The PAP Gene Family in Cotton: Impact of Genome-Wide Identification on Fiber Secondary Wall Synthesis
Abstract
1. Introduction
2. Results
2.1. Identification and Chromosomal Locations of PAP Genes in Four Cotton Species
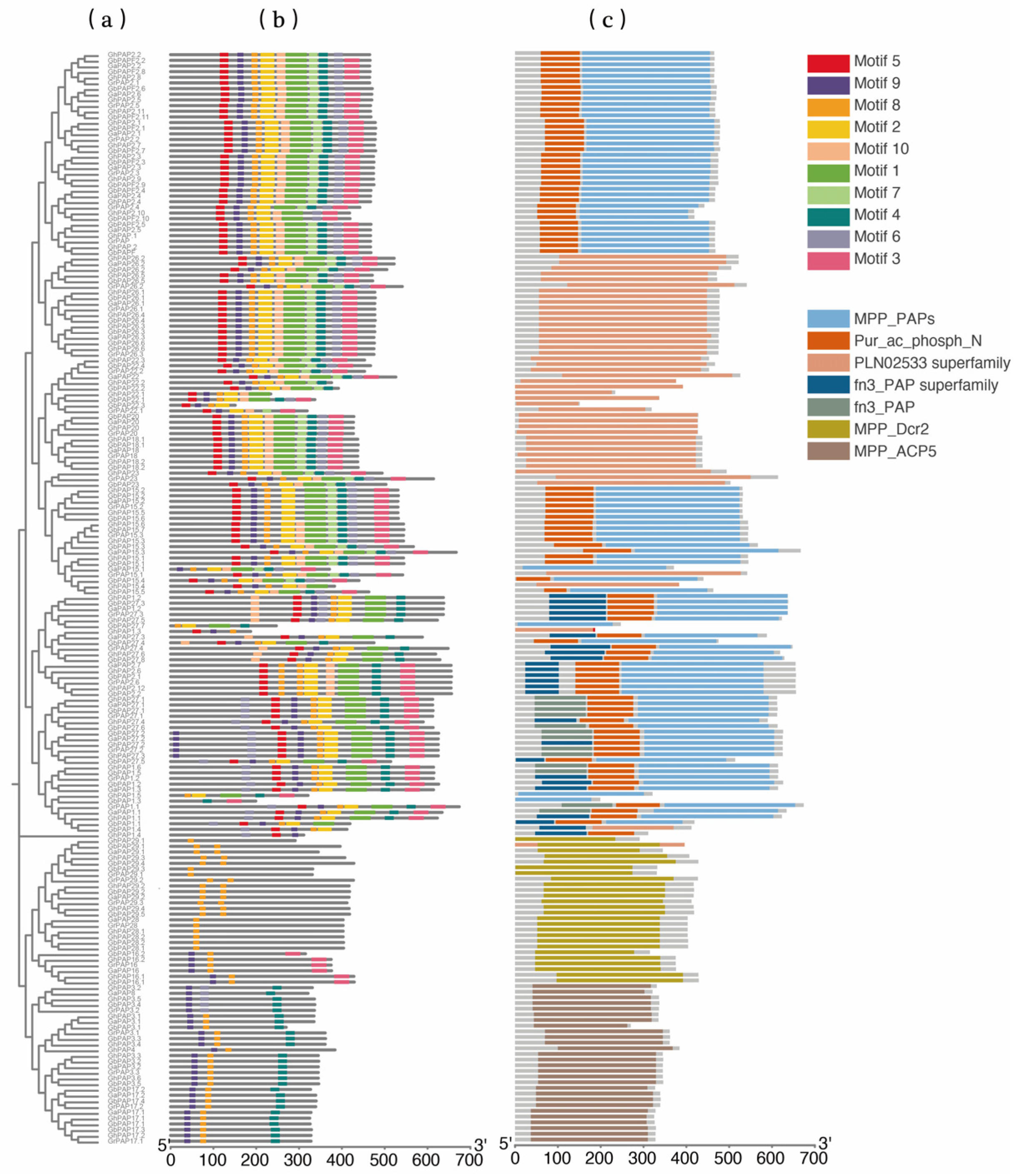
2.2. Phylogenetic Analysis of Cotton PAP Proteins
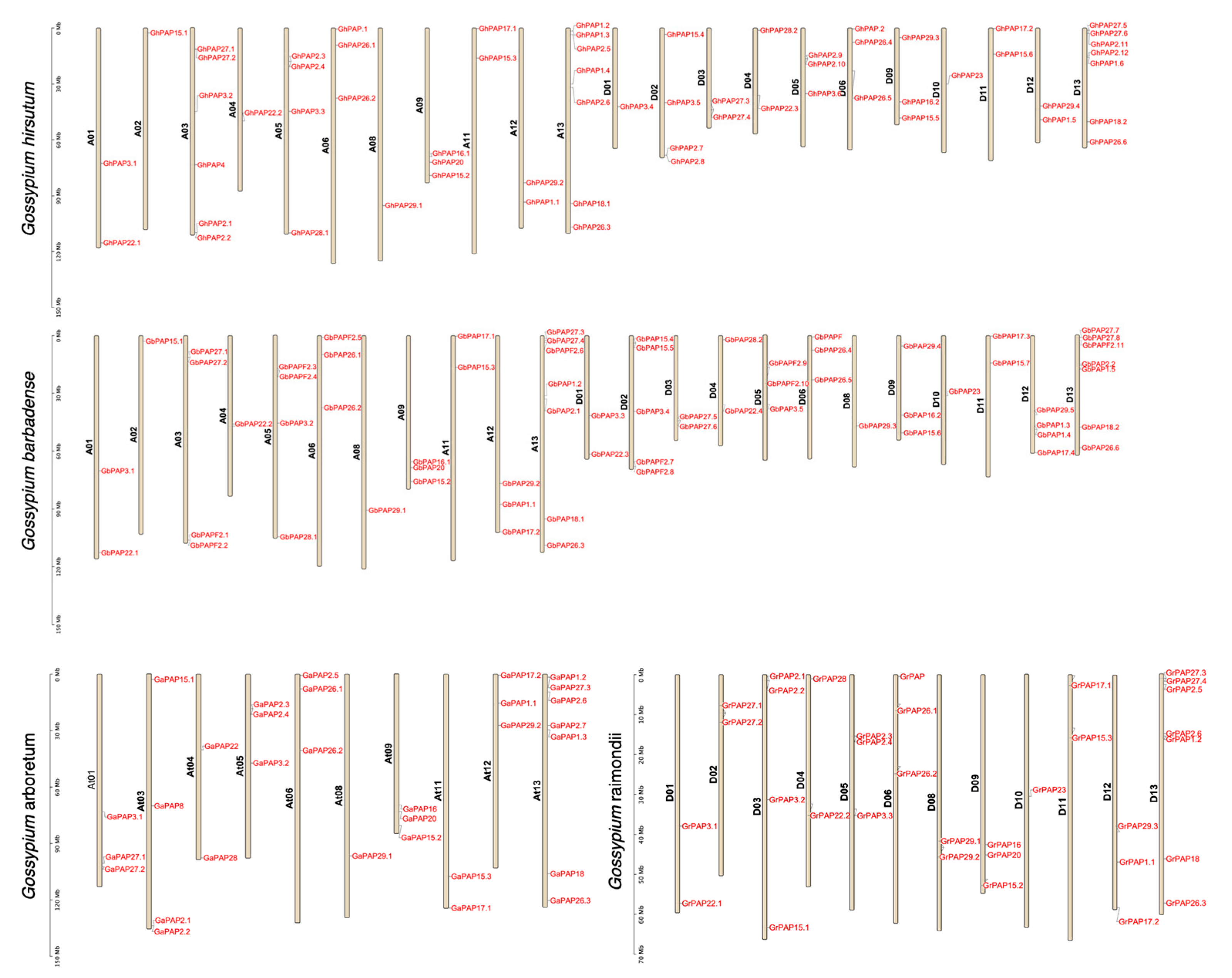
2.3. Chromosomal Locations of PAP Genes in Four Cotton Species
2.4. Multiple Sequence Alignment, Conserved Motif Analysis, and Gene Structure of PAP Genes in Cotton
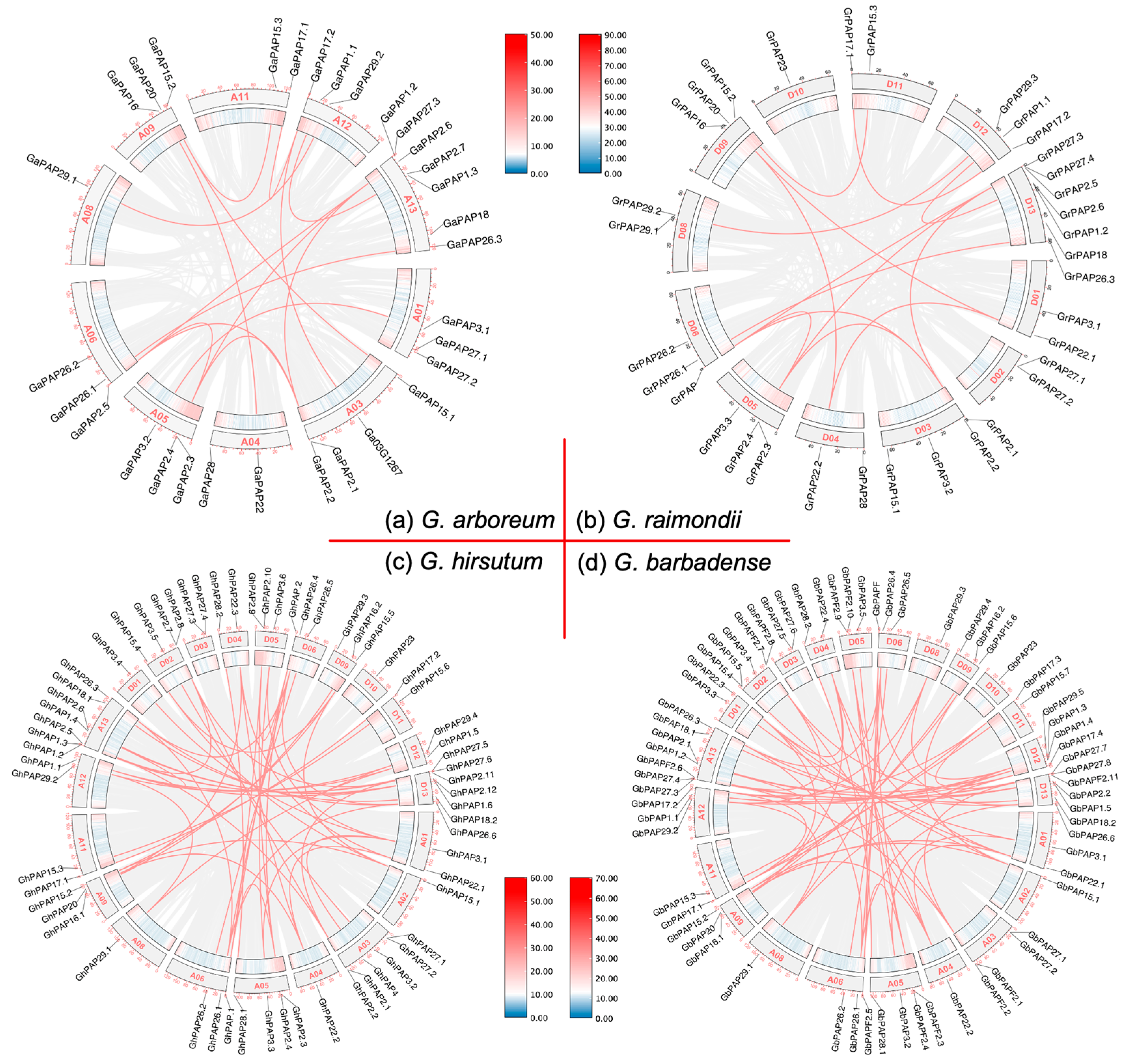
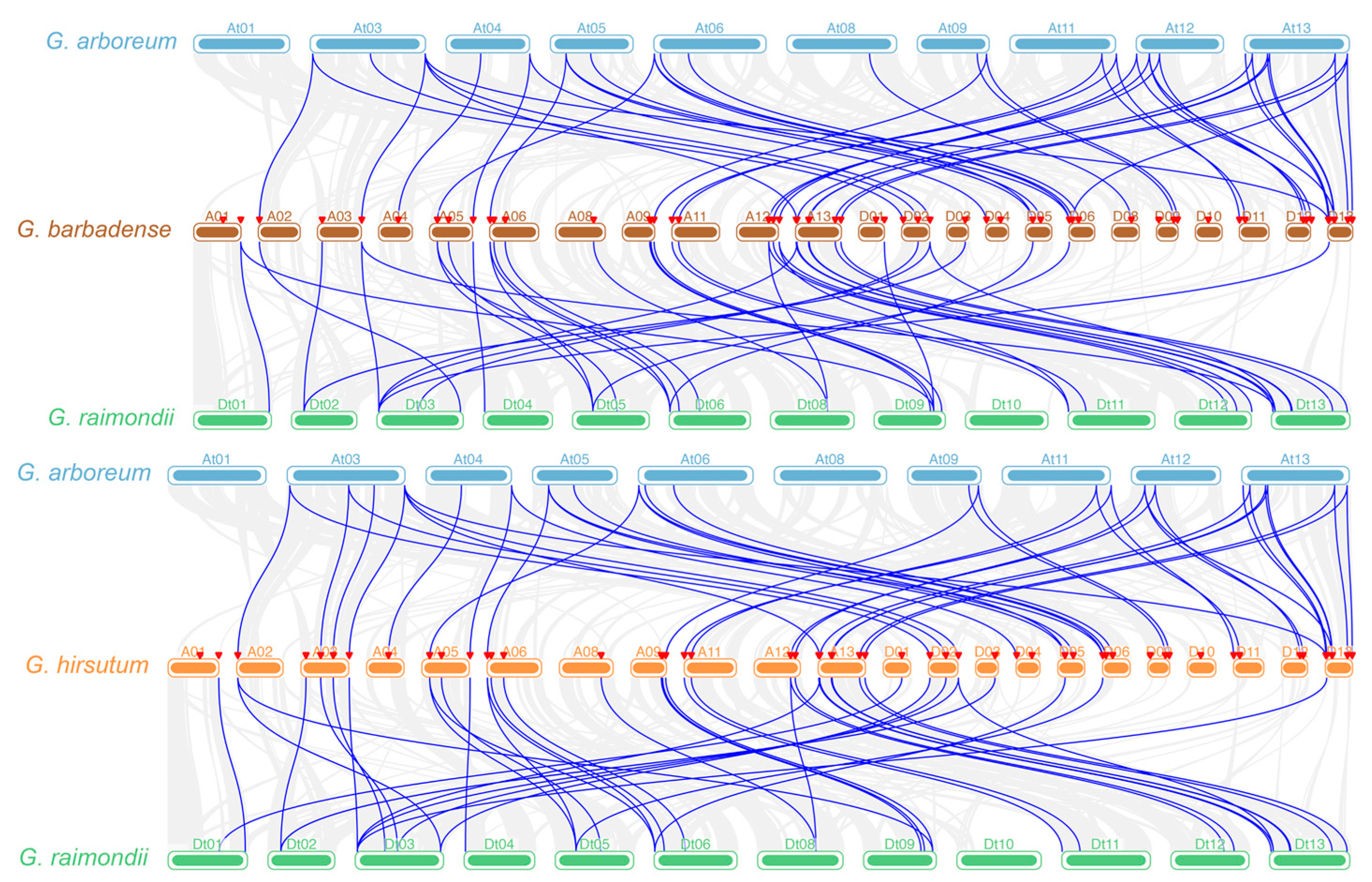
2.5. Collinearity Analysis of PAP Genes in Cotton
2.6. Cis-Element Analysis in the Promoter Regions of the PAP Genes in Cotton
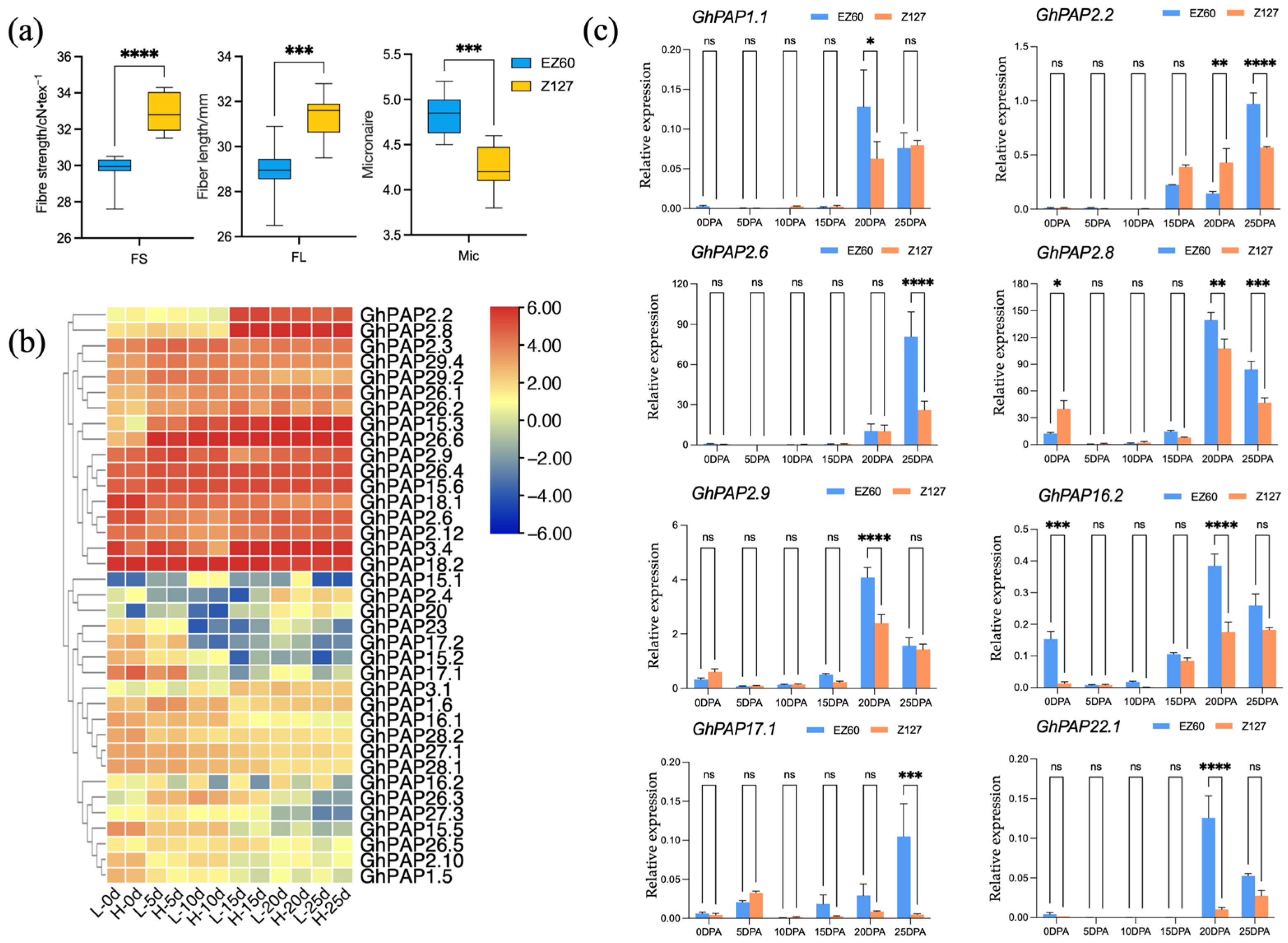
2.7. Expression Patterns of GhPAP Genes at Different Stages of Fiber Development in Two Cotton Species
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Genome-Wide Identification of the PAP Genes in Cotton
4.3. Analysis of the Conserved Domain, Motifs, and Gene Structure
4.4. Multiple Sequence Alignment and Phylogenetic Analysis of the PAP Proteins
4.5. Chromosomal Locations and Gene Collinearity Analysis
4.6. Promoter Cis-Element Analysis of PAP Genes
4.7. RNA Expression Data and Analysis of qRT–PCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jacob, J.; Lawlor, D.W. Dependence of Photosynthesis of Sunflower and Maize Leaves on Phosphate Supply, Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase Activity, and Ribulose-1,5-Bisphosphate Pool Size. Plant Physiol. 1992, 98, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Kavanová, M.; Grimoldi, A.A.; Lattanzi, F.A.; Schnyder, H. Phosphorus nutrition and mycorrhiza effects on grass leaf growth. P status- and size-mediated effects on growth zone kinematics. Plant Cell Environ. 2006, 29, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Alexova, R.; Millar, A.H. Proteomics of phosphate use and deprivation in plants. Proteomics 2013, 13, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Olczak, M.; Morawiecka, B.; Watorek, W. Plant purple acid phosphatases—genes, structures and biological function. Acta Biochim. Pol. 2003, 50, 1245–1256. [Google Scholar] [CrossRef]
- Klabunde, T.; Sträter, N.; Fröhlich, R.; Witzel, H.; Krebs, B. Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures. J. Mol. Biol. 1996, 259, 737–748. [Google Scholar] [CrossRef]
- Li, D.; Zhu, H.; Liu, K.; Liu, X.; Leggewie, G.; Udvardi, M.; Wang, D. Purple Acid Phosphatases of Arabidopsis thaliana: Comparative analysis and differential regulation by phosphate deprivation*. J. Biol. Chem. 2002, 277, 27772–27781. [Google Scholar] [CrossRef]
- Li, C.; Gui, S.; Yang, T.; Walk, T.; Wang, X.; Liao, H. Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Ann. Bot. 2011, 109, 275–285. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, C.; Tian, J.; Li, K.; Shou, H. Identification of rice purple acid phosphatases related to posphate starvation signalling. Plant Biol. 2011, 13, 7–15. [Google Scholar] [CrossRef]
- Pang, X.; Cheng, Y.; Ruan, M.; Ye, Q.; Wang, R.; Yao, Z.; Zhou, G.; Wan, H. The PAP Gene Family in Tomato: Comprehensive Comparative Analysis, Phylogenetic Relationships and Expression Profiles. Plants 2022, 11, 563. [Google Scholar] [CrossRef]
- González-Muñoz, E.; Avendaño-Vázquez, A.O.; Montes, R.A.; de Folter, S.; Andrés-Hernández, L.; Abreu-Goodger, C.; Sawers, R.J. The maize (Zea mays ssp. mays var. B73) genome encodes 33 members of the purple acid phosphatase family. Front. Plant Sci. 2015, 6, 341. [Google Scholar]
- Hou, L.; Zhang, D.; Wu, Q.; Gao, X.; Wang, J. Analysis and profiling of the purple acid phosphatase gene family in wheat (Triticum aestivum L.). Protoplasma 2024, 262, 73–86. [Google Scholar] [CrossRef]
- Venkidasamy, B.; Selvaraj, D.; Ramalingam, S. Genome-wide analysis of purple acid phosphatase (PAP) family proteins in Jatropha curcas L. Int. J. Biol. Macromol. 2019, 123, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lu, S.; Zhang, Y.; Li, Z.; Du, X.; Liu, D. Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J. Integr. Plant Biol. 2014, 56, 299–314. [Google Scholar] [CrossRef]
- Lu, L.; Qiu, W.; Gao, W.; Tyerman, S.D.; Shou, H.; Wang, C. OsPAP10c, a novel secreted acid phosphatase in rice, plays an important role in the utilization of external organic phosphorus. Plant Cell Environ. 2016, 39, 2247–2259. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Lu, L.; Li, J.; Du, Z.; Liu, T.; Li, W.; Xu, F.; Shi, L.; Shou, H.; Wang, C. Purple acid phosphatase 10c encodes a major acid phosphatase that regulates plant growth under phosphate-deficient conditions in rice. J. Exp. Bot. 2020, 71, 4321–4332. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-Y.F.; Shao, G.; Lam, H.-M. Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses. New Phytol. 2008, 178, 80–91. [Google Scholar] [CrossRef]
- Bhadouria, J.; Singh, A.P.; Mehra, P.; Verma, L.; Srivastawa, R.; Parida, S.K.; Giri, J. Identification of Purple Acid Phosphatases in Chickpea and Potential Roles of CaPAP7 in Seed Phytate Accumulation. Sci. Rep. 2017, 7, 11012. [Google Scholar] [CrossRef]
- Li, Y.; Si, Z.; Wang, G.; Shi, Z.; Chen, J.; Qi, G.; Jin, S.; Han, Z.; Gao, W.; Tian, Y.; et al. Genomic insights into the genetic basis of cotton breeding in China. Mol. Plant 2023, 16, 662–677. [Google Scholar] [CrossRef]
- Wu, Y.; Tang, J.; Ma, S.; Zhang, K.; Yan, T.; Pan, Z. A Review of Flexible Strain Sensors Based on Natural Fiber Materials. Adv. Mater. Technol. 2023, 8, 2201503. [Google Scholar] [CrossRef]
- Kim, H.J.; Triplett, B.A. Cotton Fiber Growth in Planta and in Vitro. Models for Plant Cell Elongation and Cell Wall Biogenesis. Plant Physiol. 2001, 127, 1361–1366. [Google Scholar] [CrossRef]
- Yu, Y.; Wu, S.; Nowak, J.; Wang, G.; Han, L.; Feng, Z.; Mendrinna, A.; Ma, Y.; Wang, H.; Zhang, X.; et al. Live-cell imaging of the cytoskeleton in elongating cotton fibres. Nat. Plants 2019, 5, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Kaida, R.; Satoh, Y.; Bulone, V.; Yamada, Y.; Kaku, T.; Hayashi, T.; Kaneko, T.S. Activation of β-Glucan Synthases by Wall-Bound Purple Acid Phosphatase in Tobacco Cells. Plant Physiol. 2009, 150, 1822–1830. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Z.; Qian, W.; Guo, W.; Gao, X.; Huang, L.; Wang, H.; Zhu, H.; Wu, J.-W.; Wang, D.; et al. The Arabidopsis Purple Acid Phosphatase AtPAP10 Is Predominantly Associated with the Root Surface and Plays an Important Role in Plant Tolerance to Phosphate Limitation. Plant Physiol. 2011, 157, 1283–1299. [Google Scholar] [CrossRef] [PubMed]
- Mehra, P.; Pandey, B.K.; Giri, J. Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol. J. 2017, 15, 1054–1067. [Google Scholar] [CrossRef]
- Ma, Z.; He, S.; Wang, X.; Sun, J.; Zhang, Y.; Zhang, G.; Wu, L.; Li, Z.; Liu, Z.; Sun, G.; et al. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 2018, 50, 803–813. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Fan, D.; Gong, J.; Li, S.; Gao, Y.; Liu, A.; Liu, L.; Deng, X.; Shi, Y.; et al. AAQSP increases mapping resolution of stable QTLs through applying NGS-BSA in multiple genetic backgrounds. Theor. Appl. Genet. 2022, 135, 3223–3235. [Google Scholar] [CrossRef]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K.C.; Shu, S.; Udall, J.; et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Stewart, C.N. Plant synthetic promoters and transcription factors. Curr. Opin. Biotechnol. 2016, 37, 36–44. [Google Scholar] [CrossRef]
- Ezeh, O.S.; Yamamoto, Y.Y. Combinatorial Effects of Cis-Regulatory Elements and Functions in Plants. Rev. Agric. Sci. 2024, 12, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, Z.H.; Bukowski, R.; Sun, Q.; Doebley, J.F. The Role of cis Regulatory Evolution in Maize Domestication. PLoS Genet. 2014, 10, e1004745. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Ali, S.; Hayat, K.; Iqbal, A.; Xie, L. Implications of Abscisic Acid in the Drought Stress Tolerance of Plants. Agronomy 2020, 10, 1323. [Google Scholar] [CrossRef]
- Mishra, S.; Roychowdhury, R.; Ray, S.; Hada, A.; Kumar, A.; Sarker, U.; Aftab, T.; Das, R. Salicylic acid (SA)-mediated plant immunity against biotic stresses: An insight on molecular components and signaling mechanism. Plant Stress 2024, 11, 100427. [Google Scholar] [CrossRef]
- Schenk, G.; Mitić, N.; Hanson, G.R.; Comba, P. Purple acid phosphatase: A journey into the function and mechanism of a colorful enzyme. Coord. Chem. Rev. 2013, 257, 473–482. [Google Scholar] [CrossRef]
- Ullah, A.H.; Cummins, B.J. Aspergillus ficuum extracellular pH 6.0 optimum acid phosphatase: Purification, N-terminal amino acid sequence, and biochemical characterization. Prep. Biochem. 1988, 18, 37–65. [Google Scholar]
- Ravichandran, S.; Stone, S.L.; Benkel, B.; Prithiviraj, B. Purple Acid Phosphatase5 is required for maintaining basal resistance against Pseudomonas syringae in Arabidopsis. BMC Plant Biol. 2013, 13, 107. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Suen, P.K.; Zhang, Y.; Liang, C.; Carrie, C.; Whelan, J.; Ward, J.L.; Hawkins, N.D.; Jiang, L.; Lim, B.L. A dual-targeted purple acid phosphatase in Arabidopsis thaliana moderates carbon metabolism and its overexpression leads to faster plant growth and higher seed yield. New Phytol. 2012, 194, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Voon, C.P.; Law, Y.-S.; Guan, X.; Lim, S.-L.; Xu, Z.; Chu, W.-T.; Zhang, R.; Sun, F.; Labs, M.; Leister, D.; et al. Modulating the activities of chloroplasts and mitochondria promotes adenosine triphosphate production and plant growth. Quant. Plant Biol. 2021, 2, e7. [Google Scholar] [CrossRef]
- Johnson, M.P. Photosynthesis. Essays Biochem. 2016, 60, 255–273. [Google Scholar] [CrossRef]
- Martinoia, E.; Maeshima, M.; Neuhaus, H.E. Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 2007, 58, 3–102. [Google Scholar] [CrossRef]
- Deng, L.; Chen, F.; Jiang, L.; Lam, H.-M.; Xiao, G. Ectopic expression of Gm3 enhances salt tolerance in rice by alleviating oxidative damage. Plant Breed. 2014, 133, 348–355. [Google Scholar] [CrossRef]
- Ravichandran, S.; Stone, S.; Benkel, B.; Zhang, J.; Berrue, F.; Prithiviraj, B. Optimal level of purple acid phosphatase5 is required for maintaining complete resistance to Pseudomonas syringae. Front. Plant Sci. 2015, 6, 568. [Google Scholar] [CrossRef]
- Xue, Y.; Zhuang, Q.; Zhu, S.; Xiao, B.; Liang, C.; Liao, H.; Tian, J. Genome Wide Transcriptome Analysis Reveals Complex Regulatory Mechanisms Underlying Phosphate Homeostasis in Soybean Nodules. Int. J. Mol. Sci. 2018, 19, 2924. [Google Scholar] [CrossRef]
- Li, C.; Zhou, J.; Wang, X.; Liao, H. A purple acid phosphatase, GmPAP33, participates in arbuscule degeneration during arbuscular mycorrhizal symbiosis in soybean. Plant Cell Environ. 2019, 42, 015–2027. [Google Scholar] [CrossRef]
- Haigler, C.H.; Betancur, L.; Stiff, M.R.; Tuttle, J.R. Cotton fiber: A powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 2012, 3, 104. [Google Scholar] [CrossRef]
- Huang, J.; Chen, F.; Guo, Y.; Gan, X.; Yang, M.; Zeng, W.; Persson, S.; Li, J.; Xu, W. GhMYB7 promotes secondary wall cellulose deposition in cotton fibres by regulating GhCesA gene expression through three distinct cis-elements. New Phytol. 2021, 232, 1718–1737. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, J.; Wang, Q.; Li, H.; Hu, W.; Wang, S.; Zhou, Z. Phosphorus-induced improvement of photosynthate synthesis and transport in the leaf subtending to cotton boll provided sufficient sucrose for fiber thickening during the crucial period and improved fiber bundle strength. Field Crops Res. 2024, 306, 109230. [Google Scholar] [CrossRef]
- Lei, K.; Cheng, J.Q.; An, Y.; Li, X.S.; An, G. Organ specific transcriptome analysis of upland cotton (Gossypium hirsutum) in response to low phosphorus stress during early stage of growth. Soil. Sci. Plant Nutr. 2022, 68, 463–472. [Google Scholar] [CrossRef]
- Huang, G.; Bao, Z.; Feng, L.; Zhai, J.; Wendel, J.F.; Cao, X.; Zhu, Y. A telomere-to-telomere cotton genome assembly reveals centromere evolution and a Mutator transposon-linked module regulating embryo development. Nat. Genet. 2024, 56, 1953–1963. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Shumate, A.; Wong, B.; Pertea, G.; Pertea, M. Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLoS Comput. Biol. 2022, 18, e1009730. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, M.C.; Konaté, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or Normalized Counts? A Comparative Study of Quantification Measures for the Analysis of RNA-seq Data from the NCI Patient-Derived Models Repository. J. Transl. Med. 2021, 19, 269. [Google Scholar] [CrossRef]
- Xu, S.; Li, L.; Luo, X.; Chen, M.; Tang, W.; Zhan, L.; Dai, Z.; Lam, T.T.; Guan, Y.; Yu, G. Ggtree: A serialized data object for visualization of a phylogenetic tree and annotation data. iMeta 2022, 1, e56. [Google Scholar] [CrossRef]



| G. arboreum | G. raimondii | G.hirsutum | G. barbadense | |||
|---|---|---|---|---|---|---|
| At | Dt | At | Dt | At | Dt | |
| Chr01 | 3 | 2 | 2 | 1 | 2 | 2 |
| Chr02 | 0 | 2 | 1 | 4 | 1 | 5 |
| Chr03 | 4 | 4 | 6 | 2 | 4 | 2 |
| Chr04 | 2 | 2 | 1 | 2 | 1 | 2 |
| Chr05 | 3 | 3 | 4 | 3 | 4 | 3 |
| Chr06 | 3 | 3 | 3 | 3 | 3 | 3 |
| Chr07 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chr08 | 1 | 2 | 1 | 0 | 1 | 1 |
| Chr09 | 3 | 3 | 3 | 3 | 3 | 3 |
| Chr10 | 0 | 1 | 0 | 1 | 0 | 1 |
| Chr11 | 2 | 2 | 2 | 2 | 2 | 2 |
| Chr12 | 3 | 3 | 2 | 2 | 3 | 4 |
| Chr13 | 7 | 7 | 7 | 7 | 7 | 7 |
| Total | 31 | 34 | 32 | 30 | 31 | 35 |
| Cotton Species | WGD or Segmental | Dispersed | Proximal | Tandem |
|---|---|---|---|---|
| G. arboreum | 22 | 7 | 0 | 2 |
| G. raimondii | 17 | 10 | 2 | 5 |
| G. hirsutum | 50 | 4 | 4 | 4 |
| G. barbadense | 56 | 2 | 5 | 3 |
| Steps | Procedure | Repetition | Temperature | Time |
|---|---|---|---|---|
| 1 | Initial denaturation | 1 | 95 °C | 30 s |
| 2 | Cyclic reaction | 40 | 95 °C | 10 s |
| 60 °C | 30 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, C.; Li, W.; Qi, R.; Liu, Y.; Wang, X.; Gong, J.; Gong, W.; Pan, J.; Li, Y.; Shi, Y.; et al. The PAP Gene Family in Cotton: Impact of Genome-Wide Identification on Fiber Secondary Wall Synthesis. Int. J. Mol. Sci. 2025, 26, 3944. https://doi.org/10.3390/ijms26093944
Sun C, Li W, Qi R, Liu Y, Wang X, Gong J, Gong W, Pan J, Li Y, Shi Y, et al. The PAP Gene Family in Cotton: Impact of Genome-Wide Identification on Fiber Secondary Wall Synthesis. International Journal of Molecular Sciences. 2025; 26(9):3944. https://doi.org/10.3390/ijms26093944
Chicago/Turabian StyleSun, Cong, Weijie Li, Ruiqiang Qi, Yangming Liu, Xiaoyu Wang, Juwu Gong, Wankui Gong, Jingtao Pan, Yang Li, Yuzhen Shi, and et al. 2025. "The PAP Gene Family in Cotton: Impact of Genome-Wide Identification on Fiber Secondary Wall Synthesis" International Journal of Molecular Sciences 26, no. 9: 3944. https://doi.org/10.3390/ijms26093944
APA StyleSun, C., Li, W., Qi, R., Liu, Y., Wang, X., Gong, J., Gong, W., Pan, J., Li, Y., Shi, Y., Yan, H., Shang, H., & Yuan, Y. (2025). The PAP Gene Family in Cotton: Impact of Genome-Wide Identification on Fiber Secondary Wall Synthesis. International Journal of Molecular Sciences, 26(9), 3944. https://doi.org/10.3390/ijms26093944







