Advances in Anthelmintic Target Identification
Abstract
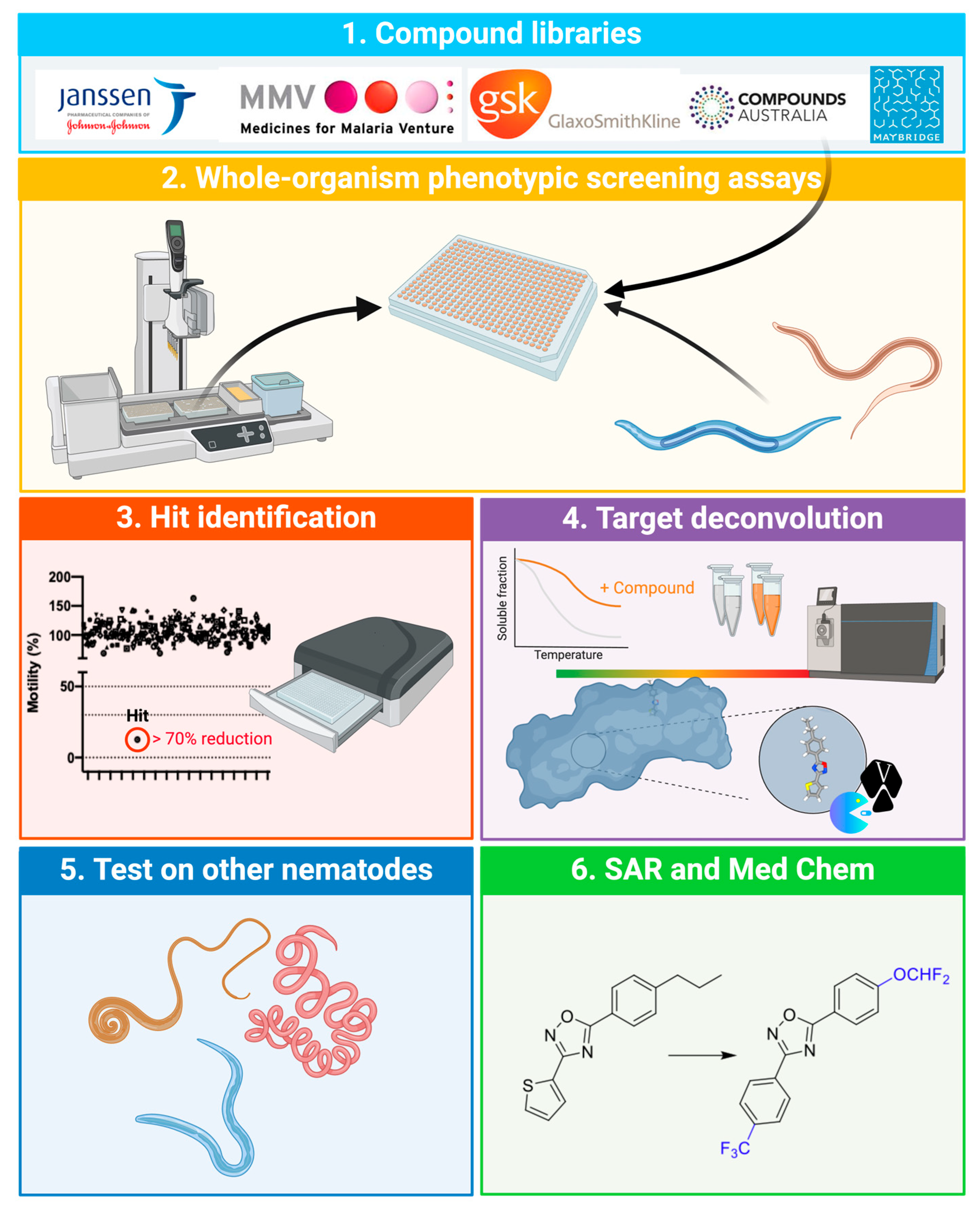
1. Importance of Parasitic Nematodes and Impetus for Drug Discovery
2. Tools for Drug Target Deconvolution
2.1. Proteomic Approaches
2.2. Chemical Probe-Based Approaches
2.3. Genetic Approaches
2.3.1. Resistance Assays
2.3.2. RNA Interference (RNAi)
2.3.3. CRISPR/Cas9
3. Challenges Associated with Using C. elegans as a Model for Nematocide Discovery and Target Deconvolution
4. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fenwick, A. The global burden of neglected tropical diseases. Public Health 2012, 126, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, J.L. Global food security: The impact of veterinary parasites and parasitologists. Vet. Parasitol. 2013, 195, 233–248. [Google Scholar] [CrossRef]
- Casulli, A. New global targets for NTDs in the WHO roadmap 2021–2030. PLoS Negl. Trop. Dis. 2021, 15, e0009373. [Google Scholar] [CrossRef] [PubMed]
- Montresor, A.; Mwinzi, P.; Mupfasoni, D.; Garba, A. Reduction in DALYs lost due to soil-transmitted helminthiases and schistosomiasis from 2000 to 2019 is parallel to the increase in coverage of the global control programmes. PLoS Negl. Trop. Dis. 2022, 16, e0010575. [Google Scholar] [CrossRef]
- Shephard, R.; Ware, J.W.; Blomfield, B.; Neithe, G. Priority List of Endemic Diseases for the Red Meat Industry—2022 Update; Meat & Livestock Australia Limited: North Sydney, Australia, 2022. [Google Scholar]
- World Health Organization. Soil-Transmitted Helminth Infections. Fact Sheet. 2023. Available online: http://who.int/data/gho/data/themes/topics/soil-transmitted-helminthiases (accessed on 19 October 2023).
- Institute for Health Metrics and Evaluation. Global Burden of Disease (GBD) Methods Appendices 2021; Institute for Health Metrics and Evaluation: Seattle, WA, USA, 2021. [Google Scholar]
- Charlier, J.; Rinaldi, L.; Musella, V.; Ploeger, H.W.; Chartier, C.; Vineer, H.R.; Hinney, B.; von Samson-Himmelstjerna, G.; Băcescu, B.; Mickiewicz, M.; et al. Initial assessment of the economic burden of major parasitic helminth infections to the ruminant livestock industry in Europe. Prev. Vet. Med. 2020, 182, 105103. [Google Scholar] [CrossRef] [PubMed]
- Selzer, P.M.; Epe, C. Antiparasitics in animal health: Quo vadis? Trends Parasitol. 2021, 37, 77–89. [Google Scholar] [CrossRef]
- Kearney, P.E.; Murray, P.J.; Hoy, J.M.; Hohenhaus, M.; Kotze, A. The ‘Toolbox’ of strategies for managing Haemonchus contortus in goats: What’s in and what’s out. Vet. Parasitol. 2016, 220, 93–107. [Google Scholar] [CrossRef]
- Waller, P.J. Towards sustainable nematode parasite control of livestock. Vet. Parasitol. 1993, 48, 295–309. [Google Scholar] [CrossRef]
- Sargison, N.D. Keys to solving health problems in small ruminants: Anthelmintic resistance as a threat to sustainable nematode control. Small Rumin. Res. 2016, 142, 11–15. [Google Scholar] [CrossRef]
- Charlier, J.; Bartley, D.J.; Sotiraki, S.; Martinez-Valladares, M.; Claerebout, E.; von Samson-Himmelstjerna, G.; Thamsborg, S.; Hoste, H.; Morgan, E.; Rinaldi, L. Anthelmintic resistance in ruminants: Challenges and solutions. Adv. Parasitol. 2022, 115, 171–227. [Google Scholar]
- Tinkler, S.H. Preventive chemotherapy and anthelmintic resistance of soil-transmitted helminths—Can we learn nothing from veterinary medicine? One Health 2020, 9, 100106. [Google Scholar] [CrossRef] [PubMed]
- Kotze, A.C.; Hunt, P.W. The current status and outlook for insecticide, acaricide, and anthelmintic resistances across the Australian ruminant livestock industries: Assessing the threat these resistances pose to the livestock sector. Aust. Vet. J. 2023, 101, 321–333. [Google Scholar] [CrossRef]
- Jiao, Y.; Preston, S.; Hofmann, A.; Taki, A.; Baell, J.; Chang, B.C.H.; Jabbar, A.; Gasser, R.B. A perspective on the discovery of selected compounds with anthelmintic activity against the barber’s pole worm—Where to from here? Adv. Parasitol. 2020, 108, 1–45. [Google Scholar] [PubMed]
- Herath, H.; Taki, A.C.; Rostami, A.; Jabbar, A.; Keiser, J.; Geary, T.G.; Gasser, R.B. Whole-organism phenotypic screening methods used in early-phase anthelmintic drug discovery. Biotechnol. Adv. 2022, 57, 107937. [Google Scholar] [CrossRef]
- Herath, H.; Taki, A.C.; Sleebs, B.E.; Hofmann, A.; Nguyen, N.; Preston, S.; Davis, R.A.; Jabbar, A.; Gasser, R.B. Advances in the discovery and development of anthelmintics by harnessing natural product scaffolds. Adv. Parasitol. 2021, 111, 203–251. [Google Scholar] [PubMed]
- Harrington, S.; Knox, J.J.; Burns, A.R.; Choo, K.L.; Au, A.; Kitner, M.; Haeberli, C.; Pyche, J.; D’amata, C.; Kim, Y.-H.; et al. Egg-laying and locomotory screens with C. elegans yield a nematode-selective small molecule stimulator of neurotransmitter release. Commun. Biol. 2022, 5, 865. [Google Scholar] [CrossRef]
- Harrington, S.; Pyche, J.; Burns, A.R.; Spalholz, T.; Ryan, K.T.; Baker, R.J.; Ching, J.; Rufener, L.; Lautens, M.; Kulke, D.; et al. Nemacol is a small molecule inhibitor of C. elegans vesicular acetylcholine transporter with anthelmintic potential. Nat. Commun. 2023, 14, 1816. [Google Scholar] [CrossRef]
- Taki, A.C.; Wang, T.; Nguyen, N.N.; Ang, C.S.; Leeming, M.G.; Nie, S.; Byrne, J.J.; Young, N.D.; Zheng, Y.; Ma, G.; et al. Thermal proteome profiling reveals Haemonchus orphan protein HCO_011565 as a target of the nematocidal small molecule UMW-868. Front. Pharmacol. 2022, 13, 1014804. [Google Scholar] [CrossRef]
- Shanley, H.T.; Taki, A.C.; Nguyen, N.; Wang, T.; Byrne, J.J.; Ang, C.S.; Leeming, M.G.; Williamson, N.; Chang, B.C.; Jabbar, A.; et al. Comparative structure-activity and target exploration of 1,2-diphenylethynes in Haemonchus contortus and Caenorhabditis elegans. Int. J. Parasitol. Drugs Drug Resist. 2024, 25, 100534. [Google Scholar] [CrossRef]
- Shanley, H.T.; Taki, A.C.; Byrne, J.J.; Jabbar, A.; Wells, T.N.C.; Samby, K.; Boag, P.R.; Nguyen, N.; Sleebs, B.E.; Gasser, R.B. A high-throughput phenotypic screen of the ‘Pandemic Response Box’ identifies a quinoline derivative with significant anthelmintic activity. Pharmaceuticals 2022, 15, 257. [Google Scholar] [CrossRef]
- Shanley, H.T.; Taki, A.C.; Nguyen, N.; Wang, T.; Byrne, J.J.; Ang, C.S.; Leeming, M.G.; Nie, S.; Williamson, N.; Zheng, Y.; et al. Structure-activity relationship and target prediction for ABX464 analogues in Caenorhabditis elegans. Bioorg. Med. Chem. 2024, 98, 117540. [Google Scholar] [CrossRef]
- Shanley, H.T.; Taki, A.C.; Nguyen, N.; Wang, T.; Byrne, J.J.; Ang, C.S.; Leeming, M.G.; Nie, S.; Williamson, N.; Zheng, Y.; et al. Structure-activity relationship and target investigation of 2-aryl quinolines with nematocidal activity. Int. J. Parasitol. Drugs Drug Resist. 2024, 24, 100522. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Song, J.; Young, N.D.; Chang, B.C.H.; Korhonen, P.K.; Campos, T.L.; Liu, H.; Gasser, R.B. ‘Bingo’—A large language model- and graph neural network-based workflow for the prediction of essential genes from protein data. Brief. Bioinform. 2023, 25, bbad472. [Google Scholar] [CrossRef]
- Wang, T.; Gasser, R.B. Prospects of using high-throughput proteomics to underpin the discovery of animal host-nematode interactions. Pathogens 2021, 10, 825. [Google Scholar] [CrossRef] [PubMed]
- Quinzo, M.J.; Perteguer, M.J.; Brindley, P.J.; Loukas, A.; Sotillo, J. Transgenesis in parasitic helminths: A brief history and prospects for the future. Parasit. Vectors 2022, 15, 110. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.A.; Wu, C.H.; Levine, J.H.; Berg, H. Levamisole-resistant mutants of the nematode Caenorhabditis elegans appear to lack pharmacological acetylcholine receptors. Neuroscience 1980, 5, 967–989. [Google Scholar] [CrossRef]
- Dent, J.A.; Davis, M.W.; Avery, L. avr-15 encodes a chloride channel subunit that mediates inhibitory glutamatergic neurotransmission and ivermectin sensitivity in Caenorhabditis elegans. EMBO J. 1997, 16, 5867–5879. [Google Scholar] [CrossRef]
- Kaminsky, R.; Ducray, P.; Jung, M.; Clover, R.; Rufener, L.; Bouvier, J.; Weber, S.S.; Wenger, A.; Wieland-Berghausen, S.; Goebel, T.; et al. A new class of anthelmintics effective against drug-resistant nematodes. Nature 2008, 452, 176–180. [Google Scholar] [CrossRef]
- Geary, T.G.; Sakanari, J.A.; Caffrey, C.R. Anthelmintic drug discovery: Into the future. J. Parasitol. 2015, 101, 125–133. [Google Scholar] [CrossRef]
- Castelletto, M.L.; Gang, S.S.; Hallem, E.A. Recent advances in functional genomics for parasitic nematodes of mammals. J. Exp. Biol. 2020, 223, jeb206482. [Google Scholar] [CrossRef]
- Ma, G.; Wang, T.; Korhonen, P.K.; Hofmann, A.; Sternberg, P.W.; Young, N.D.; Gasser, R.B. Elucidating the molecular and developmental biology of parasitic nematodes: Moving to a multi-omics paradigm. Adv. Parasitol. 2020, 108, 175–229. [Google Scholar] [PubMed]
- Nixon, S.A.; Welz, C.; Woods, D.J.; Costa-Junior, L.; Zamanian, M.; Martin, R.J. Where are all the anthelmintics? Challenges and opportunities on the path to new anthelmintics. Int. J. Parasitol. Drugs Drug Resist. 2020, 14, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Lomenick, B.; Hao, R.; Jonai, N.; Chin, R.M.; Aghajan, M.; Warburton, S.; Wang, J.; Wu, R.P.; Gomez, F.; Loo, J.A.; et al. Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 2009, 106, 21984–21989. [Google Scholar] [CrossRef] [PubMed]
- Lomenick, B.; Jung, G.; Wohlschlegel, J.A.; Huang, J. Target identification using drug affinity responsive target stability (DARTS). Curr. Protoc. Chem. Biol. 2011, 3, 163–180. [Google Scholar] [CrossRef]
- Ziegler, S.; Pries, V.; Hedberg, C.; Waldmann, H. Target identification for small bioactive molecules: Finding the needle in the haystack. Angew. Chem. Int. Ed. Engl. 2013, 52, 2744–2792. [Google Scholar] [CrossRef]
- Jafari, R.; Almqvist, H.; Axelsson, H.; Ignatushchenko, M.; Lundbäck, T.; Nordlund, P.; Molina, D.M. The cellular thermal shift assay for evaluating drug-target interactions in cells. Nat. Protoc. 2014, 9, 2100–2122. [Google Scholar] [CrossRef]
- Dziekan, J.M.; Yu, H.; Chen, D.; Dai, L.; Wirjanata, G.; Larsson, A.; Prabhu, N.; Sobota, R.M.; Bozdech, Z.; Nordlund, P. Identifying purine nucleoside phosphorylase as the target of quinine using cellular thermal shift assay. Sci. Transl. Med. 2019, 11, eaau3174. [Google Scholar] [CrossRef]
- Dziekan, J.M.; Wirjanata, G.; Dai, L.; Go, K.D.; Yu, H.; Lim, Y.T.; Chen, L.; Wang, L.C.; Puspita, B.; Prabhu, N.; et al. Cellular thermal shift assay for the identification of drug-target interactions in the Plasmodium falciparum proteome. Nat. Protoc. 2020, 15, 1881–1921. [Google Scholar] [CrossRef]
- Friman, T. Mass spectrometry-based Cellular Thermal Shift Assay (CETSA®) for target deconvolution in phenotypic drug discovery. Bioorg. Med. Chem. 2020, 28, 115174. [Google Scholar] [CrossRef]
- Zhang, M.; Li, B.; Tian, J. Mitochondrial targets exploration of epigallocatechin gallate and theaflavin in regards to differences in stress protection under different temperatures. J. Nutr. Biochem. 2023, 119, 109400. [Google Scholar] [CrossRef]
- Franken, H.; Mathieson, T.; Childs, D.; Sweetman, G.M.; Werner, T.; Tögel, I.; Doce, C.; Gade, S.; Bantscheff, M.; Drewes, G.; et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 2015, 10, 1567–1593. [Google Scholar] [CrossRef] [PubMed]
- Gurumayum, S.; Jiang, P.; Hao, X.; Campos, T.L.; Young, N.D.; Korhonen, P.K.; Gasser, R.B.; Bork, P.; Zhao, X.-M.; He, L.-J.; et al. OGEE v3: Online GEne Essentiality database with increased coverage of organisms and human cell lines. Nucleic Acids Res. 2021, 49, D998–D1003. [Google Scholar] [CrossRef]
- Yu, C.; Chen, X.; Xu, W.; Li, S.; Chai, Q.; Zhang, Y. Solvent-induced proteome profiling for proteomic quantitation and target discovery of small molecular drugs. Proteomics 2023, 23, e2200281. [Google Scholar] [CrossRef]
- Bravo, P.; Bizzarri, L.; Steinbrunn, D.; Lohse, J.; Hirsch, A.K.H.; Mäser, P.; Rottmann, M.; Hahne, H. Integral Solvent-Induced Protein Precipitation for Target-Engagement Studies in Plasmodium falciparum. ACS Infect. Dis. 2024, 10, 4073–4086. [Google Scholar] [CrossRef]
- Creek, D.; Giannangelo, C.; Challis, M.P.; Siddiqui, G.; Edgar, R.; Malcolm, T.R.; Webb, C.T.; Drinkwater, N.; Vinh, N.; MacRaild, C.; et al. Chemoproteomics validates selective targeting of Plasmodium M1 alanyl aminopeptidase as an antimalarial strategy. eLife 2024, 13, e92990. [Google Scholar]
- Ursu, A.; Waldmann, H. Hide and seek: Identification and confirmation of small molecule protein targets. Bioorg. Med. Chem. Lett. 2015, 25, 3079–3086. [Google Scholar] [CrossRef] [PubMed]
- Him, N.A.; Gillan, V.; Emes, R.D.; Maitland, K.; Devaney, E. Hsp-90 and the biology of nematodes. BMC Evol. Biol. 2009, 9, 254. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.Y.; Corson, T.W. Small molecule target identification using photo-affinity chromatography. Methods Enzymol. 2019, 622, 347–374. [Google Scholar]
- Gaetani, M.; Sabatier, P.; Saei, A.A.; Beusch, C.M.; Yang, Z.; Lundstrom, S.L.; Zubarev, R.A. Proteome Integral Solubility Alteration: A high-throughput proteomics assay for target deconvolution. J. Proteome Res. 2019, 18, 4027–4037. [Google Scholar] [CrossRef]
- Artal-Sanz, M.; de Jong, L.; Tavernarakis, N. Caenorhabditis elegans: A versatile platform for drug discovery. Biotechnol. J. 2006, 1, 1405–1418. [Google Scholar] [CrossRef]
- Burns, A.R.; Kwok, T.C.; Howard, A.; Houston, E.; Johanson, K.; Chan, A.; Cutler, S.R.; McCourt, P.; Roy, P.J. High-throughput screening of small molecules for bioactivity and target identification in Caenorhabditis elegans. Nat. Protoc. 2006, 1, 1906–1914. [Google Scholar] [CrossRef] [PubMed]
- Lesa, G.M. Isolation of Caenorhabditis elegans gene knockouts by PCR screening of chemically mutagenized libraries. Nat. Protoc. 2006, 1, 2231–2240. [Google Scholar] [CrossRef]
- Kutscher, L.M.; Shaham, S. Forward and reverse mutagenesis in C. elegans. WormBook, pp. 1–26. Available online: http://www.wormbook.org/chapters/www_frmutagenesis/frmutagenesis.html (accessed on 12 April 2025).
- Kwok, T.C.; Ricker, N.; Fraser, R.; Chan, A.W.; Burns, A.; Stanley, E.F.; McCourt, P.; Cutler, S.R.; Roy, P.J. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature 2006, 441, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Geldhof, P.; Murray, L.; Couthier, A.; Gilleard, J.S.; McLauchlan, G.; Knox, D.P.; Britton, C. Testing the efficacy of RNA interference in Haemonchus contortus. Int. J. Parasitol. 2006, 36, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Geldhof, P.; Visser, A.; Clark, D.; Saunders, G.; Britton, C.; Gilleard, J.; Berriman, M.; Knox, D. RNA interference in parasitic helminths: Current situation, potential pitfalls and future prospects. Parasitology 2007, 134, 609–619. [Google Scholar] [CrossRef]
- Kaletta, T.; Hengartner, M.O. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 2006, 5, 387–398. [Google Scholar] [CrossRef]
- Ashrafi, K.; Chang, F.Y.; Watts, J.L.; Fraser, A.G.; Kamath, R.S.; Ahringer, J.; Ruvkun, G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 2003, 421, 268–272. [Google Scholar] [CrossRef]
- Hou, B.; Hai, Y.; Buyin, B.; Hasi, S. Research progress and limitation analysis of RNA interference in Haemonchus contortus in China. Front. Vet. Sci. 2023, 10, 1079676. [Google Scholar] [CrossRef]
- Blanchard, A.; Guegnard, F.; Charvet, C.L.; Crisford, A.; Courtot, E.; Sauve, C.; Harmache, A.; Duguet, T.; O’Connor, V.; Castagnone-Sereno, P.; et al. Deciphering the molecular determinants of cholinergic anthelmintic sensitivity in nematodes: When novel functional validation approaches highlight major differences between the model Caenorhabditis elegans and parasitic species. PLoS Pathog. 2018, 14, e1006996. [Google Scholar] [CrossRef]
- Kotze, A.C.; Bagnall, N.H. RNA interference in Haemonchus contortus: Suppression of beta-tubulin gene expression in L3, L4 and adult worms in vitro. Mol. Biochem. Parasitol. 2006, 145, 101–110. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing: The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Fenk, L.A.; de Bono, M. Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res. 2013, 41, e193. [Google Scholar] [CrossRef] [PubMed]
- Waaijers, S.; Portegijs, V.; Kerver, J.; Lemmens, B.B.; Tijsterman, M.; van den Heuvel, S.; Boxem, M. CRISPR/Cas9-targeted mutagenesis in Caenorhabditis elegans. Genetics 2013, 195, 1187–1191. [Google Scholar] [CrossRef]
- Gang, S.S.; Castelletto, M.L.; Bryant, A.S.; Yang, E.; Mancuso, N.; Lopez, J.B.; Pellegrini, M.; Hallem, E.A. Targeted mutagenesis in a human-parasitic nematode. PLoS Pathog. 2017, 13, e1006675. [Google Scholar] [CrossRef]
- Liu, C.; Grote, A.; Ghedin, E.; Unnasch, T.R. CRISPR-mediated transfection of Brugia malayi. PLoS Negl. Trop. Dis. 2020, 14, e0008627. [Google Scholar] [CrossRef]
- Hagen, J.; Ghosh, S.; Sarkies, P.; Selkirk, M.E. Gene editing in the nematode parasite Nippostrongylus brasiliensis using extracellular vesicles to deliver active Cas9/guide RNA complexes. Front. Parasitol. 2023, 2, 1071738. [Google Scholar] [CrossRef] [PubMed]
- Jost, M.; Weissman, J.S. CRISPR approaches to small molecule target identification. ACS Chem. Biol. 2018, 13, 366–375. [Google Scholar] [CrossRef]
- Geary, T.G.; Thompson, D.P. Caenorhabditis elegans: How good a model for veterinary parasites? Vet. Parasitol. 2001, 101, 371–386. [Google Scholar] [CrossRef]
- Burns, A.R.; Luciani, G.M.; Musso, G.; Bagg, R.; Yeo, M.; Zhang, Y.; Rajendran, L.; Glavin, J.; Hunter, R.; Redman, E.; et al. Caenorhabditis elegans is a useful model for anthelmintic discovery. Nat. Commun. 2015, 6, 7485. [Google Scholar] [CrossRef]
- Zheng, Y.; Young, N.D.; Campos, T.L.; Korhonen, P.K.; Wang, T.; Sumanam, S.B.; Taki, A.C.; Byrne, J.J.; Chang, B.C.; Song, J.; et al. Chromosome-contiguous genome for the Haecon-5 strain of Haemonchus contortus reveals marked genetic variability and enables the discovery of essential gene candidates. Int. J. Parasitol. 2024, 54, 705–715. [Google Scholar] [CrossRef] [PubMed]
| Category | Techniques | Principle | Advantages | Disadvantages |
|---|---|---|---|---|
| (i) Proteomic approaches | ||||
| DARTS (drug affinity responsive target stability) | Measures protein stability upon drug binding via proteolysis. | No need to modify compounds (label-free); simple setup. | Requires optimisation; prone to false positives; does not work on protease-resistant proteins. | |
| Thermal shift assays: CETSA (cellular thermal shift assay) and TPP (thermal proteome profiling) | Monitors drug-induced thermal stability of proteins. | Label-free; applicable to lysed or live cells; high-throughput potential. | Need high concentrations of compound; may not detect weak binders; data analysis is complex. Prone to detecting binding of non-essential proteins. | |
| SPROX | Uses methionine oxidation to assess drug–protein interactions. | Label-free; unbiased and quantitative; proteome-wide applications. | Only proteins/binding sites with methionine residues are detected. Met oxidation can be heterogeneous, complicating analysis. | |
| iSPP (integral solvent-induced protein precipitation) | Monitors solvent-induced precipitation to detect drug–protein binding. | Label-free; can detect novel interactions; useful for parasites. | Requires careful validation; limited broad application. | |
| (ii) Chemical probe-based approaches | ||||
| Affinity purification (pull-down) | Uses modified/labelled compounds to pull down binding proteins. | Directly isolates genuine binding partners; well-established. | Need compound SAR to apply chemical label. Label prone to false positives unless stringent compound and biological controls are used. Only works on parasite lysate. | |
| Photoaffinity labelling | Uses UV-activated probes to covalently bind targets. | Captures transient interactions; applicable to live cells. | Needs SAR to label compound; UV irradiation can cause non-specific binding; requires irradiation equipment. | |
| Activity-based protein profiling (ABPP) | Uses covalent probes to label active-site residues of enzymes. | High specificity for enzyme targets; detects active proteins. | Limited to enzymes; requires designing reactive and selective probes. | |
| (iii) Genetic approaches | ||||
| Resistance assays | Identifies drug targets by analysing resistance mutations. | Unbiased; powerful for finding essential drug pathways. | Labour-intensive; typically requires chemically induced mutagenesis; mutations can be indirect or a resistance mechanism, e.g., drug efflux pump. | |
| RNA interference (RNAi) | Uses dsRNA to silence genes and assess drug effects. | Efficient in C. elegans; high-throughput screens possible. | Limited effectiveness with RNAi uptake in some parasitic nematodes. | |
| CRISPR/Cas9 | Uses gene editing to knock out or modify target genes. | Highly specific; enables functional validation. | Requires development and optimisation for most parasites. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shanley, H.T.; Wang, T.; Taki, A.C.; Byrne, J.J.; Chang, B.C.H.; Sleebs, B.E.; Gasser, R.B. Advances in Anthelmintic Target Identification. Int. J. Mol. Sci. 2025, 26, 3738. https://doi.org/10.3390/ijms26083738
Shanley HT, Wang T, Taki AC, Byrne JJ, Chang BCH, Sleebs BE, Gasser RB. Advances in Anthelmintic Target Identification. International Journal of Molecular Sciences. 2025; 26(8):3738. https://doi.org/10.3390/ijms26083738
Chicago/Turabian StyleShanley, Harrison T., Tao Wang, Aya C. Taki, Joseph J. Byrne, Bill C. H. Chang, Brad E. Sleebs, and Robin B. Gasser. 2025. "Advances in Anthelmintic Target Identification" International Journal of Molecular Sciences 26, no. 8: 3738. https://doi.org/10.3390/ijms26083738
APA StyleShanley, H. T., Wang, T., Taki, A. C., Byrne, J. J., Chang, B. C. H., Sleebs, B. E., & Gasser, R. B. (2025). Advances in Anthelmintic Target Identification. International Journal of Molecular Sciences, 26(8), 3738. https://doi.org/10.3390/ijms26083738








