Transcriptomic and Structural Insights into Leaf Variegation Development in Ilex × ‘Solar Flare’
Abstract
1. Introduction
2. Results
2.1. Varied Color Indices in VY
2.2. Suppressed Pigment Synthesis in VY
2.3. Reduced Stomatal Density and Disrupted Chloroplast Ultrastructure in VY
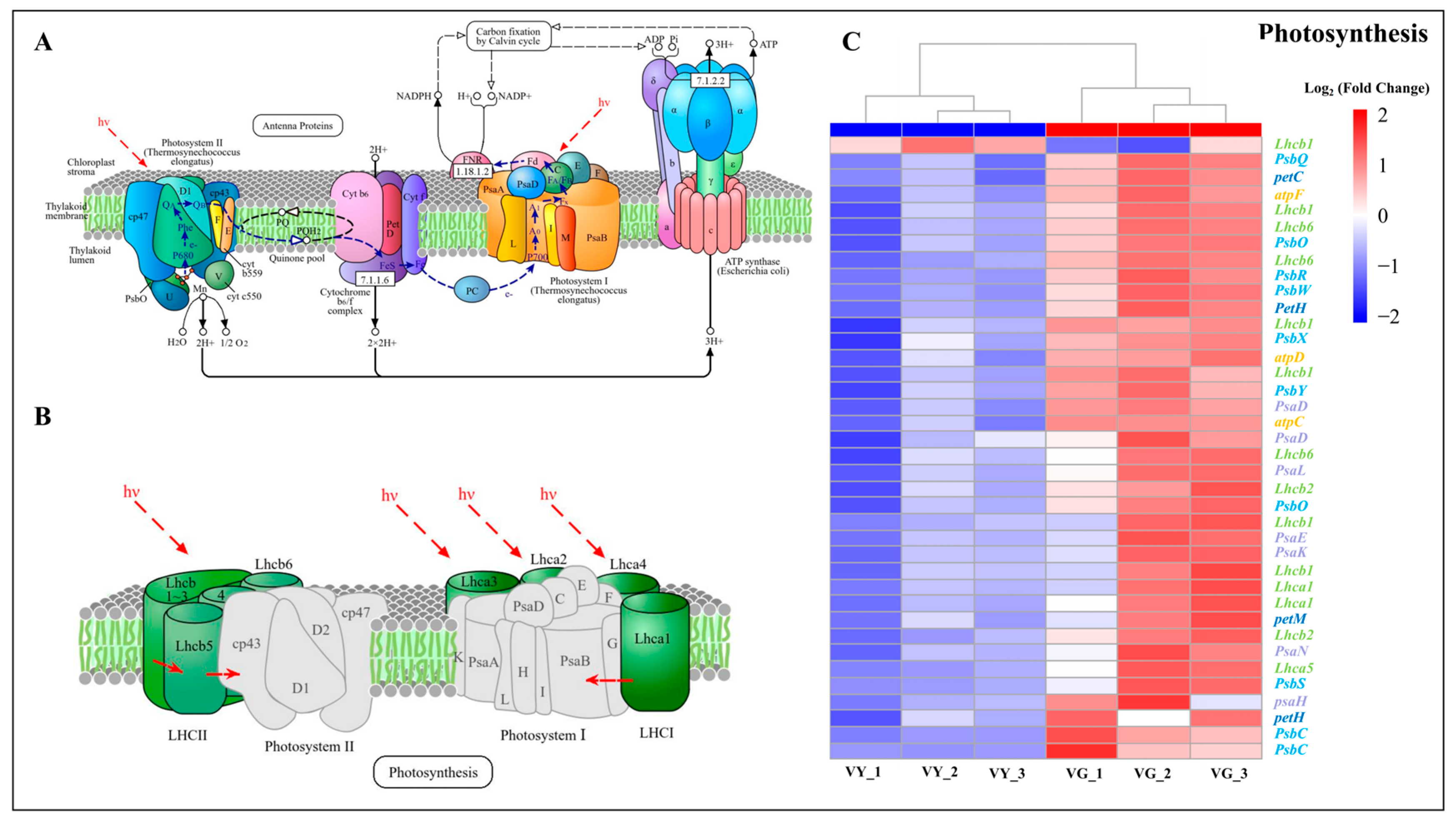
2.4. Compromised Photosynthetic Capacity in VY
2.5. Alterations in Gene Expression Related to Pigmentation and Light Harvesting Processes in VY
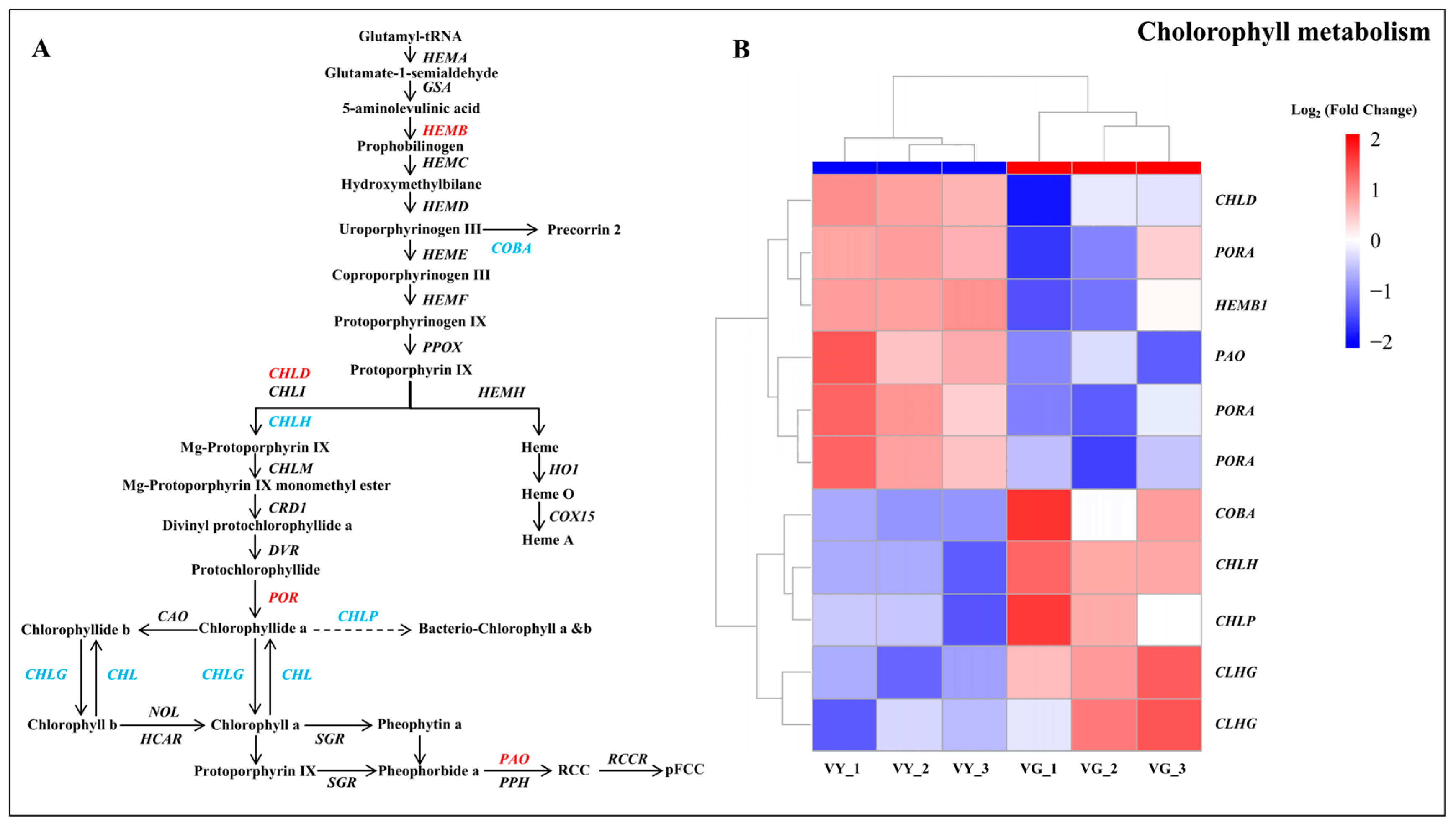
2.6. Altered Chlorophyll Biosynthesis Processes in VY
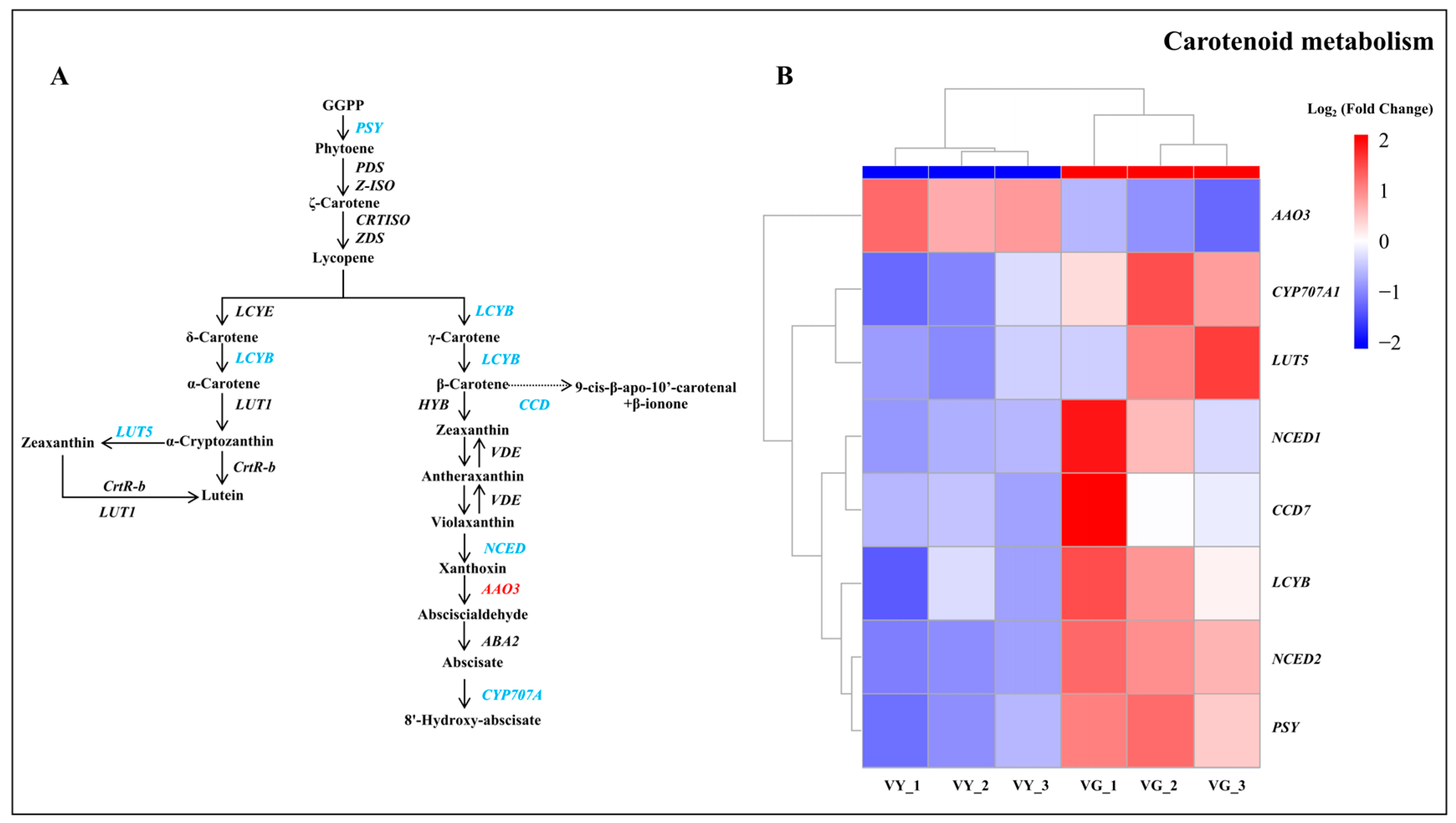
2.7. Downregulated DEGs in Carotenoid and Flavonoid Biosynthesis in VY
2.8. Downregulated DEGs in Chloroplast Development and Photosynthesis in VY
2.9. Validation of Transcriptomic Data Through Quantitative Real-Time PCR (qRT-PCR)
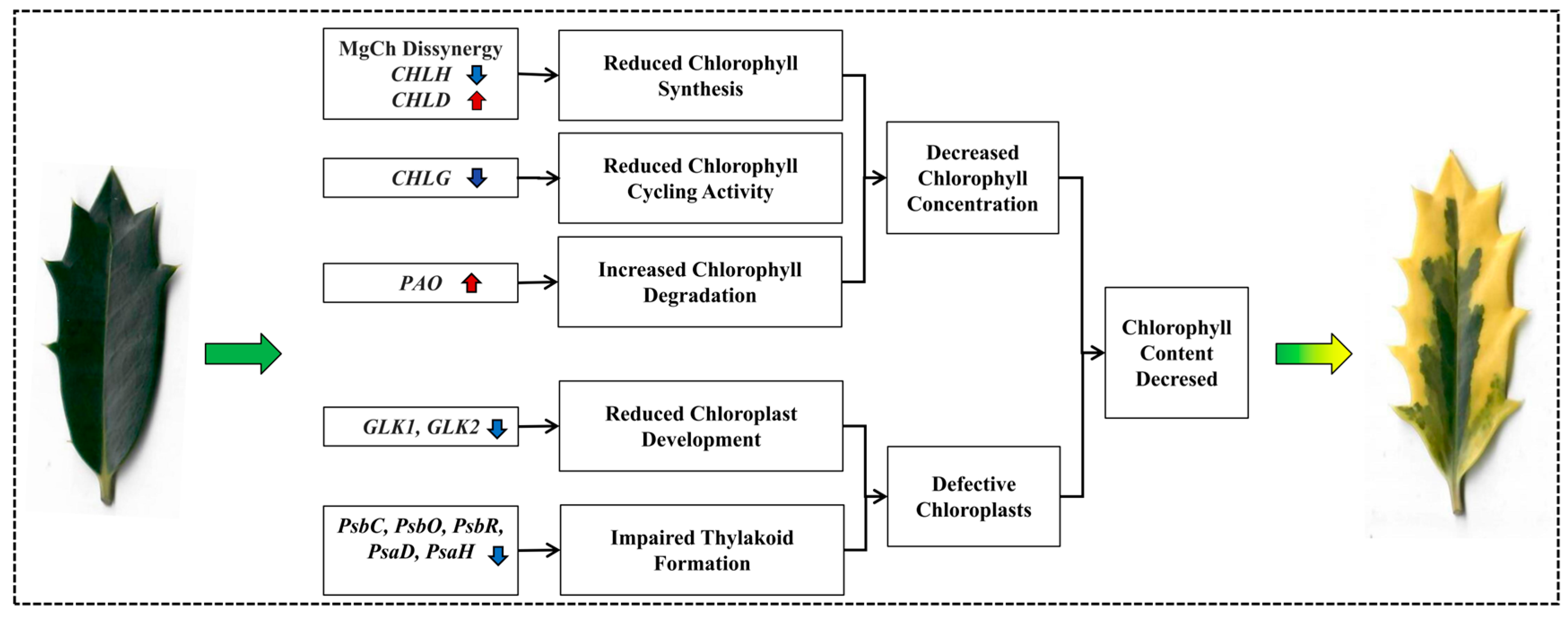
3. Discussion
3.1. I. × ‘Solar Flare’ Categorizes Chlorophyll Deficiency Type Variegation
3.2. Alterations in Pigment Metabolism DEGs May Contribute to the Leaf Variegation
3.3. Variegated Phenotype Was Linked to Abnormal Chloroplast Development
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Leaf Color Indice and SPAD
4.3. Analysis of Chlorophyll, Carotenoid, Flavonoid, and Chlorophyll Precursors Levels
4.4. Observation of Leaf Anatomical Structure
4.5. Observation of Leaf Epidermis Microstructure
4.6. Observation of Chloroplast Ultrastructure
4.7. Analysis of Photosynthetic and Chlorophyll Fluorescence
4.8. RNA Extraction and cDNA Library Preparation
4.9. Illumina Deep Sequencing and Bioinformatics Analysis
4.10. Identification and Functional Analysis of DEGs
4.11. qRT-PCR Verification
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sheue, C.R.; Pao, S.H.; Chien, L.F.; Chesson, P.; Peng, C.I. Natural foliar variegation without costs? The case of Begonia. Ann. Bot. 2012, 109, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Zeng, J.C.; Wang, X.M.; Chen, S.F.; Albach, D.C.; Li, H.Q. A revised classification of leaf variegation types. Flora 2020, 272, 151703. [Google Scholar] [CrossRef]
- Kabanov, A.V.; Khokhlacheva, Y.A. Herbaceous plants with variegated leaf color, perspective for urban landscaping. Ornam. Hortic. 2024, 30, e242728. [Google Scholar] [CrossRef]
- López-Goldar, X.; Zhang, X.N.; Hastings, A.P.; Duplais, C.; Agrawal, A.A. Plant chemical diversity enhances defense against herbivory. Proc. Natl. Acad. Sci. USA 2024, 121, e2417524121. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, C.N.; Koski, M.H. An elevational cline in leaf variegation: Testing anti-herbivory and abiotic heterogeneity hypotheses in maintaining a polymorphism. Am. J. Bot. 2024, 111, e16411. [Google Scholar] [CrossRef]
- Lin, L.W.; Huang, H.; Liu, W.W.; Sun, W.B. Cyclical drought and herbivore threats are potential causes of leaf variegation dimorphism in Cypripedium forrestii. J. Plant Ecol. 2024, 17, rtae081. [Google Scholar] [CrossRef]
- Shelef, O.; Summerfield, L.; Lev-Yadun, S.; Villamarin-Cortez, S.; Sadeh, R.; Herrmann, I.; Rachmilevitch, S. Thermal benefits from white variegation of Silybum marianum leaves. Front. Plant Sci. 2019, 10, 688. [Google Scholar] [CrossRef]
- Francomano, D.; Rodríguez González, M.I.; Valenzuela, A.E.J.; Ma, Z.; Raya Rey, A.N.; Anderson, C.B.; Pijanowski, B.C. Human-nature connection and soundscape perception: Insights from Tierra Del Fuego, Argentina. J. Nat. Conserv. 2022, 65, 126110. [Google Scholar] [CrossRef]
- Zhao, M.H.; Li, X.; Zhang, X.X.; Zhang, H.; Zhao, X.Y. Mutation mechanism of leaf color in plants: A review. Forests 2020, 11, 851. [Google Scholar] [CrossRef]
- Liu, X.Y.; Zheng, M.D.; Wang, R.; Wang, R.J.; An, L.J.; Rodermel, S.R.; Yu, F. Genetic interactions reveal that specific defects of chloroplast translation are associated with the suppression of var2-mediated leaf variegation. J. Integr. Plant Biol. 2013, 55, 979–993. [Google Scholar] [CrossRef]
- Sakamoto, W. Leaf-variegated mutations and their responsible genes in Arabidopsis thaliana. Genes Genet. Syst. 2003, 78, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Adam, Z.; Zaltsman, A.; Sinvany-Villalobo, G.; Sakamoto, W. FtsH proteases in chloroplasts and cyanobacteria. Physiol. Plant. 2005, 123, 386–390. [Google Scholar] [CrossRef]
- Yu, F.; Fu, A.; Aluru, M.; Park, S.; Xu, Y.; Liu, H.Y.; Liu, X.Y.; Foudree, A.; Nambogga, M.; Rodermel, S. Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ. 2007, 30, 350–365. [Google Scholar] [CrossRef]
- Ostermeier, M.; Garibay-Hernández, A.; Holzer, V.J.C.; Schroda, M.; Nickelsen, J. Structure, biogenesis, and evolution of thylakoid membranes. Plant Cell 2024, 36, 4014–4035. [Google Scholar] [CrossRef]
- Li, M.J.; Hensel, G.; Mascher, M.; Melzer, M.; Budhagatapalli, N.; Rutten, T.; Himmelbach, A.; Beier, S.; Korzun, V.; Kumlehn, J.; et al. Leaf variegation and impaired chloroplast development caused by a truncated CCT domain gene in albostrians barley. Plant Cell 2019, 31, 1430–1445. [Google Scholar] [CrossRef] [PubMed]
- Foudree, A.; Putarjunan, A.; Kambakam, S.; Nolan, T.; Fussell, J.; Pogorelko, G.; Rodermel, S. The mechanism of variegation in immutans provides insight into chloroplast biogenesis. Front. Plant Sci. 2012, 3, 260. [Google Scholar] [CrossRef]
- Chen, M.; Ji, M.L.; Wen, B.B.; Liu, L.; Li, S.X.; Chen, X.D.; Gao, D.S.; Li, L. Golden 2-like transcription factors of plants. Front. Plant Sci. 2016, 7, 1509. [Google Scholar] [CrossRef]
- Fitter, D.W.; Martin, D.J.; Copley, M.J.; Scotland, R.W.; Langdale, J.A. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002, 31, 713–727. [Google Scholar] [CrossRef]
- Qu, H.X.; Liang, S.; Hu, L.F.; Yu, L.; Liang, P.X.; Hao, Z.D.; Peng, Y.; Yang, J.; Shi, J.S.; Chen, J.H. Overexpression of Liriodendron hybrid LhGLK1 in Arabidopsis leads to excessive chlorophyll synthesis and improved growth. Int. J. Mol. Sci. 2024, 25, 6968. [Google Scholar] [CrossRef]
- Gang, H.X.; Li, R.H.; Zhao, Y.M.; Liu, G.F.; Chen, S.; Jiang, J. Loss of GLK1 transcription factor function reveals new insights in chlorophyll biosynthesis and chloroplast development. J. Exp. Bot. 2019, 70, 3125–3138. [Google Scholar] [CrossRef]
- Chen, J.C.; Li, Y.Y.; He, D.; Bai, M.; Li, B.; Zhang, Q.X.; Luo, L. Cytological, physiological and transcriptomic analysis of variegated leaves in Primulina pungentisepala offspring. BMC Plant Biol. 2022, 22, 419. [Google Scholar] [CrossRef] [PubMed]
- Reinbothe, S.; Reinbothe, C. The regulation of enzymes involved in chlorophyll biosynthesis. Eur. J. Biochem. 1996, 237, 323–343. [Google Scholar] [CrossRef]
- Shim, K.C.; Kang, Y.; Song, J.H.; Kim, Y.J.; Kim, J.K.; Kim, C.; Tai, T.H.; Park, I.; Ahn, S.N. A frameshift mutation in the Mg-Chelatase I subunit gene OsCHLI is associated with a lethal chlorophyll-deficient, yellow seedling phenotype in rice. Plants 2023, 12, 2831. [Google Scholar] [CrossRef] [PubMed]
- Barry, C.S. The stay-green revolution: Recent progress in deciphering the mechanisms of chlorophyll degradation in higher plants. Plant Sci. 2009, 176, 325–333. [Google Scholar] [CrossRef]
- Hörtensteiner, S.; Rodoni, S.; Schellenberg, M.; Vicentini, F.; Nandi, O.I.; Qui, Y.L.; Matile, P. Evolution of chlorophyll degradation: The significance of RCC reductase. Plant Biol. 2000, 2, 63–67. [Google Scholar] [CrossRef]
- Zhu, K.J.; Zheng, X.J.; Ye, J.L.; Huang, Y.; Chen, H.Y.; Mei, X.H.; Xie, Z.Z.; Cao, L.X.; Zeng, Y.L.; Larkin, R.M.; et al. Regulation of carotenoid and chlorophyll pools in hesperidia, anatomically unique fruits found only in Citrus. Plant Physiol. 2021, 187, 829–845. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, V.; Simkova, K.; Carrie, C.; Delannoy, E.; Giraud, E.; Whelan, J.; Small, I.D.; Apel, K.; Badger, M.R.; Pogson, B.J. The cytoskeleton and the peroxisomal-targeted SNOWY COTYLEDON3 protein are required for chloroplast development in Arabidopsis. Plant Cell 2010, 22, 3423–3438. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.F.; Yu, J.B.; Ma, R.H.; Ji, Y.Y.; Hu, Q.; Mao, Y.H.; Ding, C.Q.; Li, Z.Z.; Ge, S.B.; Deng, W.W.; et al. Chlorophyll and carotenoid metabolism varies with growth temperatures among tea genotypes with different leaf colors in Camellia sinensis. Int. J. Mol. Sci. 2024, 25, 10772. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Z.; Zhang, C.Y.; Lu, C.; Wang, M.H.; Xie, N.C.; Chen, J.J.; Li, Y.F.; Chen, J.H.; Shen, C.W. Disruption of photomorphogenesis leads to abnormal chloroplast development and leaf variegation in Camellia sinensis. Front. Plant Sci. 2021, 12, 720800. [Google Scholar] [CrossRef]
- Zhang, S.Z.; Wu, X.L.; Cui, J.; Zhang, F.; Wan, X.Q.; Liu, Q.L.; Zhong, Y.; Lin, T.T. Physiological and transcriptomic analysis of yellow leaf coloration in Populus deltoides Marsh. PLoS ONE 2019, 14, e0216879. [Google Scholar] [CrossRef]
- Galle, F.C. Hollies: The Genus Ilex; Timber Press: Portland, OR, USA, 1997. [Google Scholar]
- Yao, X.; Zhang, F.; Corlett, R.T. Utilization of the hollies (Ilex L. Spp.): A review. Forests 2022, 13, 94. [Google Scholar] [CrossRef]
- Chen, M.; Jensen, M.; Rodermel, S. The yellow variegated mutant of Arabidopsis is plastid autonomous and delayed in chloroplast biogenesis. J. Hered. 1999, 90, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, J.; Zhou, P.; Hao, M.Z.; Zhang, M. Cytological and transcriptomic analysis provide insights into the formation of variegated leaves in Ilex × altaclerensis ‘Belgica Aurea’. Plants 2021, 10, 552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, M.; Ding, Y.; Zhou, P.; Fang, Y.M. Composition of photosynthetic pigments and photosynthetic characteristics in green and yellow sectors of the variegated Aucuba japonica ‘Variegata’ leaves. Flora 2018, 240, 25–33. [Google Scholar] [CrossRef]
- Wang, T.X.; Luo, C.; Liu, Z.Y.; Zhao, Y.; Zhu, Z.X.; Song, X.Q.; Zhou, Y.; Wang, J. Comparative transcriptomic analysis to postulate the generation of variegated leaves in Bougainvillea peruviana ‘Thimma’. Ind. Crops Prod. 2024, 212, 118364. [Google Scholar] [CrossRef]
- Brzezowski, P.; Richter, A.S.; Grimm, B. Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim. Biophys. Acta BBA-Bioenerg. 2015, 1847, 968–985. [Google Scholar] [CrossRef]
- Zhang, X.L.; Zhao, Z.T.; Zhang, M.; Wang, J.; Cheng, T.R.; Zhang, Q.X.; Pan, H.T. FsHemF is involved in the formation of yellow Forsythia leaves by regulating chlorophyll synthesis in response to light intensity. Plant Physiol. Biochem. 2023, 200, 107746. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Sui, C.H.; Liu, H.M.; Chen, J.J.; Han, Z.L.; Yan, Q.; Liu, S.Y.; Liu, H.Z. Effect of chlorophyll biosynthesis-related genes on the leaf color in Hosta (Hosta plantaginea Aschers) and Tobacco (Nicotiana tabacum L.). BMC Plant Biol. 2021, 21, 45. [Google Scholar] [CrossRef]
- Long, W.H.; Long, S.F.; Jiang, X.; Xu, H.F.; Peng, Q.; Li, J.L.; Zhang, X.C.; Zhang, D.S.; Liu, X.W.; Zhu, S.S. A rice yellow-green-leaf 219 mutant lacking the divinyl reductase affects chlorophyll biosynthesis and chloroplast development. J. Plant Growth Regul. 2022, 41, 3233–3242. [Google Scholar] [CrossRef]
- Al-Karadaghi, S.; Franco, R.; Hansson, M.; Shelnutt, J.A.; Isaya, G.; Ferreira, G.C. Chelatases: Distort to select? Trends Biochem. Sci. 2006, 31, 135–142. [Google Scholar] [CrossRef]
- Willows, R.D. Biosynthesis of chlorophylls from protoporphyrin IX. Nat. Prod. Rep. 2003, 20, 327–341. [Google Scholar] [CrossRef]
- Papenbrock, J.; Mock, H.P.; Tanaka, R.; Kruse, E.; Grimm, B. Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 2000, 122, 1161–1170. [Google Scholar] [CrossRef]
- Dong, X.Y.; Huang, L.B.; Chen, Q.S.; Lv, Y.Z.; Sun, H.N.; Liang, Z.H. Physiological and anatomical differences and differentially expressed genes reveal yellow leaf coloration in shumard oak. Plants 2020, 9, 169. [Google Scholar] [CrossRef]
- Ma, Y.Y.; Shi, J.C.; Wang, D.J.; Liang, X.; Wei, F.; Gong, C.M.; Qiu, L.J.; Zhou, H.C.; Folta, K.M.; Wen, Y.Q.; et al. A point mutation in the gene encoding magnesium chelatase I subunit influences strawberry leaf color and metabolism. Plant Physiol. 2023, 192, 2737–2755. [Google Scholar] [CrossRef]
- Fan, L.Q.; Hou, Y.; Zheng, L.; Shi, H.Y.; Liu, Z.; Wang, Y.X.; Li, S.D.; Liu, L.; Guo, M.Z.; Yang, Z.R.; et al. Characterization and fine mapping of a yellow leaf gene regulating chlorophyll biosynthesis and chloroplast development in cotton (Gossypium arboreum). Gene 2023, 885, 147712. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.X.; Han, C.Y.; Jia, Y.H.; Tian, F.F.; Liang, Z.K.; Yang, J.; Zhu, K.R.; Xiao, K.X.; Mi, J.X.; Liu, Q.L.; et al. Integrating physiological and transcriptomics analysis revealed the molecular mechanisms of PdCLH regulating leaf color and growth in poplar. Ind. Crops Prod. 2024, 220, 119281. [Google Scholar] [CrossRef]
- Kuai, B.; Chen, J.; Hörtensteiner, S. The biochemistry and molecular biology of chlorophyll breakdown. J. Exp. Bot. 2018, 69, 751–767. [Google Scholar] [CrossRef] [PubMed]
- Azoulay Shemer, T.; Harpaz-Saad, S.; Belausov, E.; Lovat, N.; Krokhin, O.; Spicer, V.; Standing, K.G.; Goldschmidt, E.E.; Eyal, Y. Citrus chlorophyllase dynamics at ethylene-induced fruit color-break: A study of chlorophyllase expression, posttranslational processing kinetics, and in situ intracellular localization. Plant Physiol. 2008, 148, 108–118. [Google Scholar] [CrossRef]
- Shimoda, Y.; Ito, H.; Tanaka, A. Conversion of chlorophyll b to chlorophyll a precedes magnesium dechelation for protection against necrosis in Arabidopsis. Plant J. 2012, 72, 501–511. [Google Scholar] [CrossRef]
- Pružinská, A.; Tanner, G.; Anders, I.; Roca, M.; Hörtensteiner, S. Chlorophyll breakdown: Pheophorbide a oxygenase is a rieske-type iron–sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 2003, 100, 15259–15264. [Google Scholar] [CrossRef]
- Tian, Y.R.; Rao, S.P.; Li, Q.Q.; Xu, M.; Wang, A.K.; Zhang, H.C.; Chen, J.H.; Tian, Y.; Rao, S.; Li, Q.; et al. The coloring mechanism of a novel golden variety in Populus deltoides based on the RGB color mode. For. Res. 2021, 1, 1–13. [Google Scholar] [CrossRef]
- Shalygo, N.; Czarnecki, O.; Peter, E.; Grimm, B. Expression of chlorophyll synthase is also involved in feedback-control of chlorophyll biosynthesis. Plant Mol. Biol. 2009, 71, 425. [Google Scholar] [CrossRef] [PubMed]
- Sinijadas, K.; Paul, A.; Radhika, N.S.; Johnson, J.M.; Manju, R.V.; Anuradha, T. Piriformospora indica suppresses the symptoms produced by Banana bract mosaic virus by inhibiting its replication and manipulating chlorophyll and carotenoid biosynthesis and degradation in banana. 3 Biotech 2024, 14, 141. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Rahman, M.L.; Cho, S.H.; Kim, Y.S.; Koh, H.J.; Yoo, S.C.; Paek, N.C. The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J. 2013, 74, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.Q.; Li, Y.J.; Zhao, Y.Y.; Shi, H.Z. Physiological, transcriptome and co-expression network analysis of chlorophyll-deficient mutants in flue-cured tobacco. BMC Plant Biol. 2023, 23, 153. [Google Scholar] [CrossRef]
- Zou, Y.P.; Huang, Y.J.; Zhang, D.L.; Chen, H.; Liang, Y.W.; Hao, M.Z.; Yin, Y.L. Molecular insights into a non-lethal yellow bud mutant in Ilex × ‘Nellie R. Stevens’. Sci. Hortic. 2024, 329, 113033. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Schelbert, S.; Park, S.Y.; Han, S.H.; Lee, B.-D.; Andrès, C.B.; Kessler, F.; Hörtensteiner, S.; Paek, N.C. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 2012, 24, 507–518. [Google Scholar] [CrossRef]
- Ladygin, V.G. Reduction of the chloroplast membrane system caused by disorders in early stages of chlorophyll biosynthesis. Russ. J. Plant Physiol. 2006, 53, 10–24. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Han, S.H.; Sakuraba, Y.; Koh, H.J.; Paek, N.C. Leaf variegation in the rice zebra2 mutant is caused by photoperiodic accumulation of tetra-Cis-lycopene and singlet oxygen. Mol. Cells 2012, 33, 87–97. [Google Scholar] [CrossRef]
- Matringe, M.; Camadro, J.M.; Block, M.A.; Joyard, J.; Scalla, R.; Labbe, P.; Douce, R. Localization within chloroplasts of protoporphyrinogen oxidase, the target enzyme for diphenylether-like herbicides. J. Biol. Chem. 1992, 267, 4646–4651. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.X.; Ma, K.; Mower, J.P.; Liu, Y.; Zhou, R.C. Leaf Variegation caused by plastome structural variation: An example from Dianella Tasmanica. Hortic. Res. 2024, 11, uhae009. [Google Scholar] [CrossRef] [PubMed]
- Dechkrong, P.; Srima, S.; Sukkhaeng, S.; Utkhao, W.; Thanomchat, P.; de Jong, H.; Tongyoo, P. Mutation mapping of a variegated EMS tomato reveals an FtsH-like protein precursor potentially causing patches of four phenotype classes in the leaves with distinctive internal morphology. BMC Plant Biol. 2024, 24, 265. [Google Scholar] [CrossRef]
- Azarin, K.; Usatov, A.; Makarenko, M.; Kozel, N.; Kovalevich, A.; Dremuk, I.; Yemelyanova, A.; Logacheva, M.; Fedorenko, A.; Averina, N. A point mutation in the photosystem I P700 chlorophyll a apoprotein A1 gene confers variegation in Helianthus annuus L. Plant Mol. Biol. 2020, 103, 373–389. [Google Scholar] [CrossRef]
- Li, D.J.; Xia, K.T.; Zhang, H.; Li, Z.Q.; Xie, X.D.; Zhou, H.N.; Zhai, N.; Xu, G.Y. Silencing of PDC-E1β genes affects chloroplast development and amino acid metabolism in tobacco. Ind. Crops Prod. 2025, 225, 120488. [Google Scholar] [CrossRef]
- Wu, X.X.; Mu, W.H.; Li, F.; Sun, S.Y.; Cui, C.J.; Kim, C.; Zhou, F.; Zhang, Y. Cryo-EM structures of the plant plastid-encoded RNA polymerase. Cell 2024, 187, 1127–1144.e21. [Google Scholar] [CrossRef]
- Nevo, R.; Charuvi, D.; Tsabari, O.; Reich, Z. Composition, architecture and dynamics of the photosynthetic apparatus in higher plants. Plant J. 2012, 70, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.B.; Wu, Z.S.; Shen, W.; Zhou, H.Y.; Li, H.; He, X.H.; Li, R.B.; Qin, B.X. Disruption of the rice ALS1 localized in chloroplast causes seedling-lethal albino phenotype. Plant Sci. 2024, 338, 111925. [Google Scholar] [CrossRef]
- Zheng, M.Y.; Wang, X.Y.; Luo, J.; Ma, B.J.; Li, D.Y.; Chen, X.F. The pleiotropic functions of GOLDEN2-LIKE transcription factors in plants. Front. Plant Sci. 2024, 15, 14458750. [Google Scholar] [CrossRef]
- Yelina, N.E.; Frangedakis, E.; Wang, Z.; Schreier, T.B.; Rever, J.; Tomaselli, M.; Forestier, E.C.F.; Billakurthi, K.; Ren, S.; Bai, Y.H.; et al. Streamlined regulation of chloroplast development in the liverwort Marchantia polymorpha. Cell Rep. 2024, 43, 114696. [Google Scholar] [CrossRef]
- Hernández-Muñoz, A.; Agreda-Laguna, K.A.; Ramírez-Bernabé, I.E.; Oltehua-López, O.; Arteaga-Vázquez, M.A.; Leon, P. Marchantia polymorpha GOLDEN2-LIKE transcriptional factor; a central regulator of chloroplast and plant vegetative development. New Phytol. 2024, 243, 1406–1423. [Google Scholar] [CrossRef] [PubMed]
- Rossini, L.; Cribb, L.; Martin, D.J.; Langdale, J.A. The maize Golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell 2001, 13, 1231–1244. [Google Scholar] [CrossRef]
- Pan, Y.; Bradley, G.; Pyke, K.; Ball, G.; Lu, C.; Fray, R.; Marshall, A.; Jayasuta, S.; Baxter, C.; van Wijk, R.; et al. Network inference analysis identifies an APRR2-Like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 2013, 161, 1476–1485. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qian, J.L.; Han, Y.T.; Jia, Y.; Kuang, H.H.; Chen, J.J. Alternative splicing triggered by the insertion of a CACTA transposon attenuates LsGLK and leads to the development of pale-green leaves in lettuce. Plant J. 2022, 109, 182–195. [Google Scholar] [CrossRef]
- Zou, Y.P.; Huang, Y.J.; Zhang, D.L.; Chen, H.; Liang, Y.W.; Hao, M.Z.; Yin, Y.L. Molecular mechanisms of chlorophyll deficiency in Ilex × attenuata ‘Sunny Foster’ mutant. Plants 2024, 13, 1284. [Google Scholar] [CrossRef]
- Chen, X.; Yang, X.L.; Xie, J.; Ding, W.J.; Li, Y.L.; Yue, Y.Z.; Wang, L.G. Biochemical and comparative transcriptome analyses reveal key genes involved in major metabolic regulation related to colored leaf formation in Osmanthus fragrans ‘Yinbi Shuanghui’ during development. Biomolecules 2020, 10, 549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.L.; Zhang, J.K.; Mao, Y.F.; Yin, Y.J.; Shen, X. Physiological analysis and transcriptome sequencing of a delayed-green leaf mutant ‘Duojiao’ of ornamental crabapple (Malus sp.). Physiol. Mol. Biol. Plants 2022, 28, 1833–1848. [Google Scholar] [CrossRef]
- Li, W.X.; Yang, S.B.; Lu, Z.G.; He, Z.C.; Ye, Y.L.; Zhao, B.B.; Wang, L.; Jin, B. Cytological, physiological, and transcriptomic analyses of golden leaf coloration in Ginkgo biloba L. Hortic. Res. 2018, 5, 12. [Google Scholar] [CrossRef]
- Gonnet, J.F. Colour effects of co-pigmentation of anthocyanin revisited—3. A further description using CIELAB differences and assessment of matched colours using the CMC model. Food Chem. 2001, 75, 473–485. [Google Scholar] [CrossRef]
- Wrolstad, R.E.; Durst, R.W.; Lee, J. Tracking color and pigment changes in anthocyanin products. Trends Food Sci. Technol. 2005, 16, 423–428. [Google Scholar] [CrossRef]
- Ma, D.Y.; Sun, D.X.; Wang, C.Y.; Li, Y.G.; Guo, T.C. Expression of flavonoid biosynthesis genes and accumulation of flavonoid in wheat leaves in response to drought stress. Plant Physiol. Biochem. 2014, 80, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Qian, P.; Marklew, C.J.; Viney, J.; Davison, P.A.; Brindley, A.A.; Söderberg, C.; Al-Karadaghi, S.; Bullough, P.A.; Grossmann, J.G.; Hunter, C.N. Structure of the cyanobacterial magnesium chelatase H subunit determined by single particle reconstruction and small-angle X-ray scattering. J. Biol. Chem. 2012, 287, 4946–4956. [Google Scholar] [CrossRef] [PubMed]
- Kumachova, T.; Babosha, A.; Ryabchenko, A.; Ivanova, T.; Voronkov, A. Leaf epidermis in rosaceae: Diversity of the cuticular folding and microstructure. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2021, 91, 455–470. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Wang, S.F.; Song, S.H.; Xu, F.S.; Pan, Y.H.; Wang, H. Transcriptomic and proteomic analyses reveal new insight into chlorophyll synthesis and chloroplast structure of maize leaves under zinc deficiency stress. J. Proteomics 2019, 199, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Chong, X.R.; Wang, Y.; Xu, X.Y.; Zhang, F.; Wang, C.Y.; Zhou, Y.W.; Zhou, T.; Li, Y.L.; Lu, X.Q.; Chen, H. Efficient virus-induced gene silencing in Ilex dabieshanensis using tobacco rattle virus. Forests 2023, 14, 488. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
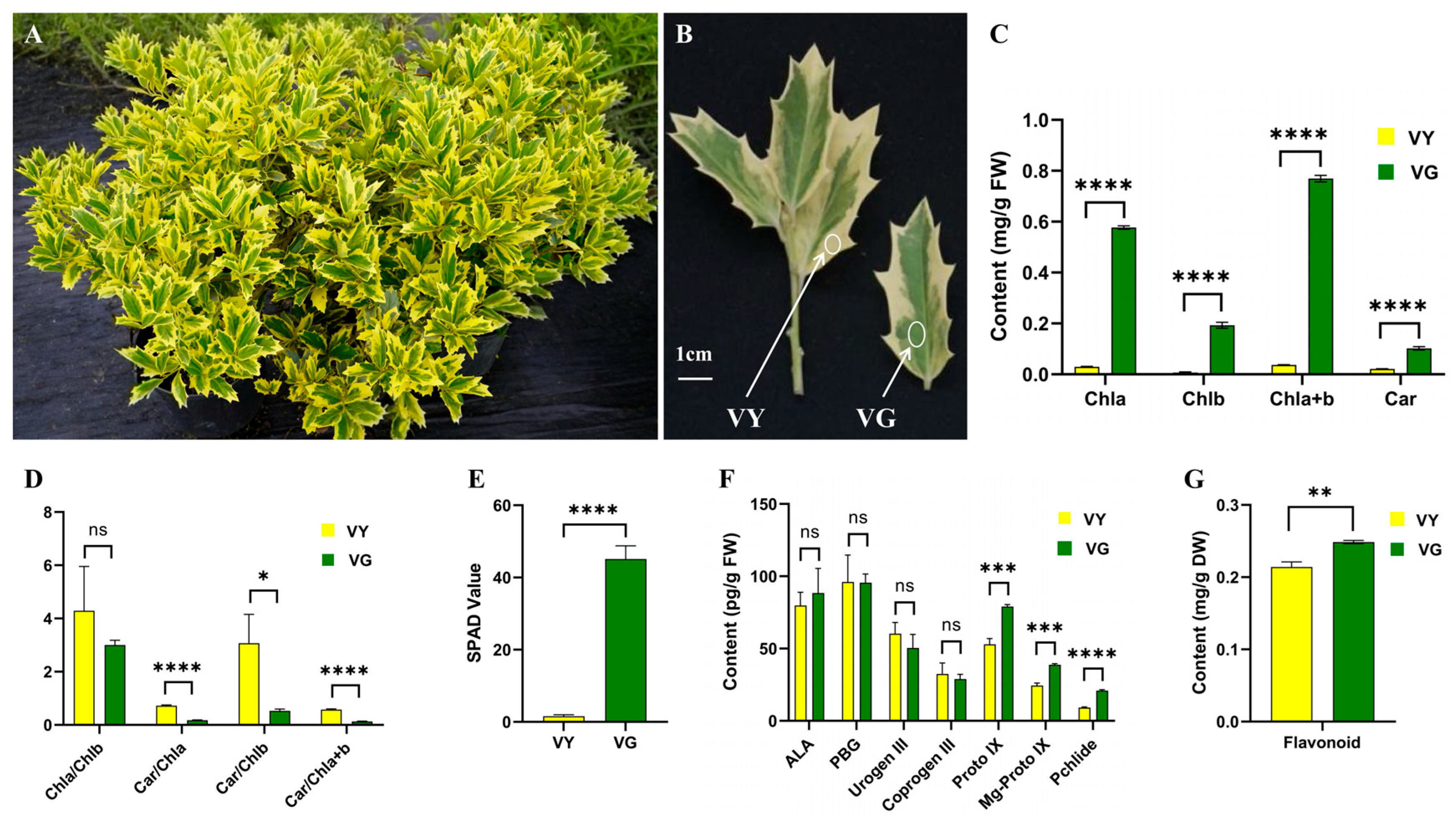
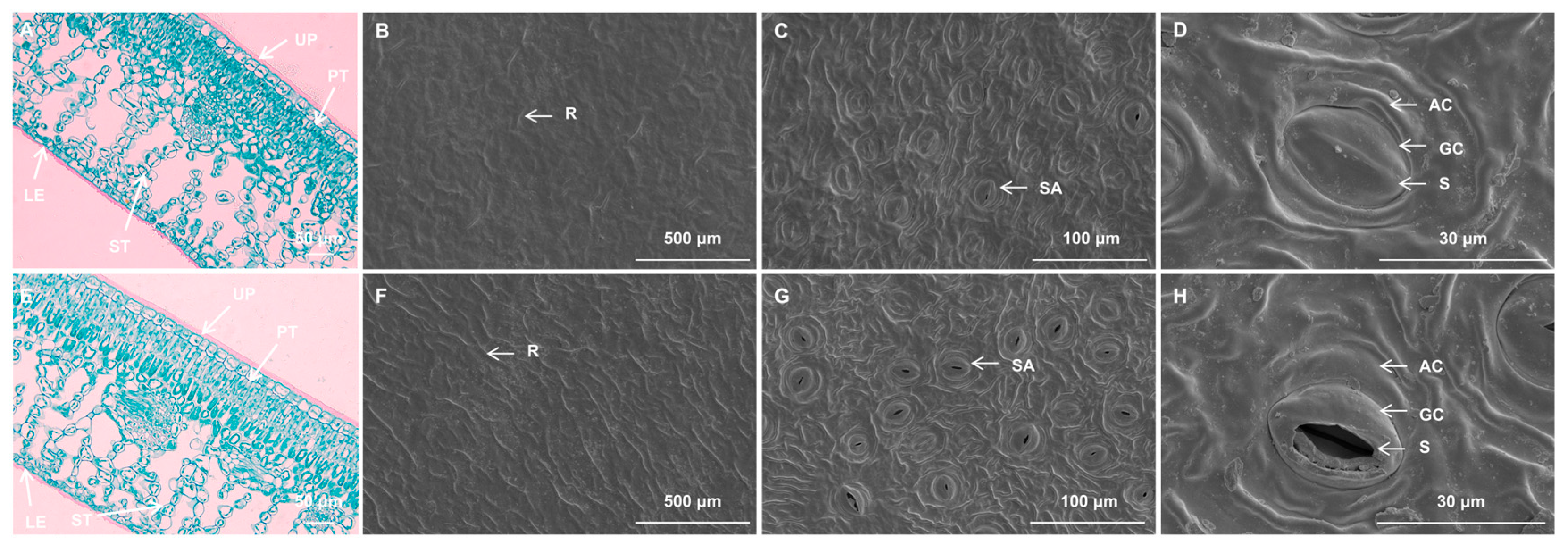
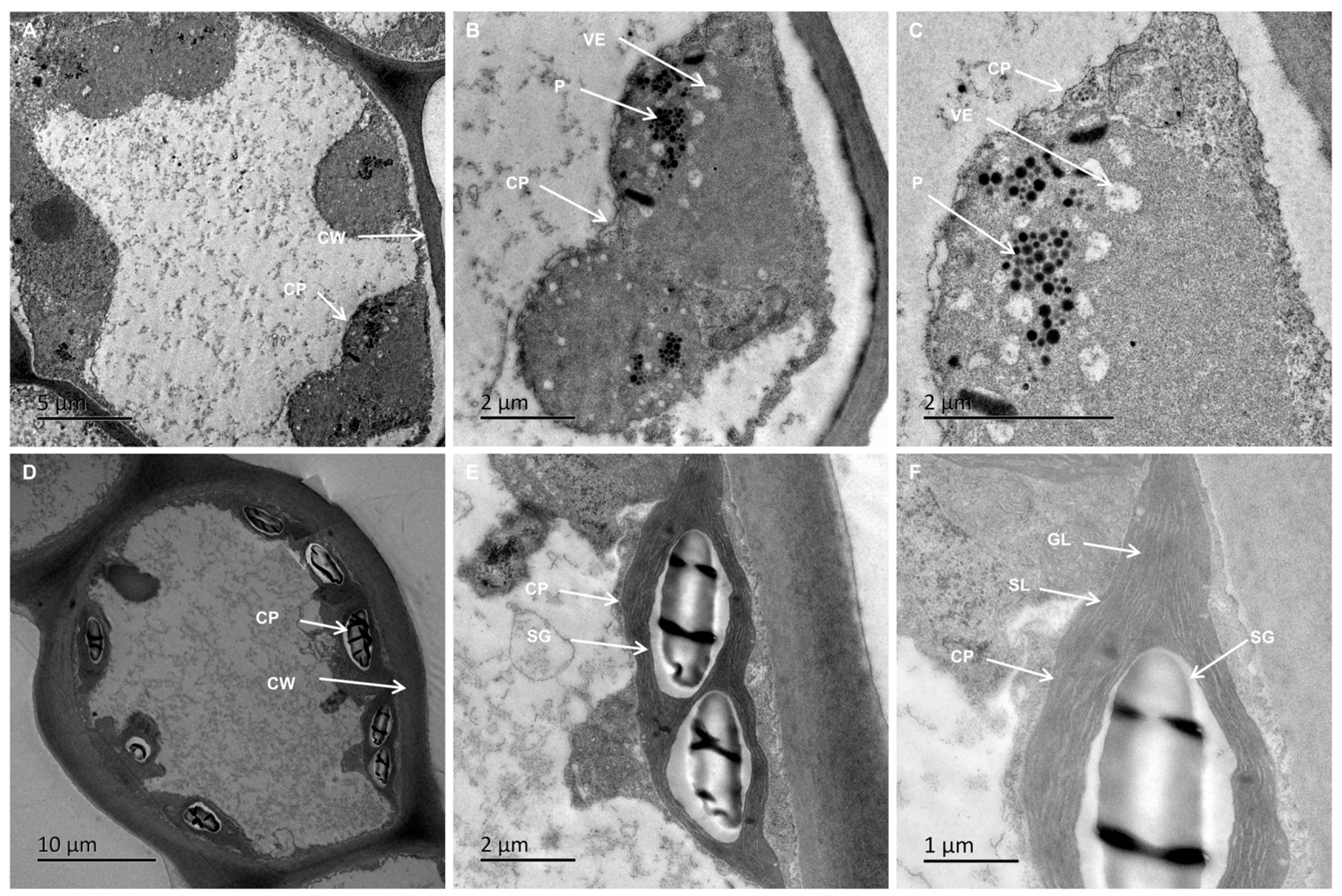
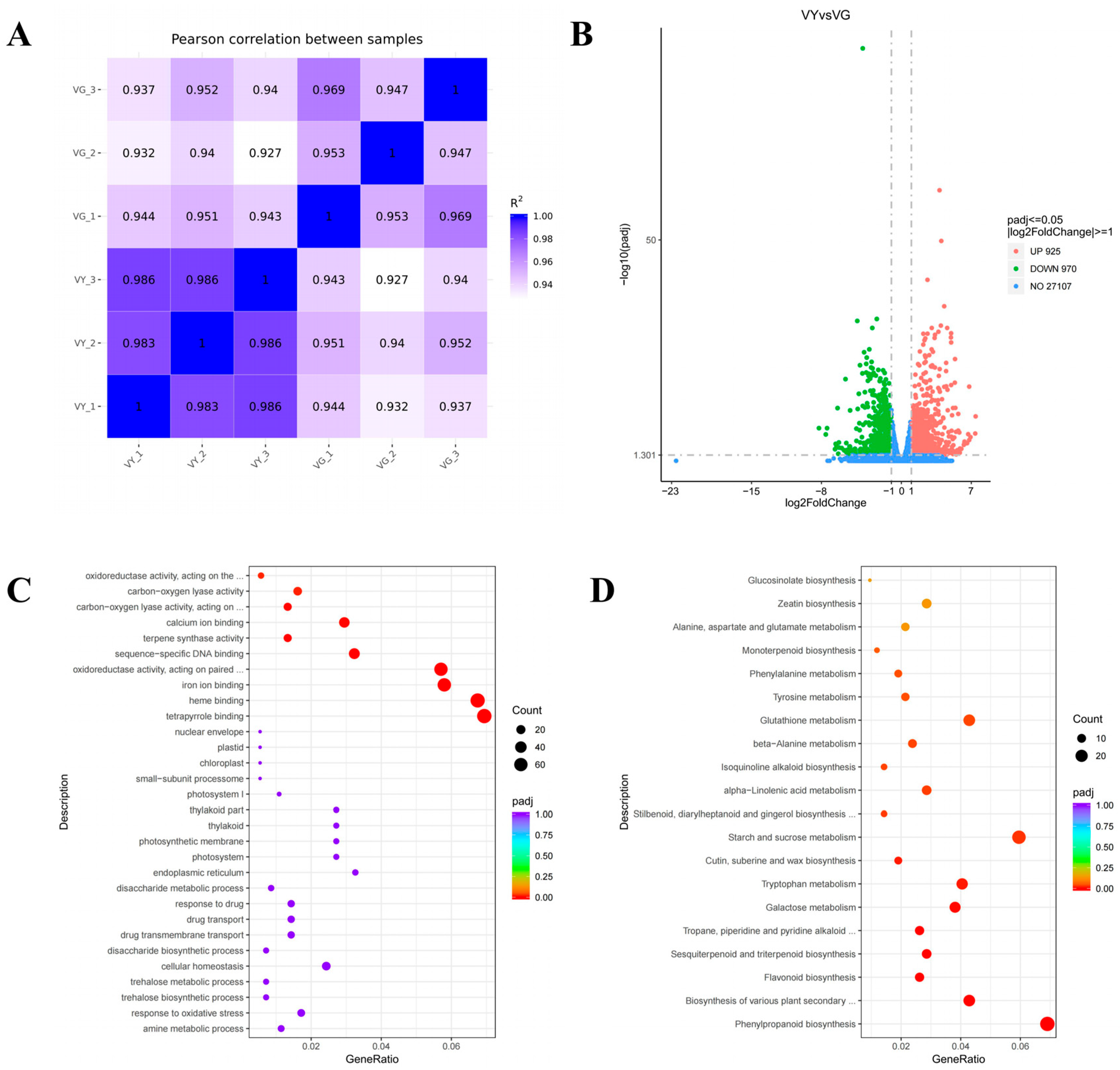
| Color Indices | VY | VG |
|---|---|---|
| L* | 75.17 ± 0.30 a | 46.76 ± 2.20 b |
| a* | 3.37 ± 1.07 a | −9.16 ± 0.61 b |
| b* | 53.99 ± 5.50 a | 27.02 ± 4.10 b |
| C | 54.10 ± 5.56 a | 28.56 ± 3.80 b |
| H° | 86.48 ± 0.76 b | 104.40 ± 11.70 a |
| Ultrastructural Parameters | VY | VG |
|---|---|---|
| Chloroplast number (per cell) | 5.00 ± 0.82 b | 8.67 ± 1.15 a |
| Chloroplast length (μm) | 4.77 ± 1.00 b | 6.78 ± 0.22 a |
| Chloroplast width (μm) | 4.29 ± 0.97 a | 4.01 ± 0.18 a |
| Chloroplast length/width | 1.12 ± 0.08 b | 1.69 ± 0.03 a |
| Starch granule number (per cell) | 1.19 ± 0.19 a | 0.00 ± 0.00 b |
| Parameters | VY | VG | |
|---|---|---|---|
| Photosynthesis | Pn [μmol(CO2) m−2 s−1] | −2.43 ± 0.25 b | 2.87 ± 0.15 a |
| E [mol(H2O) m−2 s−1] | 0.63 ± 0.32 b | 1.13 ± 0.35 a | |
| Gs [mol(H2O) m−2 s−1] | 28.33 ± 15.50 a | 47.33 ± 16.62 a | |
| Ci [μmol(CO2) mol−1] | 574.67 ± 72.14 a | 303.00 ± 43.31 b | |
| Chlorophyll fluorescence | Fv/Fm | 0.16 ± 0.07 b | 0.70 ± 0.03 a |
| Y (II) | 0.12 ± 0.08 b | 0.47 ± 0.02 a | |
| NPQ | 0.05 ± 0.01 b | 0.62 ± 0.08 a | |
| Y (NO) | 0.84 ± 0.08 a | 0.33 ± 0.02 b | |
| Y (NPQ) | 0.04 ± 0.01 b | 0.20 ± 0.02 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, Y.; Zhuo, T.; Duan, Y.; Chen, H.; Zhou, P.; Hao, M.; Yin, Y.; Zhang, D. Transcriptomic and Structural Insights into Leaf Variegation Development in Ilex × ‘Solar Flare’. Int. J. Mol. Sci. 2025, 26, 3999. https://doi.org/10.3390/ijms26093999
Zou Y, Zhuo T, Duan Y, Chen H, Zhou P, Hao M, Yin Y, Zhang D. Transcriptomic and Structural Insights into Leaf Variegation Development in Ilex × ‘Solar Flare’. International Journal of Molecular Sciences. 2025; 26(9):3999. https://doi.org/10.3390/ijms26093999
Chicago/Turabian StyleZou, Yiping, Tao Zhuo, Yan Duan, Hong Chen, Peng Zhou, Mingzhuo Hao, Yunlong Yin, and Donglin Zhang. 2025. "Transcriptomic and Structural Insights into Leaf Variegation Development in Ilex × ‘Solar Flare’" International Journal of Molecular Sciences 26, no. 9: 3999. https://doi.org/10.3390/ijms26093999
APA StyleZou, Y., Zhuo, T., Duan, Y., Chen, H., Zhou, P., Hao, M., Yin, Y., & Zhang, D. (2025). Transcriptomic and Structural Insights into Leaf Variegation Development in Ilex × ‘Solar Flare’. International Journal of Molecular Sciences, 26(9), 3999. https://doi.org/10.3390/ijms26093999


_Kim.png)






