Genome-Wide Identification and Functional Analysis of C2H2 Zinc Finger Transcription Factor Genes in the Intertidal Macroalga Pyropia haitanensis
Abstract
:1. Introduction
2. Results
2.1. Identification of Transcription Factors and Phylogenetic Analysis of C2H2 Gene Family
2.2. Physicochemical Properties and Predicted Cis-Acting Elements of PhC2H2 Genes
2.3. Structures, Motifs, and Transposons of PhC2H2 Genes
2.4. Chromosomal Localization, Replication, and Collinearity of PhC2H2 Genes
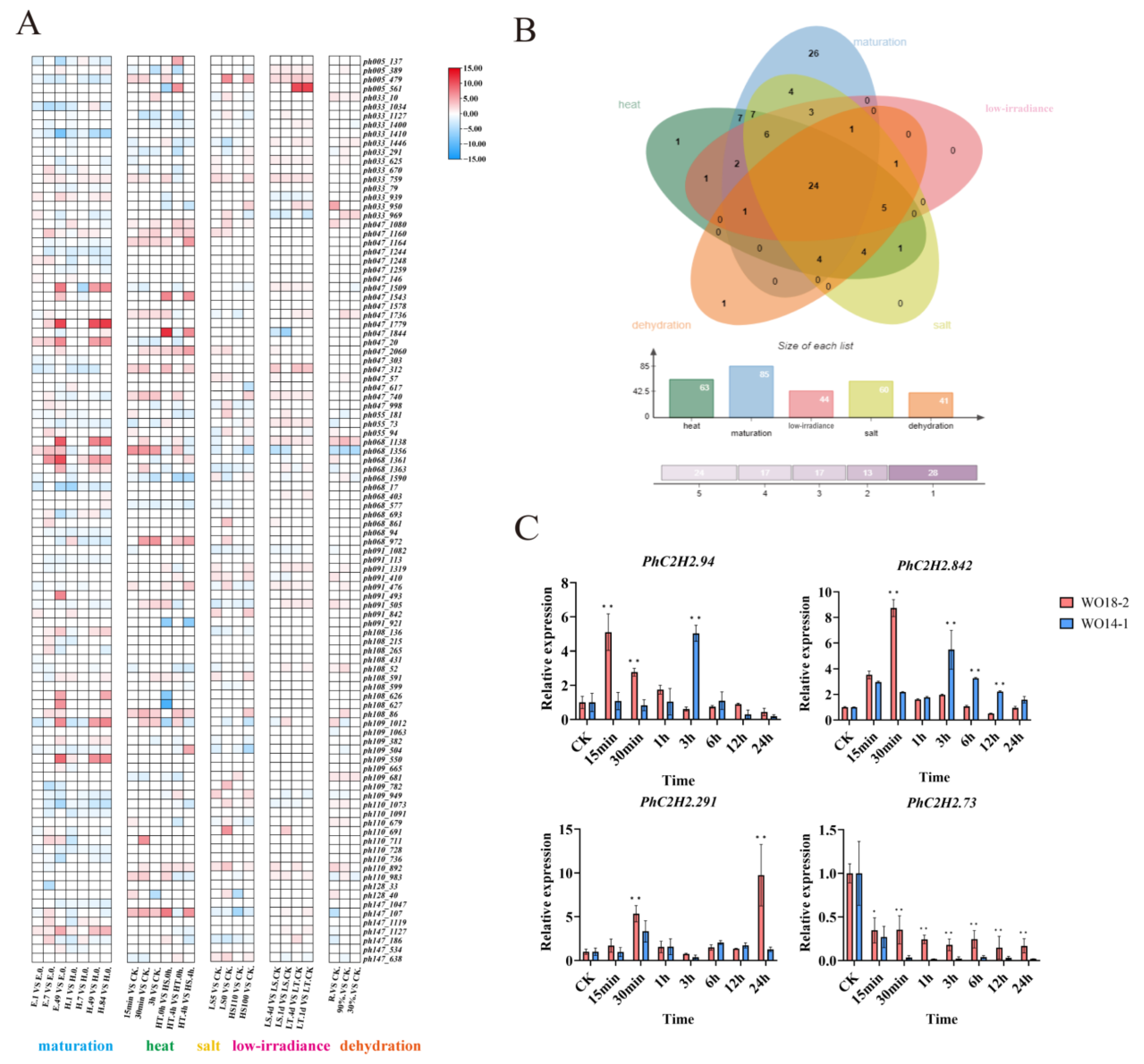
2.5. Gene Expression Pattern Analysis and qRT-PCR Verification
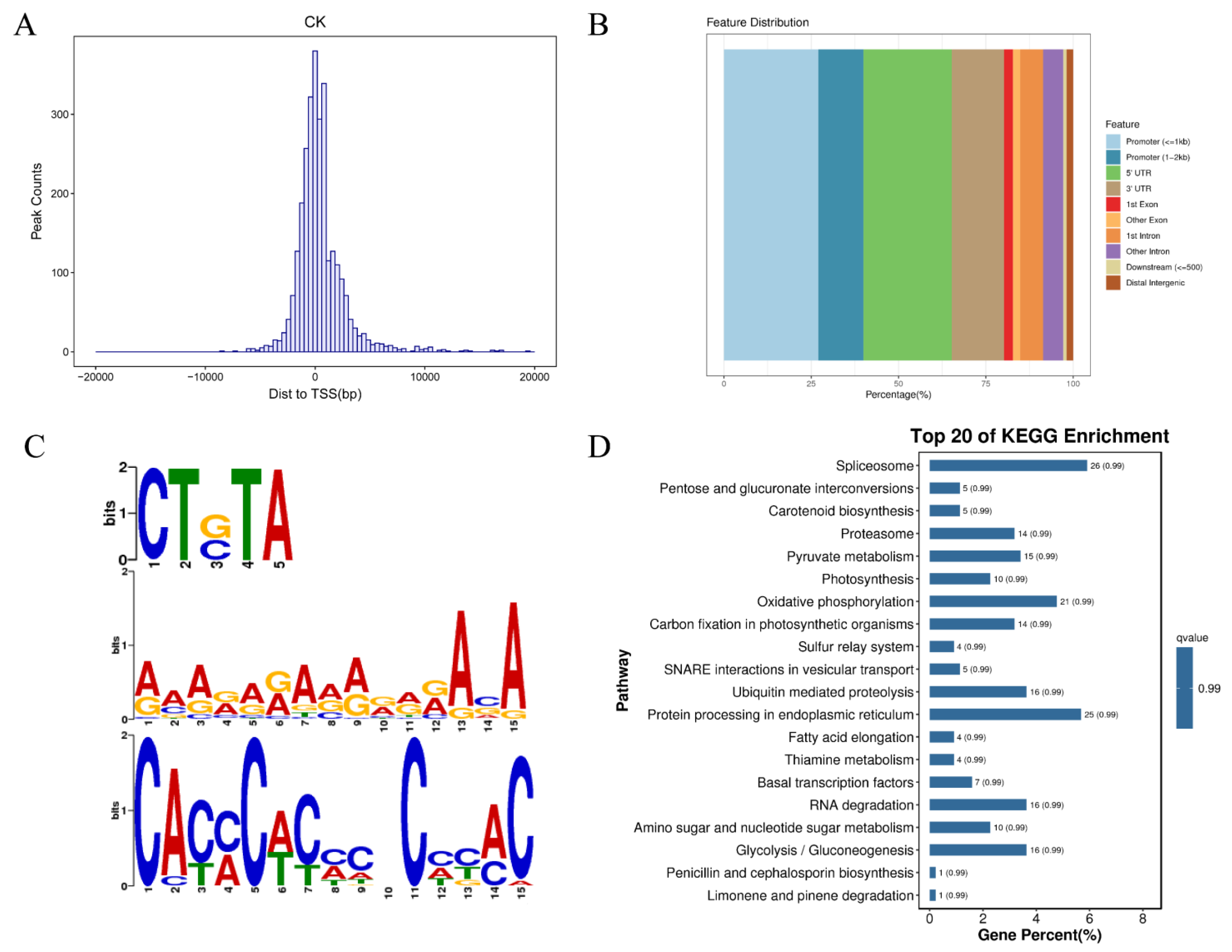
2.6. DAP-seq
2.7. Stress Resistance of Transgenic C. reinhardtii Expressing PhC2H2.94
3. Discussion
4. Materials and Methods
4.1. Identification and Phylogenetic Analysis of C2H2 Gene Family Members
4.2. Structures, Motifs, Cis-Acting Elements, and Physicochemical Properties of PhC2H2 Genes
4.3. Analyses of the Chromosomal Localization, Duplication, Collinearity, and Transposons of PhC2H2 Genes
4.4. Treatment of Experimental Materials
4.5. Isolation and Purification of Total RNA and Synthesis of cDNA
4.6. Gene Expression Pattern Analysis and Verification by qRT-PCR
4.7. Cloning and DAP-seq of PhC2H2.94
4.8. Construction of a PhC2H2.94 Expression Vector and Transformation of Chlamydomonas reinhardtii
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moreira, D.; Le Guyader, H.; Philippe, H. The origin of red algae and the evolution of chloroplasts. Nature 2000, 405, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Brawley, S.H.; Blouin, N.A.; Ficko-Blean, E.; Wheeler, G.L.; Lohr, M.; Goodson, H.V.; Jenkins, J.W.; Blaby-Haas, C.E.; Helliwell, K.E.; Chan, C.X.; et al. Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc. Natl. Acad. Sci. USA 2017, 114, E6361–E6370. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.H.; Park, S.I.; Huang, T.-Y.; Lee, Y.; Ciniglia, C.; Yadavalli, H.C.; Yang, S.W.; Bhattacharya, D.; Yoon, H.S. Genome-wide signatures of adaptation to extreme environments in red algae. Nat. Commun. 2023, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, J.; Cho, C.H.; Yoon, H.S.; Bhattacharya, D. Extremophilic red algae as models for understanding adaptation to hostile environments and the evolution of eukaryotic life on the early earth. Semin. Cell Dev. Biol. 2022, 134, 4–13. [Google Scholar] [CrossRef]
- Huang, L.-B.; Peng, L.-N.; Yan, X.-H. Multi-omics responses of red algae Pyropia haitanensis to intertidal desiccation during low tides. Algal Res. 2021, 58, 102376. [Google Scholar] [CrossRef]
- Fu, S.; Xue, S.; Chen, J.; Shang, S.; Xiao, H.; Zang, Y.; Tang, X. Effects of Different Short-Term UV-B Radiation Intensities on Metabolic Characteristics of Porphyra haitanensis. Int. J. Mol. Sci. 2021, 22, 2180. [Google Scholar] [CrossRef]
- Ma, M.J.; Wei, C.L.; Zhu, J.K.; Liu, Q.Q.; Luo, Q.J.; Chen, H.M.; Yang, R. The role of Abscisic Acid and cold adaption in improving the cryopreservation efficiency of Pyropia haitanensis conchoceils. Acta Hydrobiol. Sin. 2022, 46, 184–193. [Google Scholar]
- He, P.; Zhang, Z.; Zhang, X. Seaweed Cultivation; Science Press: Beijing, China, 2018. [Google Scholar]
- Chen, H.; Chu, J.-S.C.; Chen, J.; Luo, Q.; Wang, H.; Lu, R.; Zhu, Z.; Yuan, G.; Yi, X.; Mao, Y.; et al. Insights into the Ancient Adaptation to Intertidal Environments by Red Algae Based on a Genomic and Multiomics Investigation of Neoporphyra haitanensis. Mol. Biol. Evol. 2021, 39, 315. [Google Scholar] [CrossRef]
- Wang, Z.; Lu, C.; Chen, J.; Luo, Q.; Yang, R.; Gu, D.; Wang, T.; Zhang, P.; Chen, H. Physiological and multi-omics responses of Neoporphyra haitanensis to dehydration-rehydration cycles. BMC Plant Biol. 2022, 22, 168. [Google Scholar] [CrossRef]
- Xu, D.; Sun, N.; Xuan, S.; Wang, C.; Huang, T.; Li, C.; Zhang, J.; Yang, W. Effect of different drying methods on the physicochemical properties and phycobiliprotein structure of Porphyra haitanensis. Int. J. Food Eng. 2021, 17, 111–120. [Google Scholar] [CrossRef]
- Wang, W.; Zheng, H.; Wen, J.; Xu, K.; Xu, Y.; Ji, D.; Chen, C.; Xie, C. Early signaling events in the heat stress response of Pyropia haitanensis revealed by phosphoproteomic and lipidomic analyses. Algal Res. 2022, 67, 102837. [Google Scholar] [CrossRef]
- Zheng, H.Y.; Xu, Y.; Ji, D.; Xu, K.; Chen, C.; Wang, W.; Xie, C. Calcium-Calmodulin-Involved Heat Shock Response of Neoporphyra haitanensis. Front. Mar. Sci. 2022, 9, 875308. [Google Scholar] [CrossRef]
- Wen, J.; Xu, K.; Ji, D.; Xu, Y.; Chen, C.; Wang, W.; Xie, C. The mechanism of maintaining intracellular homeostasis in the red alga Pyropia haitanensis under hyposaline stress. Front. Mar. Sci. 2022, 7, 415. [Google Scholar]
- Zhao, R.; Wen, J.; Xu, K.; Xu, Y.; Ji, D.; Xie, C.; Wang, W. Ubiquitination-dependent protein homeostasis is critical for the thermotolerance of the economically valuable seaweed Pyropia haitanensis. Aquaculture 2023, 581, 740466. [Google Scholar] [CrossRef]
- Farnham, P.J. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009, 10, 605–616. [Google Scholar] [CrossRef]
- Zhong, C.J.; Peng, W.Y.; Wang, B. The research progress of C2H2 zinc finger protein related to plant stress response. Plant Physiol. J. 2020, 56, 2356–2366. [Google Scholar]
- Takatsuji, H. Zinc-finger transcription factors in plants. CMLS Cell Mol. Life Sci. 1998, 54, 582–596. [Google Scholar] [CrossRef]
- Zhou, C.; Li, L.Y. Advances in C2H2 zinc finger proteins. Life Sci. Res. 2004, 8, 215–220. [Google Scholar]
- Kundu, A.; Das, S.; Basu, S.; Kobayashi, Y.; Koyama, H.; Ganesan, M. GhSTOP1, a C2H2 type zinc finger transcription factor is essential for Aluminum and proton stress tolerance and lateral root initiation in cotton. Plant Biol. 2019, 21, 35–44. [Google Scholar] [CrossRef]
- Liu, Y.; Khan, A.R.; Gan, Y. C2H2 Zinc Finger Proteins Response to Abiotic Stress in Plants. Int. J. Mol. Sci. 2022, 23, 2730. [Google Scholar] [CrossRef]
- Takatsuji, H.M.M.; Benfey, P.N. Characterization of a zinc finger DNA-binding protein expressed specifically in petunia petals and seedlings. EMBO J. 1992, 11, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Muthamilarasan, M.; Bonthala, V.S.; Mishra, A.K.; Khandelwal, R.; Khan, Y.; Roy, R.; Prasad, M. C2H2 type of zinc finger transcription factors in foxtail millet define response to abiotic stresses. Funct. Integr. Genom. 2014, 14, 531–541. [Google Scholar] [CrossRef]
- Wei, K.; Pan, S.; Li, Y. Functional characterization of maize C2H2 zinc-finger gene family. Plant Mol. Biol. Report. 2015, 34, 761–776. [Google Scholar] [CrossRef]
- Yang, M.; Chao, T.; Wang, D.; Hu, J.; Wu, H.; Gong, D.; Liu, G. Genome-wide identification and expression profiling of the C2H2-type zinc finger protein transcription factor family in tobacco. Hereditas 2016, 38, 337–349. [Google Scholar]
- Yuan, S.; Li, X.; Li, R.; Wang, L.; Zhang, C.; Chen, L.; Hao, Q.; Zhang, X.; Chen, H.; Shan, Z.; et al. Genome-wide identification and classification of soybean C2H2 zinc finger proteins and their expression analysis in legume-rhizobium symbiosis. Front. Microbiol. 2018, 9, 126. [Google Scholar] [CrossRef]
- Li, W.; Zhu, Z.; Chern, M.; Yin, J.; Yang, C.; Ran, L.; Cheng, M.; He, M.; Wang, K.; Wang, J.; et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 2017, 170, 114–126. [Google Scholar] [CrossRef]
- Sakamoto, H.; Araki, T.; Meshi, T.; Iwabuchi, M. Expression of a subset of the Arabidopsis Cys2/His2 -type zinc-finger protein gene family under water stress. Genes 2000, 248, 23–32. [Google Scholar] [CrossRef]
- Puentes-Romero, A.C.; González, S.A.; González-Villanueva, E.; Figueroa, C.R.; Ruiz-Lara, S. AtZAT4, a C2H2-Type Zinc Finger Transcription Factor from Arabidopsis thaliana, Is Involved in Pollen and Seed Development. Plants 2022, 11, 1974. [Google Scholar] [CrossRef]
- Bai, Q.; Niu, Z.; Chen, Q.; Gao, C.; Zhu, M.; Bai, J.; Liu, M.; He, L.; Liu, J.; Jiang, Y.; et al. The C2H2-type zinc finger transcription factor OSIC1 positively regulates stomatal closure under osmotic stress in poplar. Plant Biotechnol. J. 2023, 21, 943–960. [Google Scholar] [CrossRef]
- Xu, Y.; Shi, Y.Q.; Zhang, W.J.; Zhu, K.K.; Feng, L.G.; Wang, J.W. C2H2 Zinc Finger Protein Family Analysis of Rosa rugosa Identified a Salt-Tolerance Regulator, RrC2H2-8. Plants 2024, 13, 3580. [Google Scholar] [CrossRef]
- Zhou, X.J.; Gao, T.; Zhang, Y.M.; Han, M.; Shen, Y.X.; Su, Y.; Feng, X.L.; Wu, Q.; Sun, G.L.; Wang, Y.L. Genome-wide identification, characterization and expression of C2H2 zinc finger gene family in Opisthopappus species under salt stress. BMC Genom. 2024, 25, 385. [Google Scholar] [CrossRef]
- Li, X.; Cao, X.B.; Li, J.L.; Niu, Q.Q.; Mo, Y.P.; Xiao, L.H. Genome-wide characterization of C2H2 zinc-finger gene family provides insight into the mechanisms and evolution of the dehydration-rehydration responses in Physcomitrium and Arabidopsis. Front. Plant Sci. 2022, 13, 953459. [Google Scholar] [CrossRef]
- Xu, Y.; Leung, S.K.K.; Li, T.M.W.; Yung, C.C.M. Hidden genomic diversity drives niche partitioning in a cosmopolitan eukaryotic picophytoplankton. ISME J. 2024, 18, 163. [Google Scholar] [CrossRef] [PubMed]
- Petroll, R.; Schreiber, M.; Finke, H.; Cock, J.M.; Gould, S.B.; Rensing, S.A. Signatures of Transcription Factor Evolution and the Secondary Gain of Red Algae Complexity. Genes 2021, 12, 1055. [Google Scholar] [CrossRef]
- Wang, W.; Ge, Q.; Wen, J.; Zhang, H.; Guo, Y.; Li, Z.; Xu, Y.; Ji, D.; Chen, C.; Guo, L.; et al. Horizontal gene transfer and symbiotic microorganisms regulate the adaptive evolution of intertidal algae, Porphyra sense lato. Commun. Biol. 2024, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Zhao, X.; Song, C.; Huang, Y.; Jia, Z.; Song, S. Research progress of plant CaMs/CMLs family. Acta Phytophysiol. Sin. 2024, 60, 1357–1366. [Google Scholar]
- Englbrecht, C.C.; Schoof, H.; Böhm, S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genom. 2004, 5, 39. [Google Scholar] [CrossRef]
- Cao, M.; Xu, K.; Yu, X.; Bi, G.; Liu, Y.; Kong, F.; Sun, P.; Tang, X.; Du, G.; Ge, Y.; et al. A chromosome-level genome assembly of Pyropia haitanensis (Bangiales, Rhodophyta). Mol. Ecol. Resour. 2019, 21, 216–227. [Google Scholar]
- Varela-Álvarez, E.; Loureiro, J.; Paulino, C.; Serrão, E.A. Polyploid lineages in the genus Porphyra. Sci. Rep. 2018, 8, 8696. [Google Scholar] [CrossRef]
- Lee, J.; Yang, E.C.; Graf, L.; Yang, J.H.; Qiu, H.; Zelzion, U.; Chan, C.X.; Stephens, T.G.; Weber, A.P.M.; Boo, G.H.; et al. Analysis of the Draft Genome of the Red Seaweed Gracilariopsis chorda Provides Insights into Genome Size Evolution in Rhodophyta. Mol. Biol. Evol. 2018, 35, 1869–1886. [Google Scholar] [CrossRef]
- Wu, S.; Han, B.; Jiao, Y. Genetic Contribution of Paleopolyploidy to Adaptive Evolution in Angiosperms. Mol. Plant 2019, 13, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Maere, S.; Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Geng, Y.; Ji, C.; Du, H.; Wong, C.E.; Zhang, Q.; Zhang, Y.; Zhang, P.; Riaz, A.; Chachar, S.; et al. Mesostigma viride Genome and Transcriptome Provide Insights into the Origin and Evolution of Streptophyta. Adv. Sci. 2019, 7, 1901850. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Li, Z.J.; Liu, J.Y.; Zhang, Y.; Ye, L.H.; Peng, Y.; Wang, H.; Diao, H.; Ma, Y.; Wang, M.; et al. Transposable elements orchestrate subgenome-convergent and -divergent transcription in common wheat. Nat. Commun. 2022, 13, 6940. [Google Scholar] [CrossRef]
- Baduel, P.; Quadrana, L.; Hunter, B.; Bomblies, K.; Colot, V. Relaxed purifying selection in autopolyploids drives transposable element over-accumulation which provides variants for local adaptation. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Groot Crego, C.; Hess, J.; Yardeni, G.; de La Harpe, M.; Priemer, C.; Beclin, F.; Saadain, S.; Cauz-Santos, L.A.; Temsch, E.M.; Weiss-Schneeweiss, H.; et al. CAM evolution is associated with gene family expansion in an explosive bromeliad radiation. Plant Cell 2024, 36, 4109–4131. [Google Scholar] [CrossRef]
- Petrillo, E. Don’t panic: An intron-centric guide to alternative splicing. Plant Cell 2023, 35, 1752–1761. [Google Scholar] [CrossRef]
- Kumari, A.; Sedehizadeh, S.; Brook, J.D.; Kozlowski, P.; Wojciechowska, M. Differential fates of introns in gene expression due to global alternative splicing. Hum. Genet. 2021, 141, 31–47. [Google Scholar] [CrossRef]
- Prochetto, S.; Reinheimer, R. Step by step evolution of Indeterminate Domain (IDD) transcriptional regulators: From algae to angiosperms. Ann. Bot. 2020, 126, 85–101. [Google Scholar] [CrossRef]
- Li, P.; Yu, A.M.; Sun, R.; Liu, A.Z. Function and Evolution of C1-2i Subclass of C2H2-Type Zinc Finger Transcription Factors in POPLAR. Genes 2022, 13, 1843. [Google Scholar] [CrossRef]
- Yoshioka, K.-I.; Fukushima, S.; Yamazaki, T.; Yoshida, M.; Takatsuji, H. The plant zinc finger protein ZPT2-2 has a unique mode of DNA interaction. J. Biol. Chem. 2001, 276, 35802–35807. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, H.; Maruyama, K.; Sakuma, Y.; Meshi, T.; Iwabuchi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004, 136, 2734–2746. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Zhu, Z.; Zhu, Z.; Yang, R.; Qian, F.; Chen, H.; Yan, X. Different responses to heat shock stress revealed heteromorphic adaptation strategy of Pyropia haitanensis (Bangiales, Rhodophyta). PLoS ONE 2014, 9, e94354. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Wang, W.; Xu, K.; Xu, Y.; Ji, D.; Chen, C.; Xie, C. Ca2+ influences heat shock signal transduction in Pyropia haitanensis. Aquaculture 2020, 516, 734618. [Google Scholar] [CrossRef]
- Hirata, R.; Uji, T.; Fukuda, S.; Mizuta, H.; Fujiyama, A.; Tabata, S.; Saga, N. Development of a nuclear transformation system with a codon-optimized selection marker and reporter genes in Pyropia yezoensis (Rhodophyta). J. Appl. Phycol. 2014, 26, 1863–1868. [Google Scholar] [CrossRef]
- De Saeger, J.; Coulembier Vandelannoote, E.; Lee, H.; Park, J.; Blomme, J. Genome editing in macroalgae: Advances and challenges. Front. Genome Ed. 2024, 6, 1380682. [Google Scholar] [CrossRef]
- Zhang, H.; Zeng, G.X.; Xie, J.J.; Zhang, Y.C.; Ji, D.; Xu, Y.; Wang, W.; Xie, X. PhbZIP2 regulates photosynthesis-related genes in an intertidal macroalgae, Pyropia haitanensis, under stress. Front. Mol. Biosci. 2024, 11, 1345585. [Google Scholar] [CrossRef]
- Wang, H.; Xie, X.J.; Gu, W.H.; Zheng, Z.B.; Zhuo, J.T.; Shao, Z.Z.; Huan, L.; Zhang, B.; Niu, J.; Gao, S.; et al. Gene editing of economic macroalga Neopyropia yezoensis (Rhodophyta) will promote its development into a model species of marine algae. New Phytol. 2024, 244, 1687–1691. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, K.; Xu, Y.; Ji, D.; Chen, C.; Wang, W.; Xie, C. Transcriptome co-expression network analysis identifies key genes regulating conchosporangia maturation of Pyropia haitanensis. Front. Genet. 2021, 12, 680120. [Google Scholar] [CrossRef]
- Wang, W.; Chang, J.; Zheng, H.; Ji, D.; Xu, Y.; Chen, C.; Xie, C. Full-length transcriptome sequences obtained by a combination of sequencing platforms applied to heat shock proteins and polyunsaturated fatty acids biosynthesis in Pyropia haitanensis. J. Appl. Phycol. 2018, 31, 1483–1492. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Chen, T.; Xing, L.; Xu, K.; Xu, Y.; Chen, C.; Xie, C. Regulatory mechanisms underlying the maintenance of homeostasis in Pyropia haitanensis under hypersaline stress conditions. Sci. Total Environ. 2019, 662, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Zhang, Y.; Zhang, B.; Xu, Y.; Xu, K.; Chen, C.; Xie, C. Investigating the Mechanisms Underlying the Low Irradiance-Tolerance of the Economically Important Seaweed Species Pyropia haitanensis. Life 2023, 13, 481. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wang, W.; Lin, Y.; Xu, K.; Xu, Y.; Ji, D.; Chen, C.; Xie, C. Insight into transketolase of Pyropia haitanensis under desiccation stress based on integrative analysis of omics and transformation. BMC Plant Biol. 2019, 9, 475. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, C.; Xu, Y.; Ji, D.; Xie, C. Validation of housekeeping genes as internal controls for studying the gene expression in Pyropia haitanensis (Bangiales, Rhodophyta) by quantitative real-time PCR. Acta Oceanol. Sin. 2014, 33, 152–159. [Google Scholar] [CrossRef]
- Wu, W.K.; Liu, C.Q.; Zhou, Z.G.; Lu, S. The Selection of Reference Genes in Chlamydomonas reinhardtii P.A. Dangeard by Real-Time Quantitative PCR. Plant Physiol. Commun. 2009, 45, 667–672. [Google Scholar]
- O’Malley, R.C.; Huang, S.S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 2016, 92, 1280–1291. [Google Scholar] [CrossRef]
- Han, S.; Hu, Z.; Wang, C. Expression of exogenous gene in chloroplast of Chlamydomonas reinhardtii. Biotechnol. Bull. 2007, 1, 89–94. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, J.; Ji, D.; Xu, Y.; Xu, K.; Xie, C.; Wang, W. Genome-Wide Identification and Functional Analysis of C2H2 Zinc Finger Transcription Factor Genes in the Intertidal Macroalga Pyropia haitanensis. Int. J. Mol. Sci. 2025, 26, 4042. https://doi.org/10.3390/ijms26094042
Xie J, Ji D, Xu Y, Xu K, Xie C, Wang W. Genome-Wide Identification and Functional Analysis of C2H2 Zinc Finger Transcription Factor Genes in the Intertidal Macroalga Pyropia haitanensis. International Journal of Molecular Sciences. 2025; 26(9):4042. https://doi.org/10.3390/ijms26094042
Chicago/Turabian StyleXie, Jiajia, Dehua Ji, Yan Xu, Kai Xu, Chaotian Xie, and Wenlei Wang. 2025. "Genome-Wide Identification and Functional Analysis of C2H2 Zinc Finger Transcription Factor Genes in the Intertidal Macroalga Pyropia haitanensis" International Journal of Molecular Sciences 26, no. 9: 4042. https://doi.org/10.3390/ijms26094042
APA StyleXie, J., Ji, D., Xu, Y., Xu, K., Xie, C., & Wang, W. (2025). Genome-Wide Identification and Functional Analysis of C2H2 Zinc Finger Transcription Factor Genes in the Intertidal Macroalga Pyropia haitanensis. International Journal of Molecular Sciences, 26(9), 4042. https://doi.org/10.3390/ijms26094042





