The Complementary Role of Morphology in Understanding Microglial Functional Heterogeneity
Abstract
:1. Introduction
2. Microglia Origin and Population Maintenance
3. Methods for Examining Microglial Morphology and Associated Challenges
| Microglia Identification Markers | Characteristics | Merits | Limitations |
|---|---|---|---|
| Ammoniacal silver carbonate staining [69,70] | Developed by Pío del Río-Hortega. It was the first staining protocol to allow visualization of microglia and their differentiation from oligodendrocytes. | Adaptations of this technique have also been used for investigating ciliate protozoan systematics and/or ciliate cortical structure and morphogenesis. | It is limited to early stages of research; lacks detailed morphological analysis. |
| Nissl technique [48] | It was developed by Franz Nissl. Using this staining protocol, he was the first to identify the rod microglia. | The staining was not only used for microglial identification but also for the visualization of neurons. | The staining does not show cellular morphological details. |
| Lectin stains [49] | Lectins are carbohydrate-binding proteins that present specificity for a particular carbohydrate. | For microglia, the most intense staining was observed with GS-1, RCA, WGA, and ConA. It is a technically easy and reliable staining method. | The degree of lectin binding depends on the stage of microglial activation, with resting microglia reacting less to lectins. |
| F4/80 [56] | A cell surface glycoprotein specific to macrophages. | It is expressed in mice (without evidence in humans) and is one of the most specific markers for macrophages and microglia. | Being a macrophage marker, it does not have microglial specificity. |
| CD40 [71,72] | Represents a member of the tumor necrosis factor receptor family and is involved in immune response. | Useful for studying the activation of microglia in various conditions, including Alzheimer’s disease. | Can also be expressed in other immune cells. |
| CD11b [50] | Forms a part of complement receptor 3 and is involved in adhesion processes and uptake of complement-coated molecules. | It is expressed both in the activation and the resting state of microglia. | It is not a specific marker for microglia. It is also present on the membranes of leukocytes. |
| CD80 [73,74] | T-lymphocyte activation antigen CD80; a co-stimulatory molecule for CD28 that is expressed on T cells. | The role of CD80 has mainly been studied in microglia–T cell interactions. | Can also be expressed in other immune cells (antigen-presenting cells, regulatory T cells). |
| TREM2 [50] | Triggering receptor expressed on myeloid cells 2. Controls toll-like receptor 4 signaling. | It has been studied particularly in Alzheimer’s pathology; its alleles represent a genetic risk factor for the development of AD. | Conflicting data in the literature, with studies showing increases in TREM2 in AD and others showing non-significant changes compared to controls. |
| Iba1 [75,76] | A calcium-binding protein involved in microglial activation and motility. | It is one of the most commonly used markers for microglia identification. Iba1 stains more microglia phenotypes such as ramified, activated, amoeboid, or dystrophic microglia. | Plays an important role, especially in activated microglia; thus, it does not distinguish between different stages of microglial activation. |
| CD68 [50] | Cluster of differentiation 68 or macrosialin. It is strongly upregulated during inflammation. | Although it is also expressed to some extent in resting microglia, it is considered a marker of activated phagocytic microglia. Increased expression of this marker has been found in the brains of AD patients. | It can also be identified in infiltrating macrophages. |
| ICAM-1 [77,78,79] | Intercellular Adhesion Molecule 1 is a cell surface glycoprotein involved in leukocyte extravasation and in the interaction of lymphocytes with antigen-presenting cells. | It is useful in the study of microglial cells in various pathological conditions such as axonal injury, the biology of psychiatric disorders, or in models of Alzheimer’s disease. | It is also expressed in the astrocytes and in the endothelial cells of the human brain. |
| Parameter | Exemplification | Research Context | Observations |
|---|---|---|---|
| Area surveilled/cell environment area [9,12,24,27,80,81,82,83] | 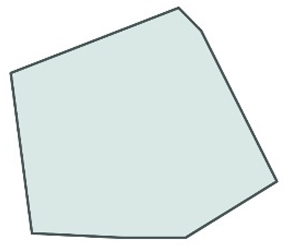 | Used to assess microglia morphology across brain regions, upon activation or in association with neuronal activity. | Cerebellar microglia have a smaller surveilled area. During the dark cycle, microglia are more ramified. An increase in microglial domain volume was observed one day following subarachnoid hemorrhage (SAH). |
| Cell area [12,25] |  | Analyzed in models of brain injury (experimental cerebral ischemia) but also in the normal aging process. | Increases with age. Activation itself increases cell area through soma enlargement. Activated microglia in the cortex have a greater cell area compared to those in the hippocampus. |
| Soma area/cell body area/cell volume [24,25,27,81,84] | 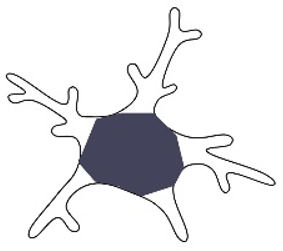 | This parameter is used to morphologically determine microglial activation (LPS-induced inflammation, response to laser lesion), as well as to study aging or inter-regional heterogeneity. | Hippocampal microglia have a smaller soma area compared to cortical microglia. Upon treatment with LPS, microglia become more homogeneous than in the control groups. Soma enlargement occurs with age and injury. Rod-like cells have a greater soma area. |
| Cytoplasm area [81] | 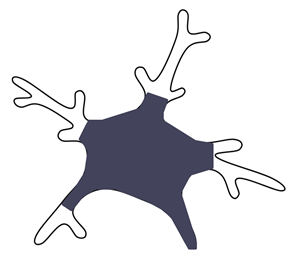 | Used to exemplify activation in the context of LPS-induced inflammation but also inter-regional differences, both in pathology and in physiological conditions. | This is the cell body area associated with the cytoplasmic area of the primary ramifications. Cerebellar microglia have a greater cytoplasm area compared to frontal cortex or striatum. |
| Total length of branch tree [11,12,23,25,54,63,80,85] | 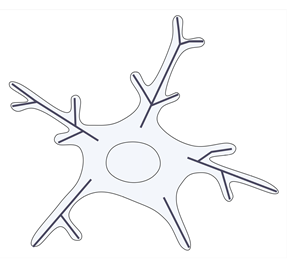 | To quantify the arborization in mouse models of Alzheimer’s disease (APP/PS1 Tg mice), after ischemic stroke and reperfusion, after microglia ablation or depletion, or to exemplify the normal aging process or the changes induced by a certain fixation method. | Decreased skeleton length is observed in activated microglia upon injury. Age differences show a sharp postnatal increase, and then a slight decline in older mice. A greater number upon plunge fixation. Rod-like cells have a similar skeleton length to ramified cells. Microglia from the Nac (nucleus accumbens) and SNr (substantia nigra pars reticulata) exhibit greater process lengths. |
| Total number of processes [12,23,80,86,87] | 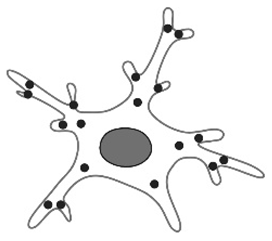 | Used to quantify microglial changes due to fixation or aging, as well as in studies involving microglial depletion or mouse models of Alzheimer’s disease (APP/PS1 Tg mice). | Microglia possess fewer branches after PFA perfusion. A drastic increase in the number of branches occurs immediately postnatally. Fewer branches in circumventricular organs (CVOs). |
| Mean branch length [12,23,80] |  | This parameter is used to quantify microglial changes due to fixation or aging or in mouse models of Alzheimer’s disease (APP/PS1 Tg mice). | Old mice possess fewer short branches. Mean branch length is greatest upon PFA perfusion. |
| Total number of primary filaments [12,23,88,89] | 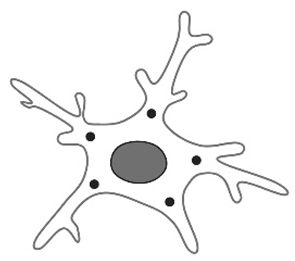 | Primary processes are the initial extensions that arise directly from the cell body (soma) of the microglia. Used to study the phenomenon of aging but also in studies that quantify immediately postnatal morphological changes. This parameter also changes according to the fixation method. | Changes are age-related, starting from immediately postnatal, with an increase in the number of primary processes from P5 to P7. Their numbers differ even within the same brain region (e.g., cerebellum). |
| Total number of secondary filaments [12,23] |  | Secondary processes are branches that emerge from the primary processes. Used to quantify microglial changes due to fixation or aging, being part of the parameters that quantify the complexity of the branching pattern. | There is a steep increase immediately postnatally. A change is also observed with the fixation methods. |
| Total number of end points [12,23,25,54,63,85] | 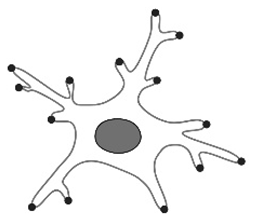 | The parameter is more commonly used as the total number of secondary filaments. Used to quantify microglial changes due to fixation or aging, being part of the parameters that quantify the complexity of the branching pattern. | There is a decrease upon injury (in activated microglia). Drastic increase immediately postnatally, followed by a decrease in aged mice. Amoeboid microglia possess the lowest number of endpoints. |
| Process or soma motility [24,90] | Direction of movement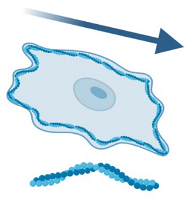 | It serves to quantify the microglial response to a pathological stimulus with age, laser injury, or in the context of neurodegenerative diseases (such as in an APPPS1 mouse model of AD). | Process motility significantly decreases with age (possibly affecting surveillance). However, soma movement increases with age, indicating a change in microglia phenotype. A delayed response to injury is observed in aged mice. Microglial motility is impaired in the presence of Aβ plaques when a focal laser lesion is induced. |
4. Molecular and Cellular Regional Differences in Microglia
4.1. Inter-Regional Morphological Changes
4.2. Intraregional Morphological Changes
| Region of the Brain | Density (Cells/mm3) ± SEM | |
|---|---|---|
| Cortical areas | Neocortex general | 6500 ± 600 [1] |
| Somatosensory cortex | 8892 ± 535 [54], 7400 ± 900 [97] | |
| Frontal cortex | 6200 ± 500 [9], 5823 ± 297 [98], 123,60 [81] | |
| Motor cortex | 8000 ± 800 [97] | |
| Occipital cortex | 6200 ± 300 [9], 5589 ± 223 [98] | |
| Parietal cortex | 6124 ± 319 [98], 6900 ± 500 [9] | |
| Cingulate cortex | 5600 ± 500 [9] | |
| Sensoriomotor cortex | 7900 ± 300 [9] | |
| Visual cortex | 7250 [27] | |
| Auditory cortex | 7500 [27] | |
| Basal ganglia | Neostriatum apical | 9100 ± 1000 [9] |
| Ventral pallidum | 16,500 ± 900 [9] | |
| Hippocampus | Dentate gyrus | 12,000 ± 700 [9] |
| Dorsal hippocampus | 5890 ± 280 [99] | |
| Ventral hippocampus | 5460 ± 300 [99] | |
| Cerrebelum | Cerebellar nuclei | 7300 ± 400 [9], 5579 ± 793 [89] |
| Molecular layer | 2200 ± 300 [9], 1387 ± 108 [89] | |
| Granular layer | 3312 ± 343 [89] | |
| White matter | 3981 ± 780 [89] | |
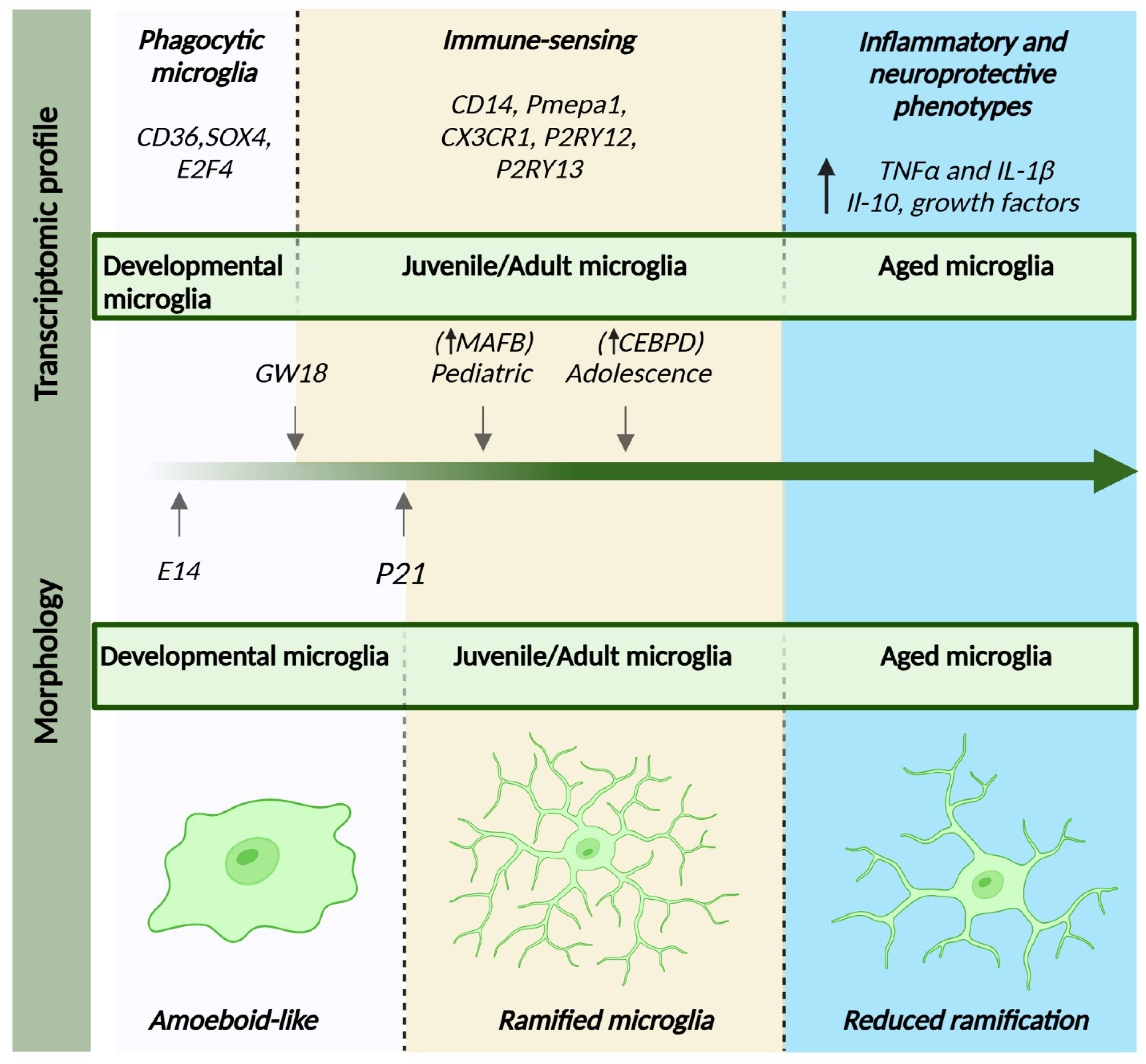
4.3. Transcriptomic Profile Changes
5. Microglia States: From Surveillant to Reactive
6. Age-Related Microglial Changes
7. Microglia in Disease
8. Conclusions
9. Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| scRNA-seq | Single-cell RNA sequencing |
| AD | Alzheimer’s disease |
| CXCL12 | C-X-C motif chemokine 12 |
| CCL2 | Linear dichroism |
| GS-1 | Griffonia simplicifolia 1 |
| RCA | Ricinus communis agglutinin |
| WGA | Wheat germ agglutinin |
| ConA | Concanavalin A |
| F4/80 | EGF module-containing mucin-like receptor |
| CD40 | Cluster of differentiation 40 |
| CD11b | Integrin αM subunit |
| CD80 | Cluster of differentiation 80 |
| TREM2 | Triggering receptor expressed on myeloid cells 2 |
| Iba1 | Ionized calcium-binding adaptor molecule 1 |
| CD68 | Cluster of differentiation 68 |
| ICAM-1 | Intercellular adhesion molecule-1 |
| CX3CR1 | C-X3-C motif chemokine receptor 1 |
| Tmem119 | Transmembrane protein 119 |
| Hexb | Hexosaminidase subunit beta |
| HFD | High-fat diet |
| NAc | Nucleus accumbens |
| SNr | Substantia nigra pars reticulata |
| VTA | Ventral tegmental area |
| SNc | Substantia nigra pars compacta |
| CA3 | Hippocampal cornu ammonis |
| CA1 | Hippocampal cornu ammonis |
| Camp | Cathelin-related antimicrobial peptide |
| Ngp | Neutrophilic granule protein |
| PAMPs | Pathogen-associated molecular patterns |
| Aβ | Amyloid β |
| TNF-α | Tumor necrosis factor-alpha |
| IL-6 | Interleukin-6 |
| IL-10 | Interleukin-10 |
| IL-13 | Interleukin-13 |
| IFN-γ | Interferon-γ |
| TGF-β | Transforming growth factor-β |
| IGF-1 | Insulin-like growth factor-1 |
| CD16 | Cluster of differentiation 16 |
| CD32 | Cluster of differentiation 32 |
| CD40 | Cluster of differentiation 40 |
| CD86 | Cluster of differentiation 86 |
| MHC II | Major histocompatibility complex Class II |
| E14 | Embryonic day 14 |
| Gpnmb | Glycoprotein nonmetastatic melanoma protein B |
| Spp1 | Osteopontin |
| Clec7a | C-type lectin domain family 7 |
| GW | Gestational week |
| P7 | Postnatal day 7 |
| MAFB | Musculoaponeurotic fibrosarcoma oncogene homolog B |
| CEBPD | CCAAT/enhancer-binding protein delta |
| ATP | Adenosine triphosphate |
| DAM | Disease-associated microglia |
| MGn | Microglia neurodegenerative phenotype |
| Trem2 | Triggering receptor expressed on myeloid cells 2 |
| Lpl | Lipoproteinlipase |
| CD33 | Sialic acid-binding Ig-like lectin 3 |
| ApoE | Apolipoprotein E |
| SOD1(G93A) | Superoxide dismutase-1 glycine 93 to alanine |
| ALS | Amyotrophic lateral sclerosis |
| BBB | Blood–brain barrier |
| CVOs | Circumventricular organs |
| CD36 | Platelet glycoprotein 4 |
| SOX4 | Sex-determining region Y-related high mobility group-BOX gene 4 |
| E2F4 | E2F Transcription Factor 4 |
| CD14 | Cluster of differentiation 14 |
| Pmepa1 | Prostate transmembrane protein androgen induced 1 |
| P2RY12 | Purinergic Receptor P2Y12 |
| P2RY13 | P2Y purinoceptor 13 |
| IL-1β | Interleukin-1β |
References
- Nimmerjahn, A.; Kirchhoff, F.; Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005, 308, 1314–1318. [Google Scholar] [CrossRef] [PubMed]
- Wake, H.; Moorhouse, A.J.; Jinno, S.; Kohsaka, S.; Nabekura, J. Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J. Neurosci. 2009, 29, 3974–3980. [Google Scholar] [CrossRef] [PubMed]
- Sierra, A.; Encinas, J.M.; Deudero, J.J.; Chancey, J.H.; Enikolopov, G.; Overstreet-Wadiche, L.S.; Tsirka, S.E.; Maletic-Savatic, M. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 2010, 7, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Paolicelli, R.C.; Gross, C.T. Microglia in development: Linking brain wiring to brain environment. Neuron Glia Biol. 2011, 7, 77–83. [Google Scholar] [CrossRef]
- Marín-Teva, J.L.; Dusart, I.; Colin, C.; Gervais, A.; van Rooijen, N.; Mallat, M. Microglia promote the death of developing Purkinje cells. Neuron 2004, 41, 535–547. [Google Scholar] [CrossRef]
- Gemma, C.; Bachstetter, A.D. The role of microglia in adult hippocampal neurogenesis. Front. Cell Neurosci. 2013, 7, 229. [Google Scholar] [CrossRef]
- Gülke, E.; Gelderblom, M.; Magnus, T. Danger signals in stroke and their role on microglia activation after ischemia. Ther. Adv. Neurol. Disord. 2018, 11, 1756286418774254. [Google Scholar] [CrossRef]
- Fu, R.; Shen, Q.; Xu, P.; Luo, J.J.; Tang, Y. Phagocytosis of microglia in the central nervous system diseases. Mol. Neurobiol. 2014, 49, 1422–1434. [Google Scholar] [CrossRef] [PubMed]
- Lawson, L.J.; Perry, V.H.; Dri, P.; Gordon, S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 1990, 39, 151–170. [Google Scholar] [CrossRef]
- Tan, Y.L.; Yuan, Y.; Tian, L. Microglial regional heterogeneity and its role in the brain. Mol. Psychiatry 2020, 25, 351–367. [Google Scholar] [CrossRef]
- De Biase, L.M.; Schuebel, K.E.; Fusfeld, Z.H.; Jair, K.; Hawes, I.A.; Cimbro, R.; Zhang, H.-Y.; Liu, Q.-R.; Shen, H.; Xi, Z.-X.; et al. Local Cues Establish and Maintain Region-Specific Phenotypes of Basal Ganglia Microglia. Neuron 2017, 95, 341–356.e6. [Google Scholar] [CrossRef] [PubMed]
- Godeanu, S.; Clarke, D.; Stopper, L.; Deftu, A.F.; Popa-Wagner, A.; Bălșeanu, A.T.; Scheller, A.; Catalin, B. Microglial morphology in the somatosensory cortex across lifespan. A quantitative study. Dev. Dyn. 2023, 252, 1113–1129. [Google Scholar] [CrossRef]
- Hammond, T.R.; Dufort, C.; Dissing-Olesen, L.; Giera, S.; Young, A.; Wysoker, A.; Walker, A.J.; Gergits, F.; Segel, M.; Nemesh, J.; et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 2019, 50, 253–271.e6. [Google Scholar] [CrossRef] [PubMed]
- Kracht, L.; Borggrewe, M.; Eskandar, S.; Brouwer, N.; Chuva de Sousa Lopes, S.M.; Laman, J.D.; Scherjon, S.A.; Prins, J.R.; Kooistra, S.M.; Eggen, B.J.L. Human fetal microglia acquire homeostatic immune-sensing properties early in development. Science 2020, 369, 530–537. [Google Scholar] [CrossRef]
- Nedelea, G.; Mușat, M.I.; Mitran, S.I.; Ciorbagiu, M.C.; Cătălin, B. Morphological Differences in Hippocampal Microglia in C57BL/6N Mice with Liver Injury and Depressive-Like Behavior. Curr. Health Sci. J. 2024, 50, 577–584. [Google Scholar] [PubMed]
- Yaqubi, M.; Groh, A.M.; Dorion, M.F.; Afanasiev, E.; Luo, J.X.; Hashemi, H.; Sinha, S.; Kieran, N.W.; Blain, M.; Cui, Q.L.; et al. Analysis of the microglia transcriptome across the human lifespan using single cell RNA sequencing. J. Neuroinflammation 2023, 20, 132. [Google Scholar] [CrossRef]
- Gerrits, E.; Heng, Y.; Boddeke, E.; Eggen, B.J.L. Transcriptional profiling of microglia; current state of the art and future perspectives. Glia 2020, 68, 740–755. [Google Scholar] [CrossRef]
- Pinosanu, L.R.; Capitanescu, B.; Glavan, D.; Godeanu, S.; Cadenas, I.F.N.; Doeppner, T.R.; Hermann, D.M.; Balseanu, A.-T.; Bogdan, C.; Popa-Wagner, A. Neuroglia Cells Transcriptomic in Brain Development, Aging and Neurodegenerative Diseases. Aging Dis. 2023, 14, 63–83. [Google Scholar] [CrossRef]
- Davalos, D.; Grutzendler, J.; Yang, G.; Kim, J.V.; Zuo, Y.; Jung, S.; Littman, D.R.; Dustin, M.L.; Gan, W.-B. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 2005, 8, 752–758. [Google Scholar] [CrossRef]
- Stopper, L.; Bălşeanu, T.A.; Cătălin, B.; Rogoveanu, O.C.; Mogoantă, L.; Scheller, A. Microglia morphology in the physiological and diseased brain—From fixed tissue to in vivo conditions. Rom. J. Morphol. Embryol. 2018, 59, 7–12. [Google Scholar]
- Surugiu, R.; Catalin, B.; Dumbrava, D.; Gresita, A.; Olaru, D.G.; Hermann, D.M.; Popa-Wagner, A. Intracortical Administration of the Complement C3 Receptor Antagonist Trifluoroacetate Modulates Microglia Reaction after Brain Injury. Neural. Plast. 2019, 2019, 1071036. [Google Scholar] [CrossRef]
- Dibaj, P.; Steffens, H.; Nadrigny, F.; Neusch, C.; Kirchhoff, F.; Schomburg, E.D. Long-lasting post-mortem activity of spinal microglia in situ in mice. J. Neurosci. Res. 2010, 88, 2431–2440. [Google Scholar] [CrossRef] [PubMed]
- Cătălin, B.; Stopper, L.; Bălşeanu, T.A.; Scheller, A. The in situ morphology of microglia is highly sensitive to the mode of tissue fixation. J. Chem. Neuroanat. 2017, 86, 59–66. [Google Scholar] [CrossRef]
- Hefendehl, J.K.; Neher, J.J.; Sühs, R.B.; Kohsaka, S.; Skodras, A.; Jucker, M. Homeostatic and injury-induced microglia behavior in the aging brain. Aging Cell 2014, 13, 60–69. [Google Scholar] [CrossRef]
- Leyh, J.; Paeschke, S.; Mages, B.; Michalski, D.; Nowicki, M.; Bechmann, I.; Winter, K. Classification of Microglial Morphological Phenotypes Using Machine Learning. Front. Cell Neurosci. 2021, 15, 701673. [Google Scholar] [CrossRef] [PubMed]
- Ling, E.A.; Wong, W.C. The origin and nature of ramified and amoeboid microglia: A historical review and current concepts. Glia 1993, 7, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, M.; Zettel, M.L.; Ison, J.R.; Allen, P.D.; Majewska, A.K. Effects of aging and sensory loss on glial cells in mouse visual and auditory cortices. Glia 2012, 60, 541–558. [Google Scholar] [CrossRef]
- Streit, W.J.; Sammons, N.W.; Kuhns, A.J.; Sparks, D.L. Dystrophic microglia in the aging human brain. Glia 2004, 45, 208–212. [Google Scholar] [CrossRef]
- Shahidehpour, R.K.; Higdon, R.E.; Crawford, N.G.; Neltner, J.H.; Ighodaro, E.T.; Patel, E.; Price, D.; Nelson, P.T.; Bachstetter, A.D. Dystrophic microglia are associated with neurodegenerative disease and not healthy aging in the human brain. Neurobiol. Aging 2021, 99, 19–27. [Google Scholar] [CrossRef]
- Taylor, S.E.; Morganti-Kossmann, C.; Lifshitz, J.; Ziebell, J.M. Rod microglia: A morphological definition. PLoS ONE 2014, 9, e97096. [Google Scholar] [CrossRef]
- Bachstetter, A.D.; Ighodaro, E.T.; Hassoun, Y.; Aldeiri, D.; Neltner, J.H.; Patel, E.; Abner, E.L.; Nelson, P.T. Rod-shaped microglia morphology is associated with aging in 2 human autopsy series. Neurobiol. Aging 2017, 52, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Holloway, O.G.; Canty, A.J.; King, A.E.; Ziebell, J.M. Rod microglia and their role in neurological diseases. Semin Cell Dev. Biol. 2019, 94, 96–103. [Google Scholar] [CrossRef]
- Spielmeyer, W. Histopathologie des Nervensystems; Springer: Berlin/Heidelberg, Germany, 1922. [Google Scholar]
- Ziebell, J.M.; Taylor, S.E.; Cao, T.; Harrison, J.L.; Lifshitz, J. Rod microglia: Elongation, alignment, and coupling to form trains across the somatosensory cortex after experimental diffuse brain injury. J. Neuroinflammation 2012, 9, 247. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Li, Y.; Fan, G.C. Tissue-Resident Macrophages in the Control of Infection and Resolution of Inflammation. Shock 2021, 55, 14–23. [Google Scholar] [CrossRef]
- Sierra, A.; de Castro, F.; Del Río-Hortega, J.; Rafael Iglesias-Rozas, J.; Garrosa, M.; Kettenmann, H. The “Big-Bang” for modern glial biology: Translation and comments on Pío del Río-Hortega 1919 series of papers on microglia. Glia 2016, 64, 1801–1840. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Gokhan, S.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010, 330, 841–845. [Google Scholar] [CrossRef]
- Ginhoux, F.; Prinz, M. Origin of microglia: Current concepts and past controversies. Cold Spring Harb. Perspect. Biol. 2015, 7, a020537. [Google Scholar] [CrossRef]
- Ginhoux, F.; Lim, S.; Hoeffel, G.; Low, D.; Huber, T. Origin and differentiation of microglia. Front. Cell Neurosci. 2013, 7, 45. [Google Scholar] [CrossRef]
- Monier, A.; Evrard, P.; Gressens, P.; Verney, C. Distribution and differentiation of microglia in the human encephalon during the first two trimesters of gestation. J. Comp. Neurol. 2006, 499, 565–582. [Google Scholar] [CrossRef]
- Monier, A.; Adle-Biassette, H.; Delezoide, A.L.; Evrard, P.; Gressens, P.; Verney, C. Entry and distribution of microglial cells in human embryonic and fetal cerebral cortex. J. Neuropathol. Exp. Neurol. 2007, 66, 372–382. [Google Scholar] [CrossRef]
- Askew, K.; Li, K.; Olmos-Alonso, A.; Garcia-Moreno, F.; Liang, Y.; Richardson, P.; Tipton, T.; Chapman, M.A.; Riecken, K.; Beccari, S.; et al. Coupled Proliferation and Apoptosis Maintain the Rapid Turnover of Microglia in the Adult Brain. Cell Rep. 2017, 18, 391–405. [Google Scholar] [CrossRef] [PubMed]
- Nikodemova, M.; Kimyon, R.S.; De, I.; Small, A.L.; Collier, L.S.; Watters, J.J. Microglial numbers attain adult levels after undergoing a rapid decrease in cell number in the third postnatal week. J. Neuroimmunol. 2015, 278, 280–288. [Google Scholar] [CrossRef]
- Bellver-Landete, V.; Bretheau, F.; Mailhot, B.; Vallières, N.; Lessard, M.; Janelle, M.E.; Vernoux, N.; Tremblay, M.; Fuehrmann, T.; Shoichet, M.S.; et al. Microglia are an essential component of the neuroprotective scar that forms after spinal cord injury. Nat. Commun. 2019, 10, 518. [Google Scholar] [CrossRef]
- Asai, H.; Ikezu, S.; Tsunoda, S.; Medalla, M.; Luebke, J.; Haydar, T.; Wolozin, B.; Butovsky, O.; Kügler, S.; Ikezu, T. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 2015, 18, 1584–1593. [Google Scholar] [CrossRef] [PubMed]
- Lampron, A.; Pimentel-Coelho, P.M.; Rivest, S. Migration of bone marrow-derived cells into the central nervous system in models of neurodegeneration. J. Comp. Neurol. 2013, 521, 3863–3876. [Google Scholar] [CrossRef]
- Del Río-Hortega Bereciartu, J. Pío del Río-Hortega: The Revolution of Glia. Anat. Rec. 2020, 303, 1232–1241. [Google Scholar] [CrossRef]
- Gomes, M.D.M. Franz Nissl (1860–1919), noted neuropsychiatrist and neuropathologist, staining the neuron, but not limiting it. Dement. Neuropsychol. 2019, 13, 352–355. [Google Scholar] [CrossRef]
- Colton, C.A.; Abel, C.; Patchett, J.; Keri, J.; Yao, J. Lectin staining of cultured CNS microglia. J. Histochem. Cytochem. 1992, 40, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Hopperton, K.E.; Mohammad, D.; Trépanier, M.O.; Giuliano, V.; Bazinet, R.P. Markers of microglia in post-mortem brain samples from patients with Alzheimer’s disease: A systematic review. Mol. Psychiatry 2018, 23, 177–198. [Google Scholar] [CrossRef]
- Ito, D.; Imai, Y.; Ohsawa, K.; Nakajima, K.; Fukuuchi, Y.; Kohsaka, S. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res. Mol. Brain Res. 1998, 57, 1–9. [Google Scholar] [CrossRef]
- Stankov, A.; Belakaposka-Srpanova, V.; Bitoljanu, N.; Cakar, L.; Cakar, Z.; Rosoklija, G. Visualisation of Microglia with the use of Immunohistochemical Double Staining Method for CD-68 and Iba-1 of Cerebral Tissue Samples in Cases of Brain Contusions. Prilozi 2015, 36, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.J.; Xie, H.; Zhang, C.Y.; Qin, H.F.; Zeng, X.W.; Lou, H.; Zhang, L.; Xu, G.-T.; Zhang, J.-F.; Xu, G.-X. Is Iba-1 protein expression a sensitive marker for microglia activation in experimental diabetic retinopathy? Int. J. Ophthalmol. 2021, 14, 200–208. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, L.; Zhang, J.; Fariss, R.N.; Ma, W.; Kretschmer, F.; Wang, M.; Qian, H.; Badea, T.C.; Diamond, J.S.; et al. Requirement for Microglia for the Maintenance of Synaptic Function and Integrity in the Mature Retina. J. Neurosci. 2016, 36, 2827–2842. [Google Scholar] [CrossRef]
- Shapiro, L.A.; Perez, Z.D.; Foresti, M.L.; Arisi, G.M.; Ribak, C.E. Morphological and ultrastructural features of Iba1-immunolabeled microglial cells in the hippocampal dentate gyrus. Brain Res. 2009, 1266, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Jurga, A.M.; Paleczna, M.; Kuter, K.Z. Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front. Cell Neurosci. 2020, 14, 198. [Google Scholar] [CrossRef]
- Jung, S.; Aliberti, J.; Graemmel, P.; Sunshine, M.J.; Kreutzberg, G.W.; Sher, A.; Littman, D.R. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol. 2000, 20, 4106–4114. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, J.; Littlewood, T.; Evan, G.I.; Soucek, L. The estrogen receptor fusion system in mouse models: A reversible switch. Cold Spring Harb. Protoc. 2015, 2015, 227–234. [Google Scholar] [CrossRef]
- Faust, T.E.; Feinberg, P.A.; O’Connor, C.; Kawaguchi, R.; Chan, A.; Strasburger, H.; Frosch, M.; Boyle, M.A.; Masuda, T.; Amann, L.; et al. A comparative analysis of microglial inducible Cre lines. Cell Rep. 2023, 42, 113031. [Google Scholar] [CrossRef]
- Kaiser, T.; Feng, G. Tmem119-EGFP and Tmem119-CreERT2 Transgenic Mice for Labeling and Manipulating Microglia. eNeuro 2019, 6. [Google Scholar] [CrossRef]
- Lee, C.Y.D.; Daggett, A.; Gu, X.; Jiang, L.L.; Langfelder, P.; Li, X.; Wang, N.; Zhao, Y.; Park, C.S.; Cooper, Y.; et al. Elevated TREM2 Gene Dosage Reprograms Microglia Responsivity and Ameliorates Pathological Phenotypes in Alzheimer’s Disease Models. Neuron 2018, 97, 1032–1048.e5. [Google Scholar] [CrossRef]
- Burgess, M.; Wicks, K.; Gardasevic, M.; Mace, K.A. Cx3CR1 Expression Identifies Distinct Macrophage Populations That Contribute Differentially to Inflammation and Repair. Immunohorizons 2019, 3, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Young, K.; Morrison, H. Quantifying Microglia Morphology from Photomicrographs of Immunohistochemistry Prepared Tissue Using ImageJ. J. Vis. Exp. 2018, 57648. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Courtney, J.M.; Morris, G.P.; Cleary, E.M.; Howells, D.W.; Sutherland, B.A. Automated Quantification of Multiple Cell Types in Fluorescently Labeled Whole Mouse Brain Sections Using QuPath. Bio. Protoc. 2022, 12, e4459. [Google Scholar] [CrossRef]
- Althammer, F.; Ferreira-Neto, H.C.; Rubaharan, M.; Roy, R.K.; Patel, A.A.; Murphy, A.; Cox, D.N.; Stern, J.E. Three-dimensional morphometric analysis reveals time-dependent structural changes in microglia and astrocytes in the central amygdala and hypothalamic paraventricular nucleus of heart failure rats. J. Neuroinflammation 2020, 17, 221. [Google Scholar] [CrossRef] [PubMed]
- York, E.M.; LeDue, J.M.; Bernier, L.P.; MacVicar, B.A. 3DMorph Automatic Analysis of Microglial Morphology in Three Dimensions from Ex Vivo and In Vivo Imaging. eNeuro 2018, 5, ENEURO.0266-18.2018. [Google Scholar] [CrossRef]
- Clarke, D.; Crombag, H.S.; Hall, C.N. An open-source pipeline for analysing changes in microglial morphology. Open Biol. 2021, 11, 210045. [Google Scholar] [CrossRef]
- Aufderheide, K.J. Refinements of and commentary on the silver staining techniques of Fernández-Galiano. Biotech. Histochem. 2016, 91, 352–356. [Google Scholar] [CrossRef]
- Tremblay, M.; Lecours, C.; Samson, L.; Sánchez-Zafra, V.; Sierra, A. From the Cajal alumni Achúcarro and Río-Hortega to the rediscovery of never-resting microglia. Front. Neuroanat. 2015, 9, 45. [Google Scholar] [CrossRef]
- Togo, T.; Akiyama, H.; Kondo, H.; Ikeda, K.; Kato, M.; Iseki, E.; Kosaka, K. Expression of CD40 in the brain of Alzheimer’s disease and other neurological diseases. Brain Res. 2000, 885, 117–121. [Google Scholar] [CrossRef]
- Ma, D.Y.; Clark, E.A. The role of CD40 and CD154/CD40L in dendritic cells. Semin Immunol. 2009, 21, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Trzupek, D.; Dunstan, M.; Cutler, A.J.; Lee, M.; Godfrey, L.; Jarvis, L.; Rainbow, D.B.; Aschenbrenner, D.; Jones, J.L.; Uhlig, H.H.; et al. Discovery of CD80 and CD86 as recent activation markers on regulatory T cells by protein-RNA single-cell analysis. Genome. Med. 2020, 12, 55. [Google Scholar] [CrossRef]
- van Meerten, T.; Hagenbeek, A. Novel antibodies against follicular non-Hodgkin’s lymphoma. Best Pract. Res. Clin. Haematol. 2011, 24, 231–256. [Google Scholar] [CrossRef] [PubMed]
- Lier, J.; Streit, W.J.; Bechmann, I. Beyond Activation: Characterizing Microglial Functional Phenotypes. Cells 2021, 10, 2236. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.G.; Lue, L.-F. Immune phenotypes of microglia in human neurodegenerative disease: Challenges to detecting microglial polarization in human brains. Alzheimer’s Res. Ther. 2015, 7, 56. [Google Scholar] [CrossRef]
- Müller, N. The Role of Intercellular Adhesion Molecule-1 in the Pathogenesis of Psychiatric Disorders. Front. Pharmacol. 2019, 10, 1251. [Google Scholar] [CrossRef]
- Werner, A.; Kloss, C.U.; Walter, J.; Kreutzberg, G.W.; Raivich, G. Intercellular adhesion molecule-1 (ICAM-1) in the mouse facial motor nucleus after axonal injury and during regeneration. J. Neurocytol. 1998, 27, 219–232. [Google Scholar] [CrossRef]
- Goswami, S.; Biswas, S.C. Intercellular adhesion molecule-1 reprograms microglia to improve cognitive functions by inhibiting ERK/STAT3 signalling pathway in a model of Alzheimer’s disease. bioRxiv 2024. [Google Scholar] [CrossRef]
- Baron, R.; Babcock, A.A.; Nemirovsky, A.; Finsen, B.; Monsonego, A. Accelerated microglial pathology is associated with Aβ plaques in mouse models of Alzheimer’s disease. Aging Cell 2014, 13, 584–595. [Google Scholar] [CrossRef]
- Verdonk, F.; Roux, P.; Flamant, P.; Fiette, L.; Bozza, F.A.; Simard, S.; Lemaire, M.; Plaud, B.; Shorte, S.L.; Sharshar, T.; et al. Phenotypic clustering: A novel method for microglial morphology analysis. J. Neuroinflammation 2016, 13, 153. [Google Scholar] [CrossRef]
- Plog, B.A.; Moll, K.M.; Kang, H.; Iliff, J.J.; Dashnaw, M.L.; Nedergaard, M.; Vates, G.E. A novel technique for morphometric quantification of subarachnoid hemorrhage-induced microglia activation. J. Neurosci. Methods 2014, 229, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Steffens, S.K.; Stenberg, T.H.; Wigren, H.M. Alterations in microglial morphology concentrate in the habitual sleeping period of the mouse. Glia 2023, 71, 366–376. [Google Scholar] [CrossRef]
- Kozlowski, C.; Weimer, R.M. An automated method to quantify microglia morphology and application to monitor activation state longitudinally in vivo. PLoS ONE 2012, 7, e31814. [Google Scholar] [CrossRef] [PubMed]
- Morrison, H.W.; Filosa, J.A. A quantitative spatiotemporal analysis of microglia morphology during ischemic stroke and reperfusion. J. Neuroinflammation 2013, 10, 4. [Google Scholar] [CrossRef]
- Takagi, S.; Murayama, S.; Torii, K.; Takemura-Morita, S.; Kurganov, E.; Nagaoka, S.; Wanaka, A.; Miyata, S. Depletion of microglia and macrophages with clodronate liposomes attenuates zymosan-induced Fos expression and hypothermia in the adult mouse. J. Neuroimmunol. 2020, 344, 577244. [Google Scholar] [CrossRef] [PubMed]
- Godeanu, S.; Mușat, M.I.; Scheller, A.; Osiac, E.; Cătălin, B. Minimal differences observed when comparing the morphological profiling of microglia obtained by confocal laser scanning and optical sectioning microscopy. Front. Neuroanat. 2024, 18, 1507140. [Google Scholar] [CrossRef]
- Arnoux, I.; Hoshiko, M.; Mandavy, L.; Avignone, E.; Yamamoto, N.; Audinat, E. Adaptive phenotype of microglial cells during the normal postnatal development of the somatosensory “Barrel” cortex. Glia 2013, 61, 1582–1594. [Google Scholar] [CrossRef]
- Vela, J.M.; Dalmau, I.; González, B.; Castellano, B. Morphology and distribution of microglial cells in the young and adult mouse cerebellum. J. Comp. Neurol. 1995, 361, 602–616. [Google Scholar] [CrossRef]
- Krabbe, G.; Halle, A.; Matyash, V.; Rinnenthal, J.L.; Eom, G.D.; Bernhardt, U.; Miller, K.R.; Prokop, S.; Kettenmann, H.; Heppner, F.L. Functional impairment of microglia coincides with Beta-amyloid deposition in mice with Alzheimer-like pathology. PLoS ONE 2013, 8, e60921. [Google Scholar] [CrossRef]
- Masuda, T.; Sankowski, R.; Staszewski, O.; Prinz, M. Microglia Heterogeneity in the Single-Cell Era. Cell Rep. 2020, 30, 1271–1281. [Google Scholar] [CrossRef]
- Gross, P.M.; Weindl, A. Peering through the windows of the brain. J. Cereb. Blood Flow Metab. 1987, 7, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Mander, T.H.; Morris, J.F. Immunophenotypic evidence for distinct populations of microglia in the rat hypothalamo-neurohypophysial system. Cell Tissue Res. 1995, 280, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro Xavier, A.L.; Kress, B.T.; Goldman, S.A.; Lacerda de Menezes, J.R.; Nedergaard, M. A Distinct Population of Microglia Supports Adult Neurogenesis in the Subventricular Zone. J. Neurosci. 2015, 35, 11848–11861. [Google Scholar] [CrossRef] [PubMed]
- Takagi, S.; Furube, E.; Nakano, Y.; Morita, M.; Miyata, S. Microglia are continuously activated in the circumventricular organs of mouse brain. J. Neuroimmunol. 2019, 331, 74–86. [Google Scholar] [CrossRef]
- Stowell, R.D.; Wong, E.L.; Batchelor, H.N.; Mendes, M.S.; Lamantia, C.E.; Whitelaw, B.S.; Majewska, A.K. Cerebellar microglia are dynamically unique and survey Purkinje neurons in vivo. Dev. Neurobiol. 2018, 78, 627–644. [Google Scholar] [CrossRef]
- Irintchev, A.; Rollenhagen, A.; Troncoso, E.; Kiss, J.Z.; Schachner, M. Structural and functional aberrations in the cerebral cortex of tenascin-C deficient mice. Cereb. Cortex. 2005, 15, 950–962. [Google Scholar] [CrossRef]
- San Jose, I.; García-Atares, N.; Pelaez, B.; Cabo, R.; Esteban, I.; Vega, J.A.; Represa, J. Reduction of glial fibrillary acidic protein-immunoreactive astrocytes in some brain areas of old hairless rhino-j mice (hr-rh-j). Neurosci. Lett. 2001, 309, 81–84. [Google Scholar] [CrossRef]
- Jinno, S.; Fleischer, F.; Eckel, S.; Schmidt, V.; Kosaka, T. Spatial arrangement of microglia in the mouse hippocampus: A stereological study in comparison with astrocytes. Glia 2007, 55, 1334–1347. [Google Scholar] [CrossRef]
- Hart, A.D.; Wyttenbach, A.; Perry, V.H.; Teeling, J.L. Age related changes in microglial phenotype vary between CNS regions: Grey versus white matter differences. Brain Behav. Immun. 2012, 26, 754–765. [Google Scholar] [CrossRef]
- Grabert, K.; Michoel, T.; Karavolos, M.H.; Clohisey, S.; Baillie, J.K.; Stevens, M.P.; Freeman, T.C.; Summers, K.M.; McColl, B.W. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 2016, 19, 504–516. [Google Scholar] [CrossRef]
- Das Sarma, J. Microglia-mediated neuroinflammation is an amplifier of virus-induced neuropathology. J. Neurovirol. 2014, 20, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Sierra, A.; Beccari, S.; Diaz-Aparicio, I.; Encinas, J.M.; Comeau, S.; Tremblay, M. Surveillance, phagocytosis, and inflammation: How never-resting microglia influence adult hippocampal neurogenesis. Neural. Plast. 2014, 2014, 610343. [Google Scholar] [CrossRef]
- Cupido, A.; Catalin, B.; Steffens, H.; Kirchhoff, F. Surgical Procedures to Study Microglial Motility in the Brain and in the Spinal Cord by In Vivo Two-Photon Laser-Scanning Microscopy. In Laser Scanning Microscopy and Quantitative Image Analysis of Neuronal Tissue; Bakota, L., Brandt, R., Eds.; Springer New York: New York, NY, USA, 2014; pp. 37–50. [Google Scholar]
- Ransohoff, R.M.; Perry, V.H. Microglial physiology: Unique stimuli, specialized responses. Annu. Rev. Immunol. 2009, 27, 119–145. [Google Scholar] [CrossRef] [PubMed]
- Hickman, S.E.; Kingery, N.D.; Ohsumi, T.K.; Borowsky, M.L.; Wang, L.C.; Means, T.K.; El Khoury, J. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 2013, 16, 1896–1905. [Google Scholar] [CrossRef]
- Hanisch, U.K. Microglia as a source and target of cytokines. Glia 2002, 40, 140–155. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.; Golebiewska, A.; Poovathingal, S.K.; Kaoma, T.; Pires-Afonso, Y.; Martina, S.; Coowar, D.; Azuaje, F.; Skupin, A.; Balling, R.; et al. Single-cell transcriptomics reveals distinct inflammation-induced microglia signatures. EMBO Rep. 2018, 19, e46171. [Google Scholar] [CrossRef] [PubMed]
- Deczkowska, A.; Keren-Shaul, H.; Weiner, A.; Colonna, M.; Schwartz, M.; Amit, I. Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 2018, 173, 1073–1081. [Google Scholar] [CrossRef]
- Gonzalez Caldito, N. Role of tumor necrosis factor-alpha in the central nervous system: A focus on autoimmune disorders. Front. Immunol. 2023, 14, 1213448. [Google Scholar] [CrossRef] [PubMed]
- Vuolteenaho, K.; Koskinen, A.; Kukkonen, M.; Nieminen, R.; Päivärinta, U.; Moilanen, T.; Moilanen, E. Leptin enhances synthesis of proinflammatory mediators in human osteoarthritic cartilage--mediator role of NO in leptin-induced PGE2, IL-6, and IL-8 production. Mediat. Inflamm. 2009, 2009, 345838. [Google Scholar] [CrossRef]
- Zöller, T.; Schneider, A.; Kleimeyer, C.; Masuda, T.; Potru, P.S.; Pfeifer, D.; Blank, T.; Prinz, M.; Spittau, B. Silencing of TGFβ signalling in microglia results in impaired homeostasis. Nat. Commun. 2018, 9, 4011. [Google Scholar] [CrossRef]
- Chabot, S.; Williams, G.; Hamilton, M.; Sutherland, G.; Yong, V.W. Mechanisms of IL-10 production in human microglia-T cell interaction. J. Immunol. 1999, 162, 6819–6828. [Google Scholar] [CrossRef] [PubMed]
- Rusin, D.; Vahl Becirovic, L.; Lyszczarz, G.; Krueger, M.; Benmamar-Badel, A.; Vad Mathiesen, C.; Schiöth, E.S.; Lambertsen, K.L.; Wlodarczyk, A. Microglia-Derived Insulin-like Growth Factor 1 Is Critical for Neurodevelopment. Cells 2024, 13, 184. [Google Scholar] [CrossRef] [PubMed]
- Petry, P.; Oschwald, A.; Kierdorf, K. Microglial tissue surveillance: The never-resting gardener in the developing and adult CNS. Eur. J. Immunol. 2023, 53, e2250232. [Google Scholar] [CrossRef]
- PR-H El “tercer elemento” de los centros nerviosos. I. La microglía en estado normal. II. Intervención de la microglía en los procesos patológicos (células en bastoncito y cuerpos gránulo-adiposos). Boletín Soc. Española Biol. 1919, 9, 68–120.
- Town, T.; Nikolic, V.; Tan, J. The microglial “activation” continuum: From innate to adaptive responses. J. Neuroinflammation 2005, 2, 24. [Google Scholar] [CrossRef]
- Orihuela, R.; McPherson, C.A.; Harry, G.J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 2016, 173, 649–665. [Google Scholar] [CrossRef]
- Bouvier, D.S.; Jones, E.V.; Quesseveur, G.; Davoli, M.A.A.; Ferreira, T.; Quirion, R.; Mechawar, N.; Murai, K.K. High Resolution Dissection of Reactive Glial Nets in Alzheimer’s Disease. Sci. Rep. 2016, 6, 24544. [Google Scholar] [CrossRef] [PubMed]
- Boboc, I.K.S.; Cojocaru, A.; Nedelea, G.; Catalin, B.; Bogdan, M.; Calina, D. Chronic Administration of Ion Channel Blockers Impact Microglia Morphology and Function in a Murine Model of Alzheimer’s Disease. Int. J. Mol. Sci. 2023, 24, 14474. [Google Scholar] [CrossRef]
- Cojocaru, A.; Burada, E.; Bălșeanu, A.T.; Deftu, A.F.; Cătălin, B.; Popa-Wagner, A.; Osiac, E. Roles of Microglial Ion Channel in Neurodegenerative Diseases. J. Clin. Med. 2021, 10, 1239. [Google Scholar] [CrossRef]
- Matcovitch-Natan, O.; Winter, D.R.; Giladi, A.; Vargas Aguilar, S.; Spinrad, A.; Sarrazin, S.; Ben-Yehuda, H.; David, E.; González, F.Z.; Perrin, P.; et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 2016, 353, aad8670. [Google Scholar] [CrossRef]
- Mastenbroek, L.J.M.; Kooistra, S.M.; Eggen, B.J.L.; Prins, J.R. The role of microglia in early neurodevelopment and the effects of maternal immune activation. Semin Immunopathol. 2024, 46, 1. [Google Scholar] [CrossRef]
- Shikha, D.; Mahish, C.; Sing, R.; Chattopadhyay, S.; Goswami, C. Modulation of TRPM8 alters the phagocytic activity of microglia and induces changes in sub-cellular organelle functions. Biochem. Biophys. Res. Commun. 2023, 682, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Cheng, Z.; Zhou, L.; Darmanis, S.; Neff, N.F.; Okamoto, J.; Gulati, G.; Bennett, M.L.; Sun, L.O.; Clarke, L.E.; et al. Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 2019, 101, 207–223.e10. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Jiang, J.; Tan, Y.; Chen, S. Microglia in neurodegenerative diseases: Mechanism and potential therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 359. [Google Scholar]
- Tau, G.Z.; Peterson, B.S. Normal Development of Brain Circuits. Neuropsychopharmacology 2010, 35, 147–168. [Google Scholar] [CrossRef]
- Lai, H.Y.; Hsu, L.W.; Tsai, H.H.; Lo, Y.C.; Yang, S.H.; Liu, P.Y.; Wang, J.-M. CCAAT/enhancer-binding protein delta promotes intracellular lipid accumulation in M1 macrophages of vascular lesions. Cardiovasc. Res. 2017, 113, 1376–1388. [Google Scholar] [CrossRef]
- Damani, M.R.; Zhao, L.; Fontainhas, A.M.; Amaral, J.; Fariss, R.N.; Wong, W.T. Age-related alterations in the dynamic behavior of microglia. Aging Cell 2011, 10, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.S.; Ma, J.; Jegathees, T.; Goldsbury, C. Microglia show altered morphology and reduced arborization in human brain during aging and Alzheimer’s disease. Brain Pathol 2017, 27, 795–808. [Google Scholar] [CrossRef]
- Franceschi, C.; Capri, M.; Monti, D.; Giunta, S.; Olivieri, F.; Sevini, F.; Panourgia, M.P.; Invidia, L.; Celani, L.; Scurti, M.; et al. Inflammaging and anti-inflammaging: A systemic perspective on aging and longevity emerged from studies in humans. Mech. Ageing Dev. 2007, 128, 92–105. [Google Scholar] [CrossRef]
- Iburg, K.M.; Charalampous, P.; Allebeck, P.; Stenberg, E.J.; O’Caoimh, R.; Monasta, L.; Peñalvo, J.L.; Pereira, D.M.; Wyper, G.M.A.; Niranjan, V.; et al. Burden of disease among older adults in Europe—Trends in mortality and disability, 1990–2019. Eur. J. Public Health 2022, 33, 121–126. [Google Scholar] [CrossRef]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef]
- Cunningham, C.L.; Martínez-Cerdeño, V.; Noctor, S.C. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci. 2013, 33, 4216–4233. [Google Scholar] [CrossRef]
- Koenigsknecht-Talboo, J.; Landreth, G.E. Microglial phagocytosis induced by fibrillar beta-amyloid and IgGs are differentially regulated by proinflammatory cytokines. J. Neurosci. 2005, 25, 8240–8249. [Google Scholar] [CrossRef] [PubMed]
- Savage, J.C.; Carrier, M.; Tremblay, M. Morphology of Microglia Across Contexts of Health and Disease. Methods Mol. Biol. 2019, 2034, 13–26. [Google Scholar] [PubMed]
- Dal Bianco, A.; Bradl, M.; Frischer, J.; Kutzelnigg, A.; Jellinger, K.; Lassmann, H. Multiple sclerosis and Alzheimer’s disease. Ann. Neurol. 2008, 63, 174–183. [Google Scholar] [CrossRef]
- Angelova, D.M.; Brown, D.R. Microglia and the aging brain: Are senescent microglia the key to neurodegeneration? J. Neurochem. 2019, 151, 676–688. [Google Scholar] [CrossRef]
- D’Andrea, M.R.; Cole, G.M.; Ard, M.D. The microglial phagocytic role with specific plaque types in the Alzheimer disease brain. Neurobiol. Aging 2004, 25, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Brawek, B.; Schwendele, B.; Riester, K.; Kohsaka, S.; Lerdkrai, C.; Liang, Y.; Garaschuk, O. Impairment of in vivo calcium signaling in amyloid plaque-associated microglia. Acta Neuropathol. 2014, 127, 495–505. [Google Scholar] [CrossRef]
- Costa, J.; Martins, S.; Ferreira, P.A.; Cardoso, A.M.S.; Guedes, J.R.; Peça, J.; Cardoso, A.L. The old guard: Age-related changes in microglia and their consequences. Mech. Ageing Dev. 2021, 197, 111512. [Google Scholar] [CrossRef]
- Norden, D.M.; Muccigrosso, M.M.; Godbout, J.P. Microglial priming and enhanced reactivity to secondary insult in aging, and traumatic CNS injury, and neurodegenerative disease. Neuropharmacology 2015, 96, 29–41. [Google Scholar] [CrossRef]
- Thomas, A.L.; Lehn, M.A.; Janssen, E.M.; Hildeman, D.A.; Chougnet, C.A. Naturally-aged microglia exhibit phagocytic dysfunction accompanied by gene expression changes reflective of underlying neurologic disease. Sci. Rep. 2022, 12, 19471. [Google Scholar] [CrossRef] [PubMed]
- Keren-Shaul, H.; Spinrad, A.; Weiner, A.; Matcovitch-Natan, O.; Dvir-Szternfeld, R.; Ulland, T.K.; David, E.; Baruch, K.; Lara-Astaiso, D.; Toth, B.; et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 2017, 169, 1276–1290.e17. [Google Scholar] [CrossRef]
- Alexianu, M.E.; Kozovska, M.; Appel, S.H. Immune reactivity in a mouse model of familial ALS correlates with disease progression. Neurology 2001, 57, 1282–1289. [Google Scholar] [CrossRef]
- Migliarini, S.; Scaricamazza, S.; Valle, C.; Ferri, A.; Pasqualetti, M.; Ferraro, E. Microglia Morphological Changes in the Motor Cortex of hSOD1(G93A) Transgenic ALS Mice. Brain Sci. 2021, 11, 807. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.; Arachchige, A. Depletion of dopamine in Parkinson’s disease and relevant therapeutic options: A review of the literature. AIMS Neurosci. 2023, 10, 200–231. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Patel, T.; Sugandh, F.; Dev, J.; Kumar, U.; Adeeb, M.; Kachhadia, M.P.; Puri, P.; Prachi, F.; Zaman, M.U.; et al. Innovative Approaches and Therapies to Enhance Neuroplasticity and Promote Recovery in Patients with Neurological Disorders: A Narrative Review. Cureus 2023, 15, e41914. [Google Scholar] [CrossRef]
- Sango, K.; Yamanaka, S.; Hoffmann, A.; Okuda, Y.; Grinberg, A.; Westphal, H.; McDonald, M.P.; Crawley, J.N.; Sandhoff, K.; Suzuki, K.; et al. Mouse models of Tay-Sachs and Sandhoff diseases differ in neurologic phenotype and ganglioside metabolism. Nat. Genet. 1995, 11, 170–176. [Google Scholar] [CrossRef]
- Krasemann, S.; Madore, C.; Cialic, R.; Baufeld, C.; Calcagno, N.; El Fatimy, R.; Beckers, L.; O’Loughlin, E.; Xu, Y.; Fanek, Z.; et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 2017, 47, 566–581.e9. [Google Scholar] [CrossRef]
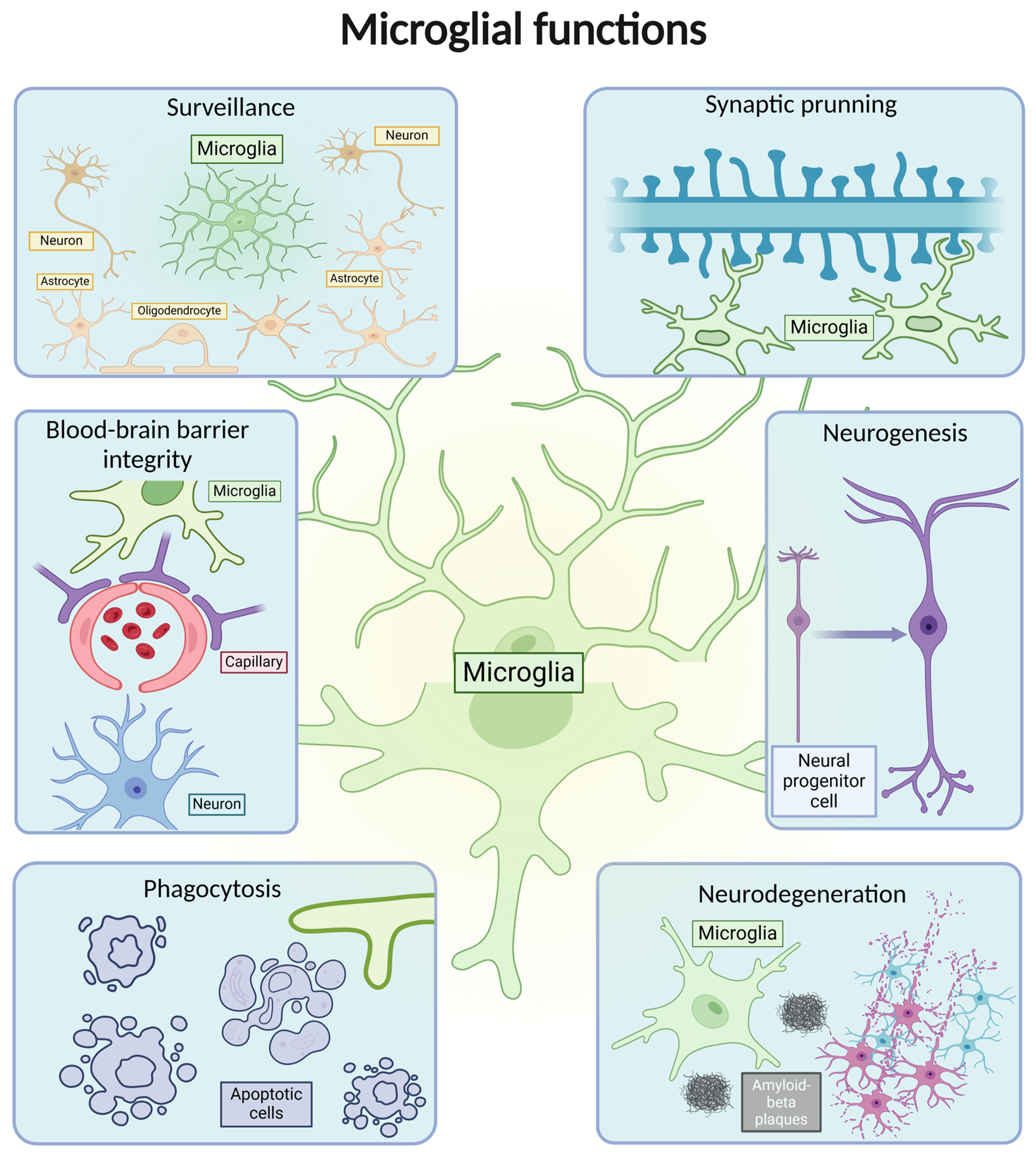

| Microglial Phenotypes | General Aspect | Publications |
|---|---|---|
Ramified microglia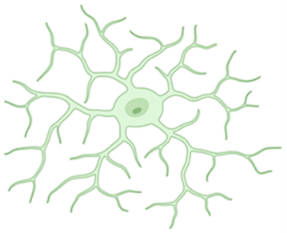 | A small number of primary branches dichotomize up to the level of the terminal branches, reaching a complex arborization. | [12,19,24] |
Amoeboid microglia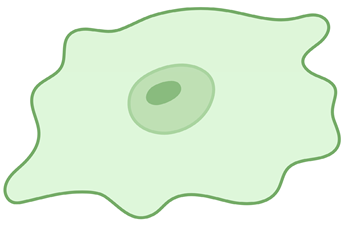 | Can be considered precursors to activation; display the smallest values for general morphological parameters. They are also found in regions with an incomplete BBB, such as the median eminence, the circumventricular organs, and the subventricular zone. | [25,26] |
Activated microglia | Have a smaller branching index, smaller cell perimeters, greater circularity of the soma, and cytoplasmic hypertrophy. | [19,24] |
Aged microglia | Reduced process length, branching, and arborized area. They possess reduced baseline process motility but increased soma motility. Upon activation, a reduced speed of microglial processes approaching the lesion was observed. | [12,24,27] |
Dystrophic microglia | Fragmented and beaded processes, increased tortuosity, swellings distinct from activation. They have been encountered in the aged brain, and furthermore, in the case of neurodegenerative disorders, are considered the morphological expression of disease-associated microglia. | [28,29] |
Rod microglia | A narrow, elongated cell body with polarized processes. They are currently considered a particular form of microglial activation, occurring only in diseases affecting the CNS. | [30,31,32,33,34] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Godeanu, S.; Cătălin, B. The Complementary Role of Morphology in Understanding Microglial Functional Heterogeneity. Int. J. Mol. Sci. 2025, 26, 3811. https://doi.org/10.3390/ijms26083811
Godeanu S, Cătălin B. The Complementary Role of Morphology in Understanding Microglial Functional Heterogeneity. International Journal of Molecular Sciences. 2025; 26(8):3811. https://doi.org/10.3390/ijms26083811
Chicago/Turabian StyleGodeanu, Sânziana, and Bogdan Cătălin. 2025. "The Complementary Role of Morphology in Understanding Microglial Functional Heterogeneity" International Journal of Molecular Sciences 26, no. 8: 3811. https://doi.org/10.3390/ijms26083811
APA StyleGodeanu, S., & Cătălin, B. (2025). The Complementary Role of Morphology in Understanding Microglial Functional Heterogeneity. International Journal of Molecular Sciences, 26(8), 3811. https://doi.org/10.3390/ijms26083811





