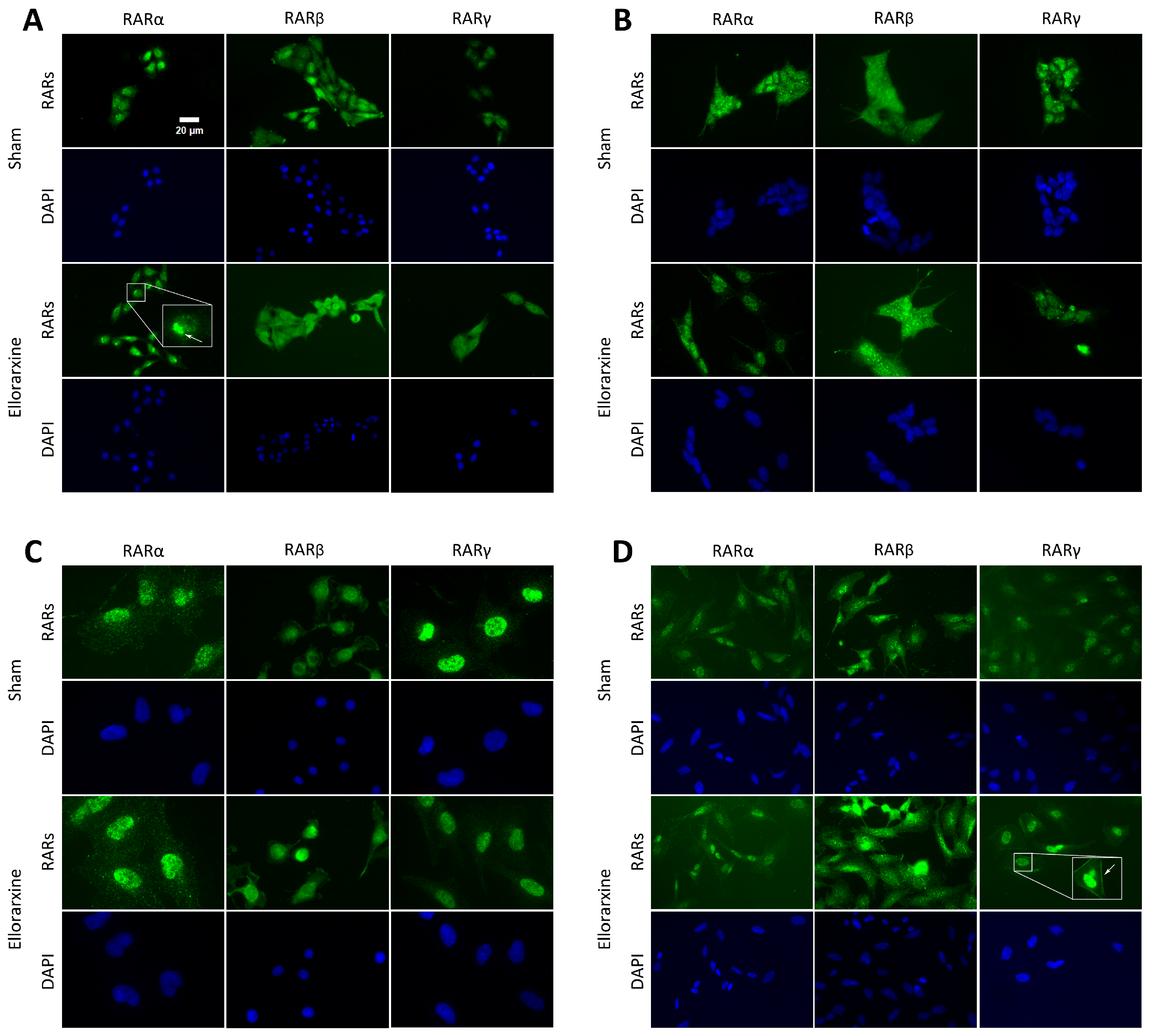
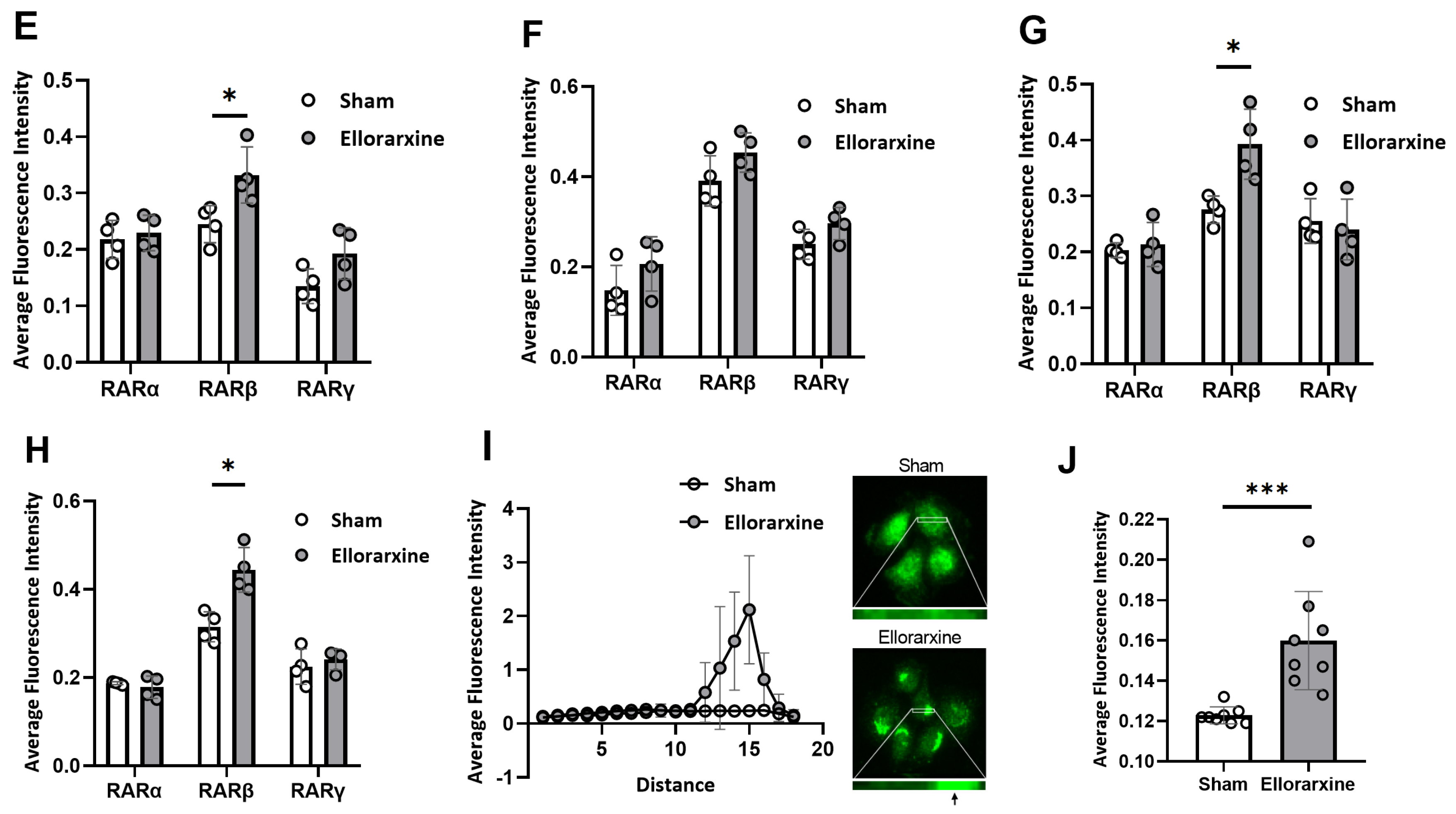
2.1. Ellorarxine Upregulates the Expression of Cyp26b1 and RARβ
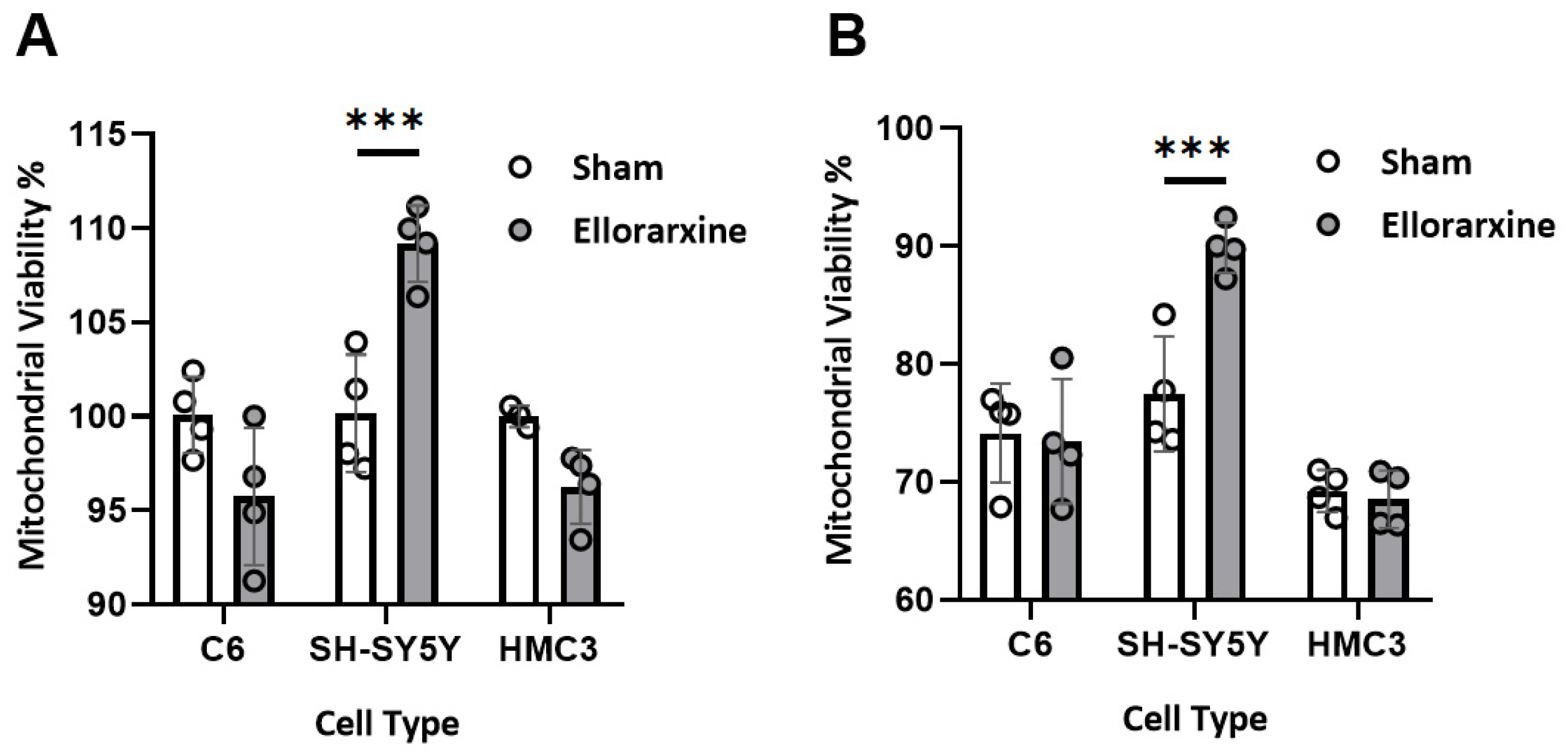
2.2. Ellorarxine Pretreatment Alleviates Mitochondrial Dysfunction
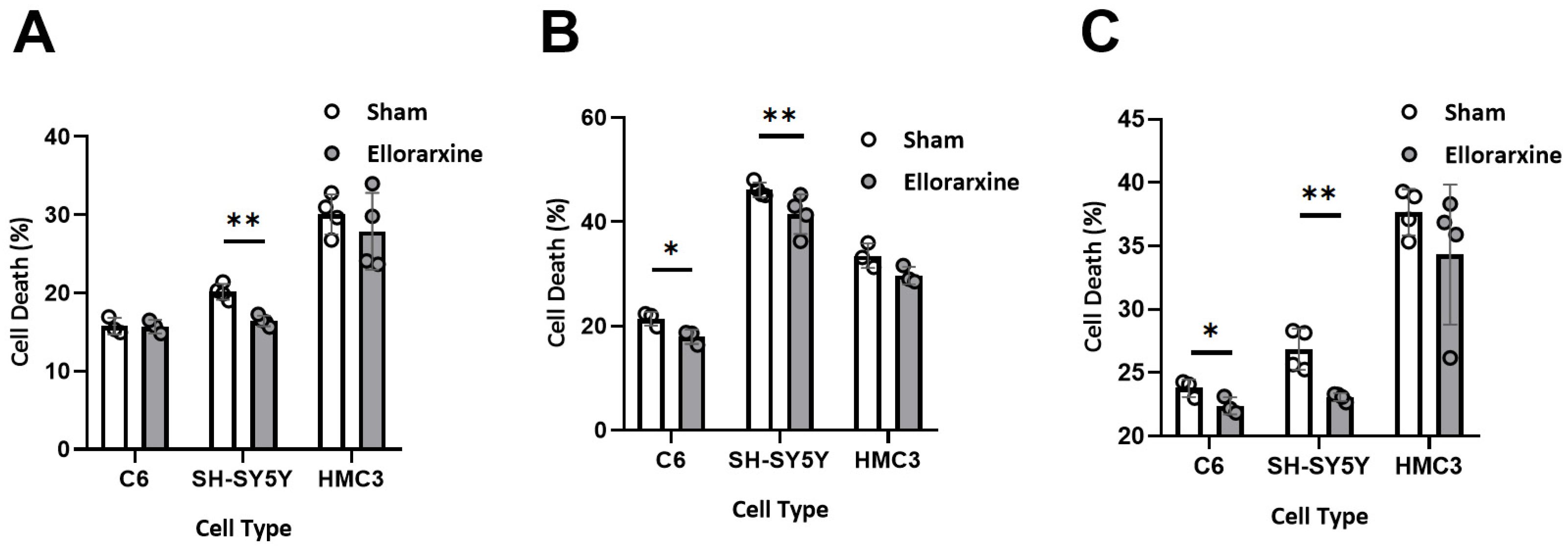
2.3. Ellorarxine Pretreatment Reduced Cell Death
2.4. Ellorarxine Pretreatment Modulated Inflammatory Cytokine Release in HMC3 Microglia
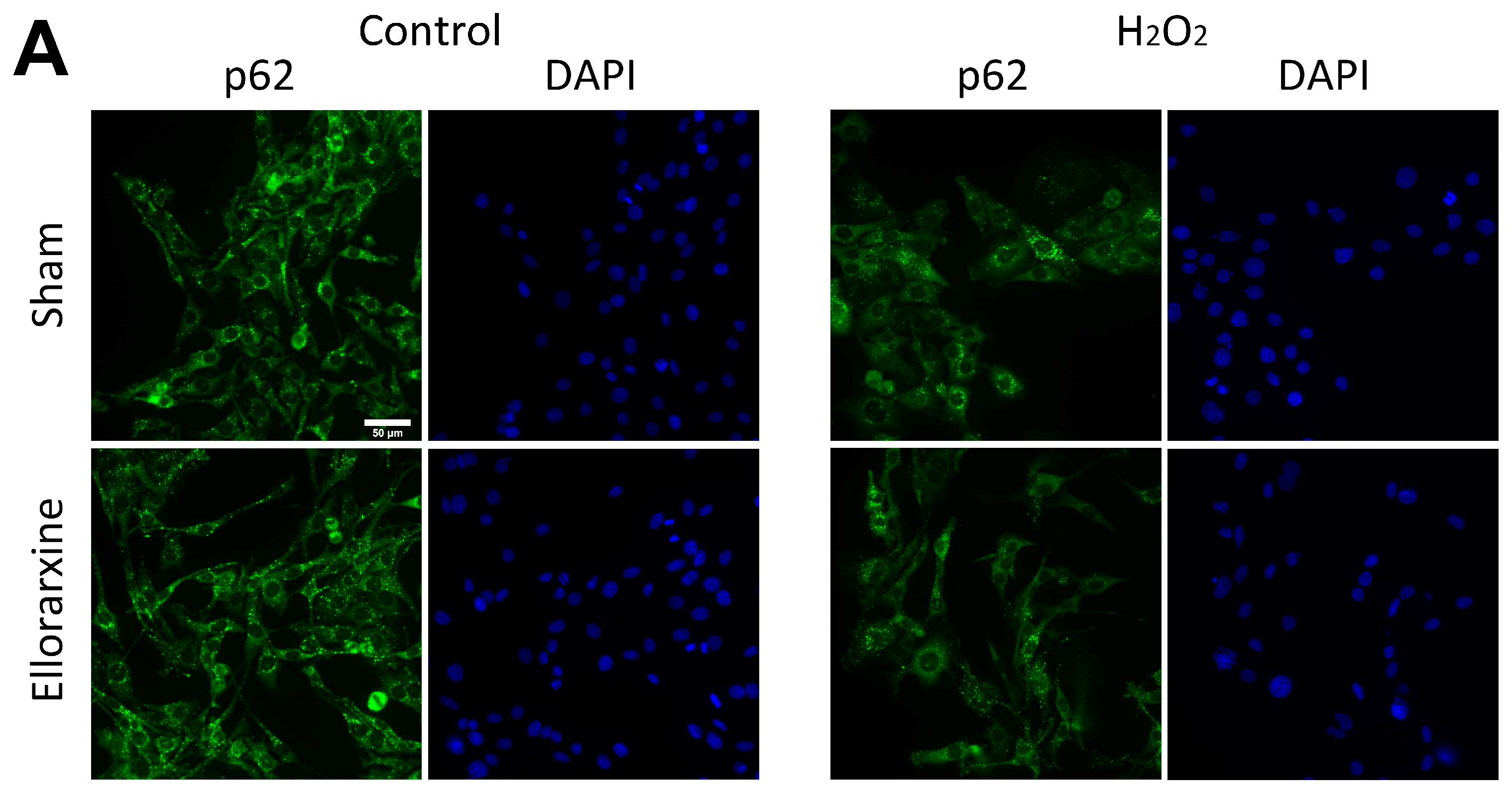
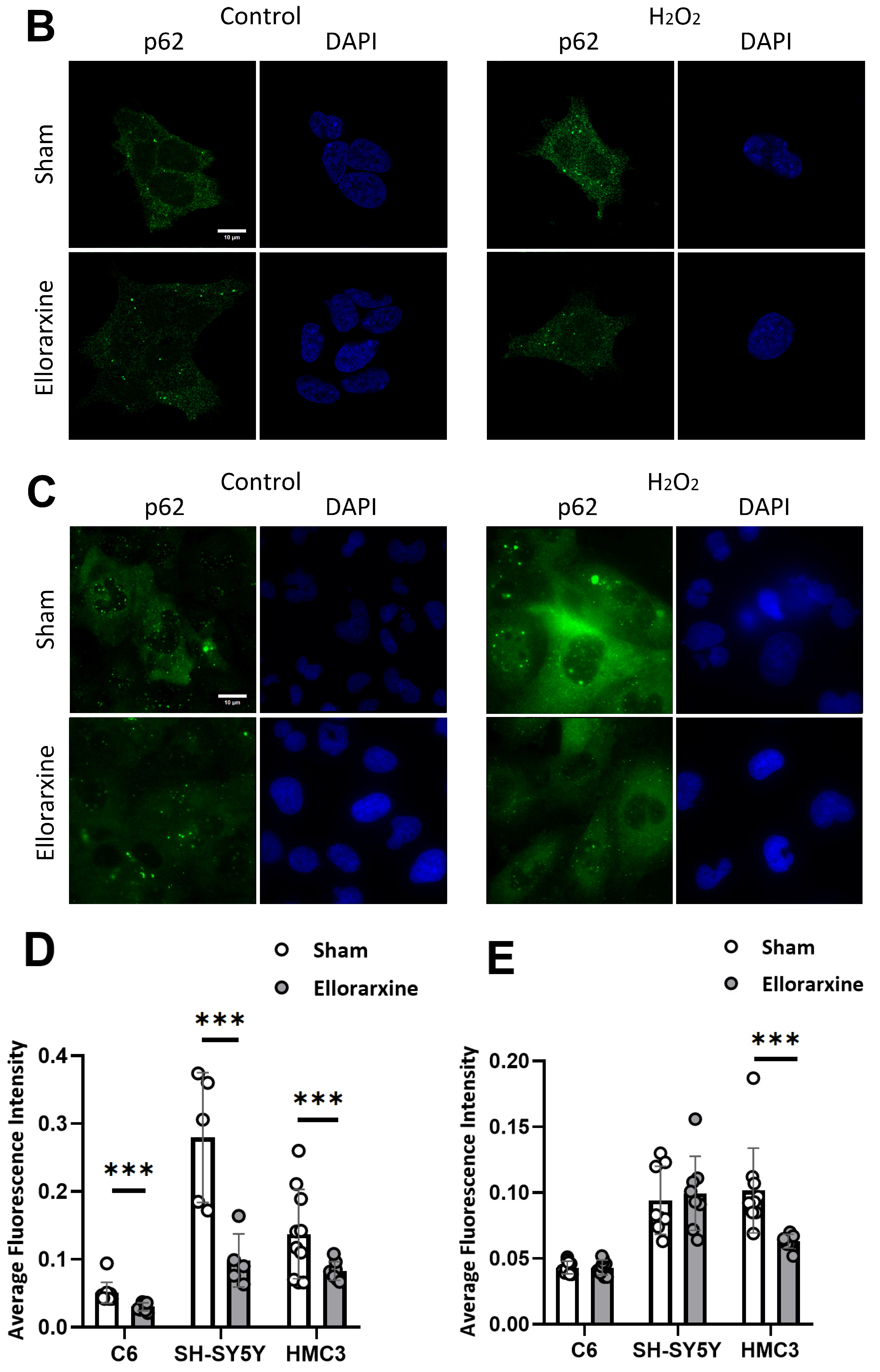
2.5. Ellorarxine Treatment Regulated Cellular Autophagy
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture
4.2. Cell Differentiation
4.3. Preparation of Ellorarxine
4.4. Pretreatments
4.5. Methyl Thiazolyl Diphenyl Tetrazolium Bromide (MTT) Assay [32]
4.6. Lactate Dehydrogenase (LDH) Release Assay [33]
4.7. Enzyme-Linked Immunosorbent Assay (ELISA) [34]
4.8. Immunocytochemistry Staining [37,38]
4.9. Immunofluorescence Staining
4.10. Quantification and Statistical Analysis
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Recombinant Anti-Retinoic Acid Receptor alpha antibody [EPR23871-271] (ab275745) | Abcam (Cambridge, UK) | https://www.abcam.com/products/primary-antibodies/retinoic-acid-receptor-alpha-antibody-epr23871-271-ab275745.html, accessed on 1 March 2025 |
| Anti-Retinoic Acid Receptor beta antibody (ab5792) | Abcam (Cambridge, UK) | https://www.abcam.com/products/primary-antibodies/retinoic-acid-receptor-beta-antibody-ab5792.html, accessed on 1 March 2025 |
| Anti-Retinoic Acid Receptor gamma antibody (ab97569) | Abcam (Cambridge, UK) | https://www.abcam.com/products/primary-antibodies/retinoic-acid-receptor-gamma-antibody-ab97569.html, accessed on 1 March 2025 |
| Anti-Cyp26B1 antibody (ab113236) | Abcam (Cambridge, UK) | https://www.abcam.com/products/primary-antibodies/cyp26b1-antibody-ab113236.html, accessed on 1 March 2025 |
| Goat Anti-Mouse IgG H&L (Alexa Fluor® 488) (ab150113) | Abcam (Cambridge, UK) | https://www.abcam.com/products/secondary-antibodies/goat-mouse-igg-hl-alexa-fluor-488-ab150113.html, accessed on 1 March 2025 |
| Anti-SQSTM1/p62 antibody [EPR23101-103] | Abcam (Cambridge, UK) | https://www.abcam.com/en-us/search?productSorting=relevance&resourceSorting=relevance&keywords=sqstm1-p62-antibody-2c11-bsa-and-azide-free-ab56416&utm_source=google&utm_medium=cpc&gad_source=1&gclid=CjwKCAjwktO_BhBrEiwAV70jXmFhWo8-FX1QCxX-PhJIrI8dVqZnbe-AAwAeTuxAXQ896rDmkyFkKRoCdYQQAvD_BwE&gclsrc=aw.ds&productcode=ab56416&view=quickview, accessed on March 2025 |
| LC3B Polyclonal Antibody, Invitrogen™ | Thermo-Fisher (Altrincham, Cheshire) | https://www.fishersci.com/shop/products/lc3b-polyclonal-antibody-invitrogen-2/PA146286?searchHijack=true&searchTerm=PA1-46286&searchType=RAPID&matchedCatNo=PA1-46286, accessed on 1 March 2025 |
| Chemicals, peptides and recombinant proteins | ||
| Phosphate-buffered saline (P5368-10pak) | Merck Life Science UK Limited. (Gillingham, Dorset, UK) | https://www.sigmaaldrich.com/GB/en/search/p5368-10pak?focus=products&page=1&perpage=30&sort=relevance&term=p5368-10pak&type=product, accessed on 1 March 2025 |
| Triton X-100 | ||
| BSA | ||
| Tween 20 | ||
| Thiazolyl Blue Tetrazolium Bromide | Merck Life Science UK Limited. (Gillingham, Dorset, UK) | https://www.sigmaaldrich.com/GB/en/search/m2128?focus=products&page=1&perpage=30&sort=relevance&term=m2128&type=product, accessed on 1 March 2025 |
| H2O2 | ||
| LPS | ||
| Critical commercial assays | ||
| CytoTox 96® Non-Radioactive Cytotoxicity Assay | Promega (Chilworth, Southampton, UK) | https://www.promega.co.uk/products/cell-health-assays/cell-viability-and-cytotoxicity-assays/cytotox-96-non_radioactive-cytotoxicity-assay/?catNum=G1780, accessed on 1 March 2025 |
| Human IL-6 ELISA kit (ab178013) | Abcam (Cambridge, UK) | https://www.abcam.com/products/elisa/human-il-6-elisa-kit-ab178013.html, accessed on 1 March 2025 |
| Human TNF alpha ELISA kit (ab46087) | Abcam (Cambridge, UK) | https://www.abcam.com/products/elisa/human-tnf-alpha-elisa-kit-ab46087.html, accessed on 1 March 2025 |
| VECTASTAIN® Elite® ABC-HRP Kit (Peroxidase, Universal) (PK-6200) | VECTASTAIN (2BScientific, Kirtlington, UK) | https://vectorlabs.com/products/vectastain-elite-abc-hrp-kit-universal, accessed on 1 March 2025 |
| ImmPACT® DAB Substrate Kit, Peroxidase (HRP) (SK-4105) | VECTASTAIN (2BScientific, Kirtlington, UK) | https://vectorlabs.com/products/immpact-dab-hrp-substrate, accessed on 1 March 2025 |
| Experimental models: cell lines | ||
| Rat glioma C6 cells | ||
| Human neuroblastoma SH-SY5Y cells | ||
| Human microglia clone 3 HMC3 cells | ||
| Software and algorithms | ||
| ZEN software | Zeiss (Cambridge, UK) | https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html, accessed on 1 March 2025 |
| Prism8 | GraphPad, Prism, (Boston, MA, USA) | https://www.graphpad.com/, accessed on 1 March 2025 |
| ImageJ | LOCI | https://imagej.net/, accessed on 1 March 2025 |
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lane, M.A.; Bailey, S.J. Role of Retinoid Signalling in the Adult Brain. Prog. Neurobiol. 2005, 75, 275–293. [Google Scholar] [CrossRef] [PubMed]
- Janesick, A.; Wu, S.C.; Blumberg, B. Retinoic Acid Signaling and Neuronal Differentiation. Cell. Mol. Life Sci. 2015, 72, 1559–1576. [Google Scholar] [CrossRef]
- Riancho, J.; Berciano, M.T.; Ruiz-Soto, M.; Berciano, J.; Landreth, G.; Lafarga, M. Retinoids and Motor Neuron Disease: Potential Role in Amyotrophic Lateral Sclerosis. J. Neurol. Sci. 2016, 360, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Gürbüz, M.; Aktaç, Ş. Understanding the Role of Vitamin A and Its Precursors in the Immune System. Nutr. Clin. Metab. 2022, 36, 89–98. [Google Scholar] [CrossRef]
- McCaffery, P.J.; Adams, J.; Maden, M.; Rosa-Molinar, E. Too Much of a Good Thing: Retinoic Acid as an Endogenous Regulator of Neural Differentiation and Exogenous Teratogen. Eur. J. Neurosci. 2003, 18, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Giguère, V. Retinoic Acid Receptors and Cellular Retinoid Binding Proteins: Complex Interplay in Retinoid Signaling. Endocr. Rev. 1994, 15, 61–79. [Google Scholar] [CrossRef]
- Lavudi, K.; Nuguri, S.M.; Olverson, Z.; Dhanabalan, A.K.; Patnaik, S.; Kokkanti, R.R. Targeting the Retinoic Acid Signaling Pathway as a Modern Precision Therapy against Cancers. Front. Cell Dev. Biol. 2023, 11, 1254612. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Cai, S.Y.; Boyer, J.L. The Role of the Retinoid Receptor, RAR/RXR Heterodimer, in Liver Physiology. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166085. [Google Scholar] [CrossRef]
- Liao, Y.P.; Ho, S.Y.; Liou, J.C. Non-Genomic Regulation of Transmitter Release by Retinoic Acid at Developing Motoneurons in Xenopus Cell Culture. J. Cell Sci. 2004, 117, 2917–2924. [Google Scholar] [CrossRef]
- Chen, L.; Lau, A.G.; Sarti, F. Synaptic Retinoic Acid Signaling and Homeostatic Synaptic Plasticity. Neuropharmacology 2014, 78, 3–12. [Google Scholar] [CrossRef]
- Corcoran, J.P.T.; Po, L.S.; Maden, M. Disruption of the Retinoid Signalling Pathway Causes a Deposition of Amyloid β in the Adult Rat Brain. Eur. J. Neurosci. 2004, 20, 896–902. [Google Scholar] [CrossRef] [PubMed]
- Etchamendy, N.; Enderlin, V.; Marighetto, A.; Pallet, V.; Higueret, P.; Jaffard, R. Vitamin A Deficiency and Relational Memory Deficit in Adult Mice: Relationships with Changes in Brain Retinoid Signalling. Behav. Brain Res. 2003, 145, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Ciancia, M.; Rataj-Baniowska, M.; Zinter, N.; Baldassarro, V.A.; Fraulob, V.; Charles, A.L.; Alvarez, R.; Muramatsu, S.; de Lera, A.R.; Geny, B.; et al. Retinoic Acid Receptor Beta Protects Striatopallidal Medium Spiny Neurons from Mitochondrial Dysfunction and Neurodegeneration. Prog. Neurobiol. 2022, 212, 102246. [Google Scholar] [CrossRef]
- Chiang, M.Y.; Misner, D.; Kempermann, G.; Schikorski, T.; Giguère, V.; Sucov, H.M.; Gage, F.H.; Stevens, C.F.; Evans, R.M. An Essential Role for Retinoid Receptors RARβ and RXRγ in Long-Term Potentiation and Depression. Neuron 1998, 21, 1353–1361. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C. Regulating Retinoic Acid Availability during Development and Regeneration: The Role of the CYP26 Enzymes. J. Dev. Biol. 2020, 8, 6. [Google Scholar] [CrossRef]
- Walters, B.J.; Josselyn, S.A. Retinoic Acid Receptor Plays Both Sides of Homeostatic Plasticity. Proc. Natl. Acad. Sci. USA 2019, 116, 6528–6530. [Google Scholar] [CrossRef]
- Sanjay; Kim, J.Y. Anti-Inflammatory Effects of 9-Cis-Retinoic Acid on β-Amyloid Treated Human Microglial Cells. Eur. J. Inflamm. 2022, 20, 1721727X221143651. [Google Scholar] [CrossRef]
- Magalingam, K.B.; Somanath, S.D.; Md, S.; Haleagrahara, N.; Fu, J.Y.; Selvaduray, K.R.; Radhakrishnan, A.K. Tocotrienols Protect Differentiated SH-SY5Y Human Neuroblastoma Cells against 6-Hydroxydopamine-Induced Cytotoxicity by Ameliorating Dopamine Biosynthesis and Dopamine Receptor D2 Gene Expression. Nutr. Res. 2022, 98, 27–40. [Google Scholar] [CrossRef]
- Pedersen, W.A.; Berse, B.; Schüler, U.; Wainer, B.H.; Blusztajn, J.K. All-trans- and 9-cis-Retinoic Acid Enhance the Cholinergic Properties of a Murine Septal Cell Line: Evidence That the Effects Are Mediated by Activation of Retinoic Acid Receptor-α. J. Neurochem. 1995, 65, 50–58. [Google Scholar] [CrossRef]
- Lee, H.P.; Casadesus, G.; Zhu, X.; Lee, H.G.; Perry, G.; Smith, M.A.; Gustaw-Rothenberg, K.; Lerner, A. All-Trans Retinoic Acid as a Novel Therapeutic Strategy for Alzheimer’s Disease. Expert Rev. Neurother. 2009, 9, 1615–1621. [Google Scholar] [CrossRef]
- Fukasawa, H.; Nakagomi, M.; Yamagata, N.; Katsuki, H.; Kawahara, K.; Kitaoka, K.; Miki, T.; Shudo, K. Tamibarotene: A Candidate Retinoid Drug for Alzheimer’s Disease. Biol. Pharm. Bull. 2012, 35, 1206–1212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Subbanna, S.; Williams, C.R.O.; Canals-Baker, S.; Smiley, J.F.; Wilson, D.A.; Das, B.C.; Saito, M. Anti-Inflammatory Action of BT75, a Novel RARα Agonist, in Cultured Microglia and in an Experimental Mouse Model of Alzheimer’s Disease. Neurochem. Res. 2023, 48, 1958–1970. [Google Scholar] [CrossRef]
- Hewson, Q.C.; Lovat, P.E.; Pearson, A.D.J.; Redfern, C.P.F. Retinoid Signalling and Gene Expression in Neuroblastoma Cells: RXR Agonist and Antagonist Effects on CRABP-II and RARβ Expression. J. Cell. Biochem. 2002, 87, 284–291. [Google Scholar] [CrossRef]
- Huang, P.; Chandra, V.; Rastinejad, F. Retinoic Acid Actions through Mammalian Nuclear Receptors. Chem. Rev. 2014, 114, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Storer, P.D.; Chavis, J.A.; Racke, M.K.; Drew, P.D. Agonists for the Peroxisome Proliferator-Activated Receptor-α and the Retinoid X Receptor Inhibit Inflammatory Responses of Microglia. J. Neurosci. Res. 2005, 81, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Galland, F.; Seady, M.; Taday, J.; Smaili, S.S.; Gonçalves, C.A.; Leite, M.C. Astrocyte Culture Models: Molecular and Function Characterization of Primary Culture, Immortalized Astrocytes and C6 Glioma Cells. Neurochem. Int. 2019, 131, 104538. [Google Scholar] [CrossRef]
- Shipley, M.M.; Mangold, C.A.; Szpara, M.L. Differentiation of the SH-SY5Y Human Neuroblastoma Cell Line. J. Vis. Exp. 2016, 2016, 53193. [Google Scholar] [CrossRef]
- Xu, L.; He, D.; Bai, Y. Microglia-Mediated Inflammation and Neurodegenerative Disease. Mol. Neurobiol. 2016, 53, 6709–6715. [Google Scholar] [CrossRef]
- Lepiarz, I.; Olajide, O. The Human Microglia (HMC-3) as a Cellular Model of Neuroinflammation. IBRO Rep. 2019, 6, S92. [Google Scholar] [CrossRef]
- Fontenete, S.; Carvalho, D.; Lourenço, A.; Guimarães, N.; Madureira, P.; Figueiredo, C.; Azevedo, N.F. FISHji: New ImageJ Macros for the Quantification of Fluorescence in Epifluorescence Images. Biochem. Eng. J. 2016, 112, 61–69. [Google Scholar] [CrossRef]
- Niewiadomska-Cimicka, A.; Krzyżosiak, A.; Ye, T.; Podleśny-Drabiniok, A.; Dembélé, D.; Dollé, P.; Krężel, W. Genome-Wide Analysis of RARβ Transcriptional Targets in Mouse Striatum Links Retinoic Acid Signaling with Huntington’s Disease and Other Neurodegenerative Disorders. Mol. Neurobiol. 2017, 54, 3859–3878. [Google Scholar] [CrossRef] [PubMed]
- Marie, A.; Darricau, M.; Touyarot, K.; Parr-Brownlie, L.C.; Bosch-Bouju, C. Role and Mechanism of Vitamin A Metabolism in the Pathophysiology of Parkinson’s Disease. J. Park. Dis. 2021, 11, 949–970. [Google Scholar] [CrossRef] [PubMed]
- Dheen, S.T.; Jun, Y.; Yan, Z.; Tay, S.S.W.; Ling, E.A. Retinoic Acid Inhibits Expression of TNF-α and INOS in Activated Rat Microglia. Glia 2005, 50, 21–31. [Google Scholar] [CrossRef]
- Almaguer, J.; Hindle, A.; Lawrence, J.J. The Contribution of Hippocampal All-Trans Retinoic Acid (ATRA) Deficiency to Alzheimer’s Disease: A Narrative Overview of ATRA-Dependent Gene Expression in Post-Mortem Hippocampal Tissue. Antioxidants 2023, 12, 1921. [Google Scholar] [CrossRef] [PubMed]
- Angelova, P.R.; Abramov, A.Y. Role of Mitochondrial ROS in the Brain: From Physiology to Neurodegeneration. FEBS Lett. 2018, 592, 692–702. [Google Scholar] [CrossRef]
- Dey, K.; Bazala, M.A.; Kuznicki, J. Targeting Mitochondrial Calcium Pathways as a Potential Treatment against Parkinson’s Disease. Cell Calcium 2020, 89, 102216. [Google Scholar] [CrossRef]
- Kolarcik, C.L.; Bowser, R. Retinoid Signaling Alterations in Amyotrophic Lateral Sclerosis. Am. J. Neurodegener. Dis. 2012, 1, 173.8. [Google Scholar] [CrossRef]
- Medina, D.X.; Chung, E.P.; Teague, C.D.; Bowser, R.; Sirianni, R.W. Intravenously Administered, Retinoid Activating Nanoparticles Increase Lifespan and Reduce Neurodegeneration in the SOD1G93A Mouse Model of ALS. Front. Bioeng. Biotechnol. 2020, 8, 224. [Google Scholar] [CrossRef]
- Riancho, J.; Ruiz-Soto, M.; Berciano, M.T.; Berciano, J.; Lafarga, M. Neuroprotective Effect of Bexarotene in the SOD1G93A Mouse Model of Amyotrophic Lateral Sclerosis. Front. Cell. Neurosci. 2015, 9, 250. [Google Scholar] [CrossRef]
- Isoherranen, N.; Zhong, G. Biochemical and Physiological Importance of the CYP26 Retinoic Acid Hydroxylases. Pharmacol. Ther. 2019, 204, 107400. [Google Scholar] [CrossRef]
- Okano, J.; Lichti, U.; Mamiya, S.; Aronova, M.; Zhang, G.; Yuspa, S.H.; Hamada, H.; Sakai, Y.; Morasso, M.I. Increased Retinoic Acid Levels through Ablation of Cyp26b1 Determine the Processes of Embryonic Skin Barrier Formation and Peridermal Development. J. Cell Sci. 2012, 125, 1827–1836. [Google Scholar] [CrossRef] [PubMed]
- Topletz, A.R.; Thatcher, J.E.; Zelter, A.; Lutz, J.D.; Tay, S.; Nelson, W.L.; Isoherranen, N. Comparison of the Function and Expression of CYP26A1 and CYP26B1, the Two Retinoic Acid Hydroxylases. Biochem. Pharmacol. 2012, 83, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Spoorendonk, K.M.; Peterson-Maduro, J.; Renn, J.; Trowe, T.; Kranenbarg, S.; Winkler, C.; Schulte-Merker, S. Retinoic Acid and Cyp26b1 Are Critical Regulators of Osteogenesis in the Axial Skeleton. Development 2008, 135, 3765–3774. [Google Scholar] [CrossRef]
- Stevison, F.; Jing, J.; Tripathy, S.; Isoherranen, N. Role of Retinoic Acid-Metabolizing Cytochrome P450s, CYP26, in Inflammation and Cancer. In Advances in Pharmacology; Academic Press: Cambridge, MA, USA, 2015; Volume 74. [Google Scholar]
- Rea, I.M.; Gibson, D.S.; McGilligan, V.; McNerlan, S.E.; Denis Alexander, H.; Ross, O.A. Age and Age-Related Diseases: Role of Inflammation Triggers and Cytokines. Front. Immunol. 2018, 9, 586. [Google Scholar] [CrossRef]
- Kevin Howcroft, T.; Campisi, J.; Louis, G.B.; Smith, M.T.; Wise, B.; Wyss-Coray, T.; Augustine, A.D.; McElhaney, J.E.; Kohanski, R.; Sierra, F. The Role of Inflammation in Age-Related Disease. Aging 2013, 5, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhang, Y.; Chen, Y.; Zhu, J.; Yang, Y.; Zhang, H.L. Role of Microglia in Neurological Disorders and Their Potentials as a Therapeutic Target. Mol. Neurobiol. 2017, 54, 7567–7584. [Google Scholar] [CrossRef]
- Escudier, O.; Zhang, Y.; Whiting, A.; Chazot, P. Evaluation of a Synthetic Retinoid, Ellorarxine, in the NSC-34 Cell Model of Motor Neuron Disease. Int. J. Mol. Sci. 2024, 25, 9764. [Google Scholar] [CrossRef]
- Nixon, R.A. The Role of Autophagy in Neurodegenerative Disease. Nat. Med. 2013, 19, 983–997. [Google Scholar] [CrossRef]
- Takalo, M.; Salminen, A.; Soininen, H.; Hiltunen, M.; Haapasalo, A. Protein Aggregation and Degradation Mechanisms in Neurodegenerative Diseases. Am. J. Neurodegener. Dis. 2013, 2, 1–14. [Google Scholar]
- Lin, M.; Yu, H.; Xie, Q.; Xu, Z.; Shang, P. Role of Microglia Autophagy and Mitophagy in Age-Related Neurodegenerative Diseases. Front. Aging Neurosci. 2022, 14, 1100133. [Google Scholar] [CrossRef]
- Hwang, H.J.; Kim, Y.K. The Role of LC3B in Autophagy as an RNA-Binding Protein. Autophagy 2023, 19, 1028–1030. [Google Scholar] [CrossRef] [PubMed]
- Visnjic, D.; Dembitz, V.; Lalic, H. The Role of AMPK/MTOR Modulators in the Therapy of Acute Myeloid Leukemia. Curr. Med. Chem. 2018, 26, 2208–2229. [Google Scholar] [CrossRef] [PubMed]
- Khatib, T.; Marini, P.; Nunna, S.; Chisholm, D.R.; Whiting, A.; Redfern, C.; Greig, I.R.; McCaffery, P. Genomic and Non-Genomic Pathways Are Both Crucial for Peak Induction of Neurite Outgrowth by Retinoids. Cell Commun. Signal. 2019, 17, 40. [Google Scholar] [CrossRef] [PubMed]
- Kouchmeshky, A.; Whiting, A.; McCaffery, P. Neuroprotective Effects of Ellorarxine in Neuronal Models of Degeneration. Front. Neurosci. 2024, 18, 1422294. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Gailloud, L.; Shin, A.; Fewkes, J.; Pinckney, R.; Whiting, A.; Chazot, P. A Comparative Study of a Potent CNS-Permeable RARβ-Modulator, Ellorarxine, in Neurons, Glia and Microglia Cells In Vitro. Int. J. Mol. Sci. 2025, 26, 3551. https://doi.org/10.3390/ijms26083551
Zhang Y, Gailloud L, Shin A, Fewkes J, Pinckney R, Whiting A, Chazot P. A Comparative Study of a Potent CNS-Permeable RARβ-Modulator, Ellorarxine, in Neurons, Glia and Microglia Cells In Vitro. International Journal of Molecular Sciences. 2025; 26(8):3551. https://doi.org/10.3390/ijms26083551
Chicago/Turabian StyleZhang, Yunxi, Lilie Gailloud, Alexander Shin, Jessica Fewkes, Rosella Pinckney, Andrew Whiting, and Paul Chazot. 2025. "A Comparative Study of a Potent CNS-Permeable RARβ-Modulator, Ellorarxine, in Neurons, Glia and Microglia Cells In Vitro" International Journal of Molecular Sciences 26, no. 8: 3551. https://doi.org/10.3390/ijms26083551
APA StyleZhang, Y., Gailloud, L., Shin, A., Fewkes, J., Pinckney, R., Whiting, A., & Chazot, P. (2025). A Comparative Study of a Potent CNS-Permeable RARβ-Modulator, Ellorarxine, in Neurons, Glia and Microglia Cells In Vitro. International Journal of Molecular Sciences, 26(8), 3551. https://doi.org/10.3390/ijms26083551







