A Comprehensive Analysis on the Regulatory Network Underlying Callus Induction and Adventitious Organogenesis Process in Stem of Populus Alba L.
Abstract
1. Introduction
2. Results
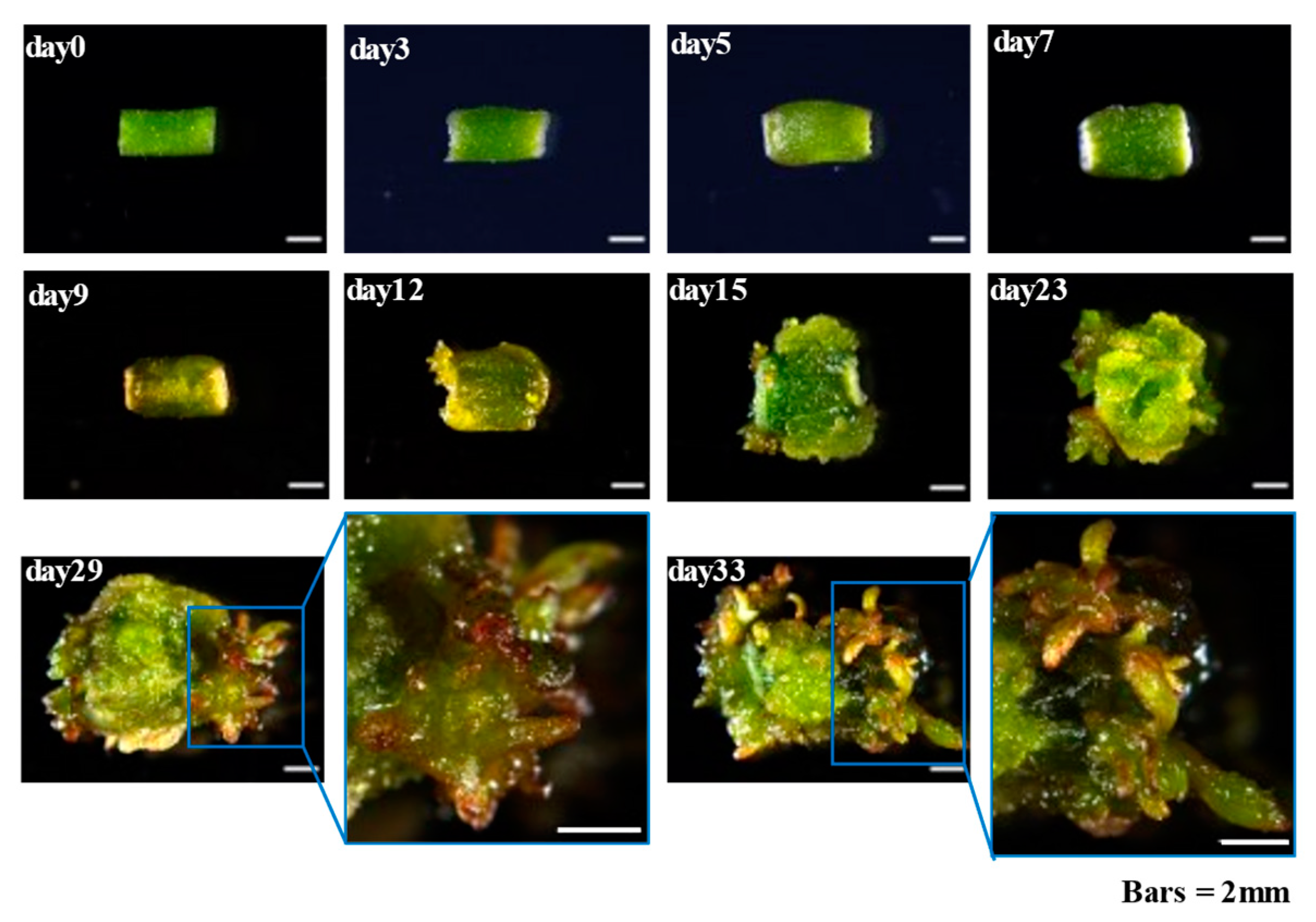
2.1. Callus Induction and Adventitious Organogenesis of Populus alba
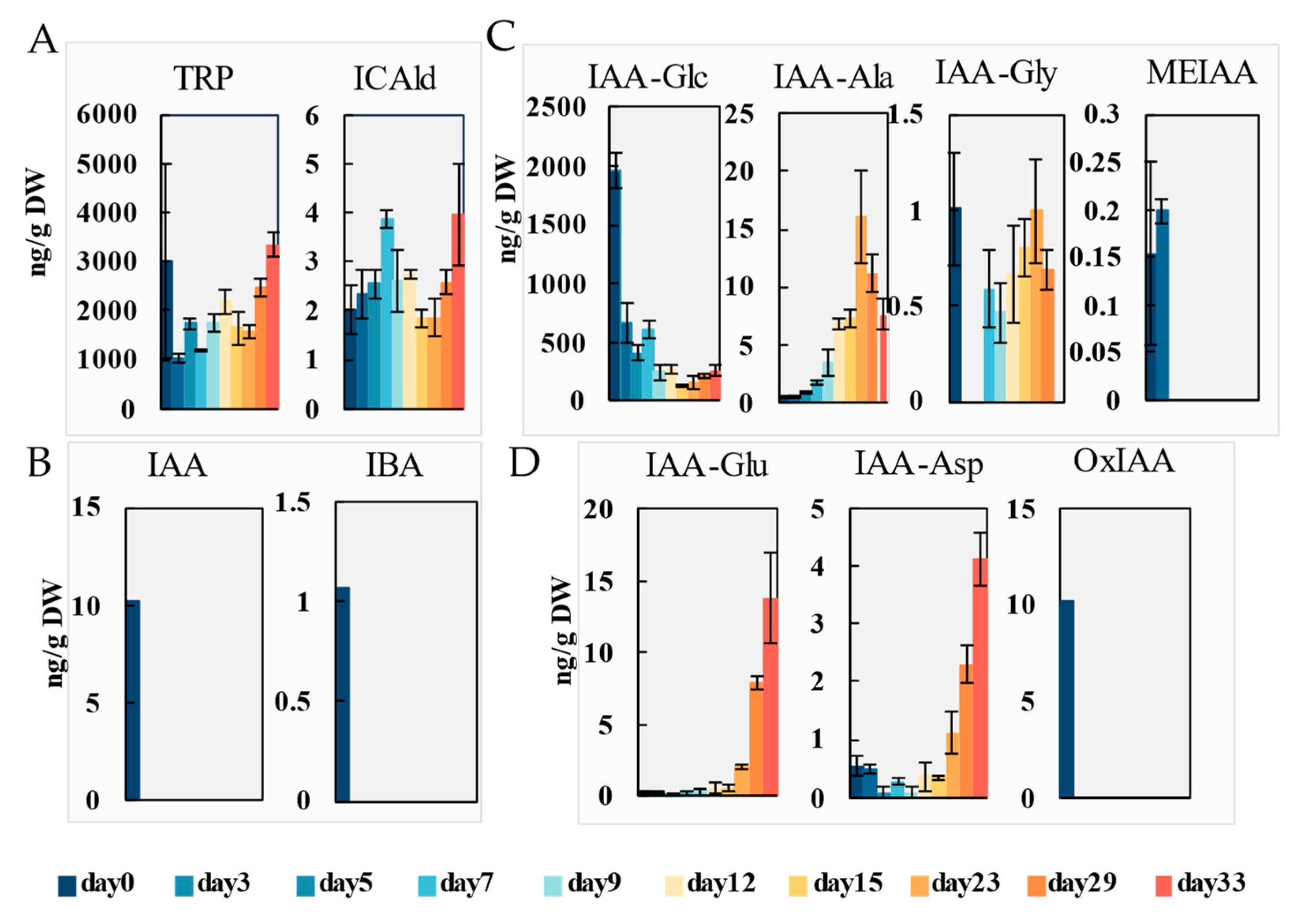
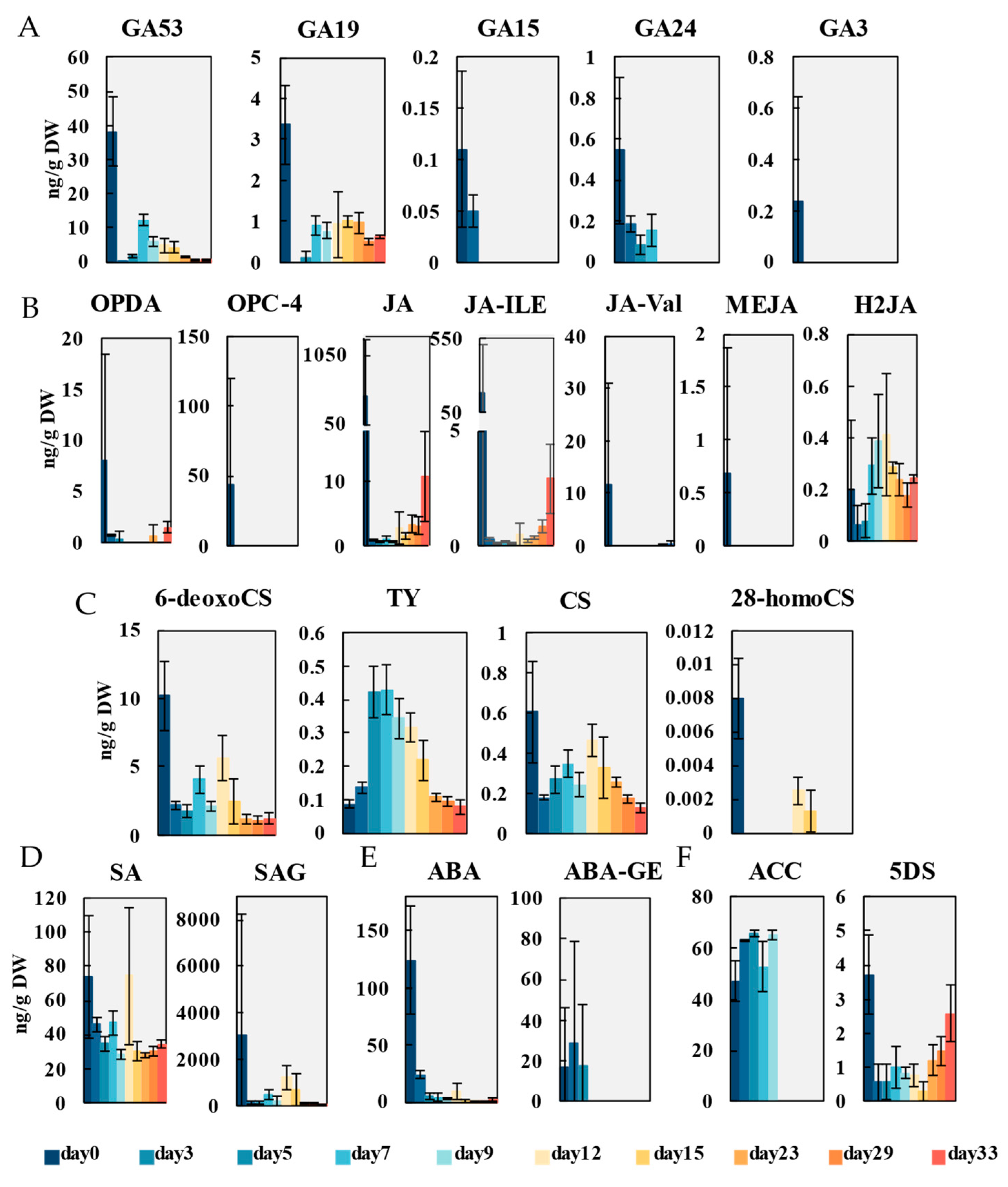
2.2. Dynamics of Endogenous Hormones During Callus Induction and Adventitious Organogenesis
2.2.1. Auxins
2.2.2. Cytokinins
2.2.3. Gibberellins
2.2.4. Jasmonic Acid
2.2.5. Brassinolide
2.2.6. Other Hormones
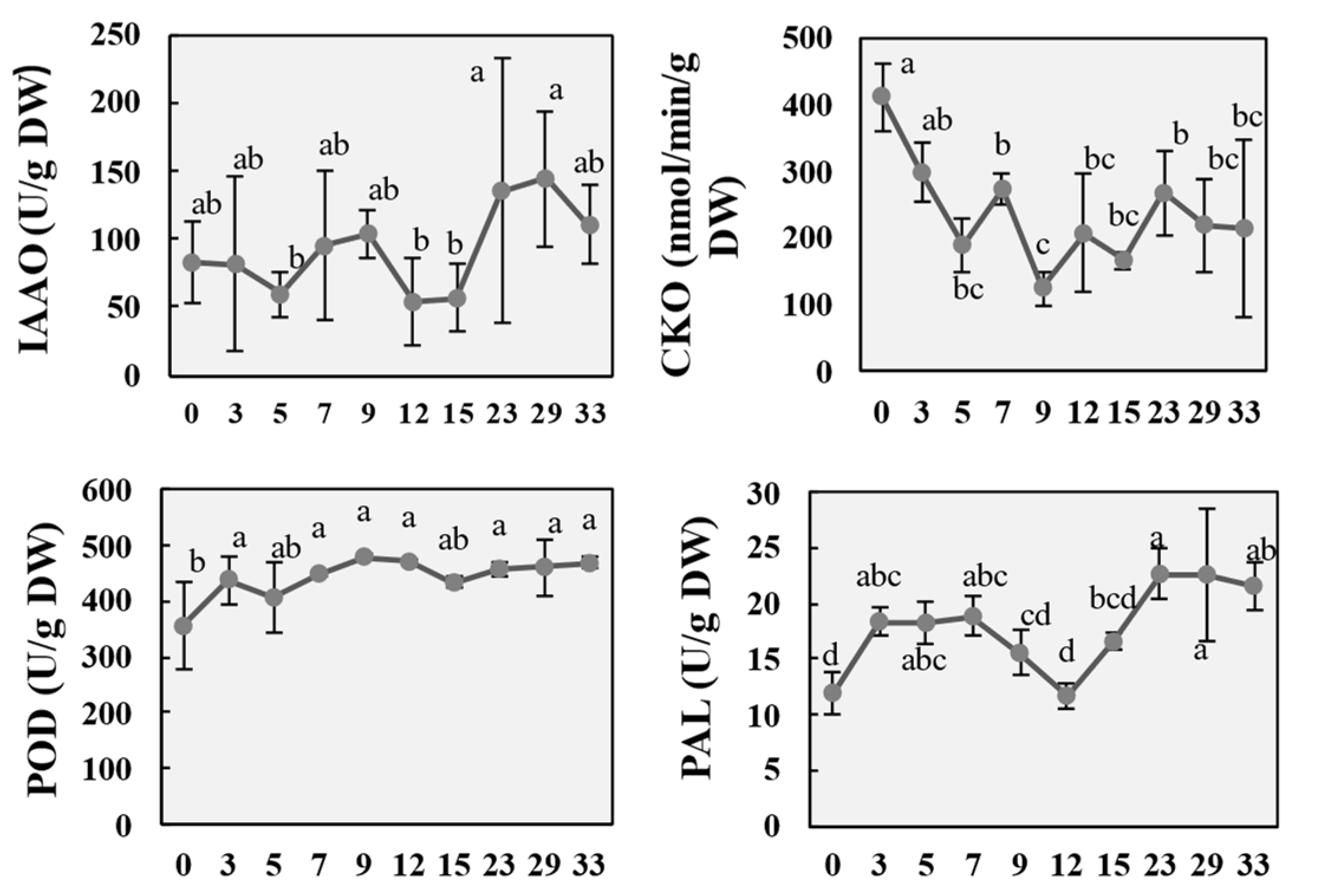
2.3. Changes in Metabolic Enzyme Activity During the Regeneration Process of P. alba
2.4. Transcriptional Regulation Callus Induction and Adventitious Organogenesis of P. alba
2.5. Corresponding Hormone Synthesis and Metabolism Pathways
3. Discussion
3.1. Synergistic Effects of Multiple Hormones During Callus Induction in Stem of Populus alba
3.2. Synergistic Induction of Adventitious Shoots by Multiple Hormones in Stem of P. alba
3.3. Auxin- and Cytokinin-Mediated Regulation in Callus Induction and Adventitious Organogenesis of P. alba
4. Materials and Methods
4.1. Plant Material and Induction of Callus and Shoot Regeneration
4.2. Quantitative Analysis of Plant Hormone
4.3. Metabolic Enzyme Activity
4.4. RNA-Seq Analysis
4.5. Identification of Hormones and Development-Related Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TRP | L-tryptophan |
| IAA | Indole-3-acetic acid |
| IAA-Ala | IAA-alanine |
| IAA-Asp | IAA-L-aspartate |
| IAA-Glc | IAA-glucose |
| IAA-Glu | IAA-L-glutamate |
| IAA-Gly | IAA-glycine |
| IBA | Indole-3-butyric acid |
| ICAld | Indole-3-carboxaldehyde |
| MEIAA | Methylindole-3-acetic acid |
| OxIAA | 2-oxindole-3-acetic acid |
| BAPR | 6-Benzyladenosine |
| IPR | N6-isopentenyladenosine |
| 2MeScZR | 2-Methylthio-cis-zeatin riboside |
| BAP | 6-Benzyladenine |
| BAP7G | N6-Benzyladenine-7-glucoside |
| BAP9G | N6-Benzyladenine-9-glucoside |
| cZ | cis-Zeatin |
| cZR | cis-Zeatin riboside |
| cZROG | cis-Zeatin-O-glucoside riboside |
| DHZROG | Dihydrozeatin-O-glucoside riboside |
| IP | N6-isopentenyladenine |
| K | Kinetin |
| K9G | Kinetin-9-glucoside |
| mT | Meta-Topolin |
| mT9G | meta-Topolin-9-glucoside |
| mTR | meta-Topolin riboside |
| oT | ortho-Topolin |
| oT9G | ortho-Topolin-9-glucoside |
| oTR | ortho-Topolin riboside |
| pT | para-Topolin |
| pT9G | para-Topolin-9-glucoside |
| pTR | para-Topolin riboside |
| tZ | trans-Zeatin |
| GA3 | Gibberellin A3 |
| GA15 | Gibberellin A15 |
| GA19 | Gibberellin A19 |
| GA24 | Gibberellin A24 |
| GA53 | Gibberellin A53 |
| H2JA | Dihydrojasmonic acid |
| JA | Jasmonic acid |
| JA-ILE | Jasmonoyl-L-isoleucine |
| JA-Val | N-[(-)-Jasmonoyl]-(L)-valine |
| MEJA | Methyl jasmonate |
| OPC-4 | 3-oxo-2-(2-(Z)-Pentenyl) cyclopentane-1-butyric acid |
| OPDA | 12-oxophytodienoic acid |
| 28-hpmpCS | 28-homocastasterone |
| 6-deoxoCS | 6-deoxocastasterone |
| CS | Castasterone |
| TY | Typhasterol |
| SA | Salicylic acid |
| SAG | Salicylic acid 2-O-β-glucoside |
| ABA | Abscisic acid |
| ABA-GE | ABA-glucosyl ester |
| ACC | 1-Aminocyclopropanecarboxylic acid |
| 5DS | 5-Deoxystrigol |
| CKO | Cytokinin oxidase/dehydrogenase |
| IAAO | Indole-3-acetic acid oxidase |
| PAL | Phenylalanine oxidase |
| POD | Peroxidase |
References
- Du, X.M.; Fang, T.; Liu, Y.; Huang, L.Y.; Zang, M.S.; Wang, G.Y.; Liu, Y.J.; Fu, J.J. Transcriptome profiling predicts new genes to promote maize callus formation and transformation. Front. Plant Sci. 2019, 10, 1633. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Gordon, S.P.; Meyerowitz, E.M. Regeneration in plants and animals: Dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 2011, 21, 212–218. [Google Scholar] [CrossRef]
- Sugiyama, M. Historical review of research on plant cell dedifferentiation. J. Plant Res. 2015, 128, 349–359. [Google Scholar] [CrossRef]
- Xu, L.; Huang, H. Genetic and epigenetic controls of plant regeneration. Curr. Top. Dev. Biol. 2014, 108, 1–33. [Google Scholar] [PubMed]
- Chandler, J.W. Founder cell specification. Trends Plant Sci. 2011, 16, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Atta, R.; Laurens, L.; Boucheron-Dubuisson, E.; Guivarc’h, A.; Carnero, E.; Giraudat-Pautot, V.; Rech, P.; Chriqui, D. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 2009, 57, 626–644. [Google Scholar] [CrossRef]
- Yang, W.; Choi, M.H.; Noh, B.; Noh, Y.S. De novo shoot regeneration controlled by HEN1 and TCP3/4 in Arabidopsis. Plant Cell Physiol. 2020, 61, 1600–1613. [Google Scholar] [CrossRef]
- Zhang, T.Q.; Lian, H.; Zhou, C.M.; Xu, L.; Jiao, Y.L.; Wang, J.W. A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 2017, 29, 1073–1087. [Google Scholar] [CrossRef]
- Duclercq, J.; Sangwan-Norreel, B.; Catterou, M.; Sangwan, R.S. De novo shoot organogenesis: From art to science. Trends Plant Sci. 2011, 16, 597–606. [Google Scholar] [CrossRef]
- Liu, J.; Hu, X.M.; Qin, P.; Prasad, K.; Hu, Y.X.; Xu, L. The WOX11-LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol. 2018, 59, 739–748. [Google Scholar] [CrossRef]
- Fan, M.Z.; Xu, C.Y.; Xu, K.; Hu, Y.X. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB domain proteins: Beyond lateral organ boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef]
- Xu, C.; Cao, H.; Zhang, Q.; Wang, H.; Xin, W.; Xu, E.; Zhang, S.; Yu, R.; Yu, D.; Hu, Y. Control of auxin-induced callus formation by bZIP59-LBD complex in Arabidopsis regeneration. Nat. Plants 2018, 4, 108–115. [Google Scholar] [CrossRef]
- Huang, K.L.; Ma, G.J.; Zhang, M.L.; Xiong, H.; Wu, H.; Zhao, C.Z.; Liu, C.S.; Jia, H.X.; Chen, L.; Kjorven, J.O.; et al. The ARF7 and ARF19 transcription factors positively regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis Roots. Plant Physiol. 2018, 178, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.K.; Lee, H.W.; Kim, M.J.; Cho, C.; Oh, E.; Kim, J. LBD18 uses a dual mode of a positive feedback loop to regulate ARF expression and transcriptional activity in Arabidopsis. Plant J. 2018, 95, 233–251. [Google Scholar] [CrossRef]
- Zhang, S.; Yu, R.; Yu, D.; Chang, P.; Guo, S.; Yang, X.; Liu, X.; Xu, C.; Hu, Y. The calcium signaling module CaM-IQM destabilizes IAA-ARF interaction to regulate callus and lateral root formation. Proc. Natl. Acad. Sci. USA 2022, 119, e2202669119. [Google Scholar] [CrossRef] [PubMed]
- Hofhuis, H.; Laskowski, M.; Du, Y.J.; Prasad, K.; Grigg, S.; Pinon, V.; Scheres, B. Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr. Biol. 2013, 23, 956–962. [Google Scholar] [CrossRef]
- Pinon, V.; Prasad, K.; Grigg, S.P.; Sanchez-Perez, G.F.; Scheres, B. Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 1107–1112. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Ogawa, Y.; Iwase, A.; Sugimoto, K. Plant regeneration: Cellular origins and molecular mechanisms. Development 2016, 143, 1442–1451. [Google Scholar] [CrossRef]
- Iwase, A.; Mitsuda, N.; Koyama, T.; Hiratsu, K.; Kojima, M.; Arai, T.; Inoue, Y.; Seki, M.; Sakakibara, H.; Sugimoto, K.; et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 2011, 21, 508–514. [Google Scholar] [CrossRef]
- Pulianmackal, A.J.; Kareem, A.V.K.; Durgaprasad, K.; Trivedi, Z.B.; Prasad, K. Competence and regulatory interactions during regeneration in plants. Front. Plant Sci. 2014, 5, 142. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Lopes, F.L.; Galvan-Ampudia, C.; Landrein, B. WUSCHEL in the shoot apical meristem: Old player, new tricks. J. Exp. Bot. 2021, 72, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.J.; Cheng, Z.J.; Sang, Y.L.; Zhang, M.M.; Rong, X.F.; Wang, Z.W.; Tang, Y.Y.; Zhang, X.S. Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 2017, 29, 1357–1372. [Google Scholar] [CrossRef]
- Motte, H.; Vercauteren, A.; Depuydt, S.; Landschoot, S.; Geelen, D.; Werbrouck, S.; Goormachtig, S.; Vuylsteke, M.; Vereecke, D. Combining linkage and association mapping identifies RECEPTOR-LIKE PROTEIN KINASE1 as an essential Arabidopsis shoot regeneration gene. Proc. Natl. Acad. Sci. USA 2014, 111, 8305–8310. [Google Scholar] [CrossRef]
- Cheon, J.; Park, S.Y.; Schulz, B.; Choe, S. Arabidopsis brassinosteroid biosynthetic mutant dwarf7-1 exhibits slower rates of cell division and shoot induction. BMC. Plant Biol. 2010, 10, 270. [Google Scholar] [CrossRef]
- Shin, J.; Bae, S.; Seo, P.J. De novo shoot organogenesis during plant regeneration. J. Exp. Bot 2020, 71, 63–72. [Google Scholar] [CrossRef]
- Bidabadi, S.S.; Jain, S.M. Cellular, molecular, and physiological aspects of in vitro plant regeneration. Plants 2020, 9, 702. [Google Scholar] [CrossRef]
- Bao, Y.; Dharmawardhana, P.; Mockler, T.C.; Strauss, S.H. Genome scale transcriptome analysis of shoot organogenesis in Populus. BMC. Plant Biol. 2009, 9, 132. [Google Scholar] [CrossRef]
- Wan, Q.; Zhai, N.; Xie, D.; Liu, W.; Xu, L. WOX11: The founder of plant organ regeneration. Cell Regen. 2023, 12, 1. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, J.; Yang, Z.; Matsui, A.; Seki, M.; Li, S.; Yan, X.; Kohnen, M.V.; Gu, L.; Prasad, K.; et al. PtWOX11 acts as master regulator conducting the expression of key transcription factors to induce de novo shoot organogenesis in poplar. Plant Mol. Biol. 2018, 98, 389–406. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef]
- Liu, H.; Wang, C.; Li, C.; Zhao, Z.; Wei, L.; Liu, Z.; Hu, D.; Liao, W. Nitric oxide is involved in hydrogen sulfide-induced adventitious rooting in tomato (Solanum lycopersicum). Funct. Plant Biol. 2022, 49, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Nisler, J. Beyond expectations: The development and biological activity of cytokinin oxidase/dehydrogenase inhibitors. Biochem. Soc. Trans. 2024, 52, 2297–2306. [Google Scholar] [CrossRef]
- Wang, B.; Bian, B.; Wang, C.; Li, C.; Fang, H.; Zhang, J.; Huang, D.; Huo, J.; Liao, W. Hydrogen gas promotes the adventitious rooting in cucumber under cadmium stress. PLoS ONE 2019, 14, e0212639. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Yang, G.; Wu, Y.; Rao, H.; Li, X.; Li, M.; Qian, P. Effects of light intensity on associated enzyme activity and gene expression during callus formation of Vitis vinifera. Chin. J. Biotechnol. 2015, 31, 1219–1229. [Google Scholar]
- Mok, D.W.S.; Mok, M.C. Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 89–118. [Google Scholar] [CrossRef]
- Iwase, A.; Harashima, H.; Ikeuchi, M.; Rymen, B.; Ohnuma, M.; Komaki, S.; Morohashi, K.; Kurata, T.; Nakata, M.; Ohme-Takagi, M.; et al. WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell 2017, 29, 54–69. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Sugimoto, K.; Iwase, A. Plant callus: Mechanisms of induction and repression. Plant Cell 2013, 25, 3159–3173. [Google Scholar] [CrossRef]
- De Marco, M.A.; Curatti, L.; Martínez-Nol, G.M.A. High auxin disrupts expression of cell-cycle genes, arrests cell division and promotes accumulation of starch in Chlamydomonas reinhardtii. Algal Res. 2024, 78, 103419. [Google Scholar] [CrossRef]
- Gordon, S.P.; Heisler, M.G.; Reddy, G.V.; Ohno, C.; Das, P.; Meyerowitz, E.M. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 2007, 134, 3539–3548. [Google Scholar] [CrossRef]
- Zhai, N.; Xu, L. Pluripotency acquisition in the middle cell layer of callus is required for organ regeneration. Nat. Plants 2021, 7, 1453–1460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, Y.; Wang, J.; Li, G.; Li, S.; Ma, J.; Peng, X.; Yin, J.; Liu, Y.; Zhu, Y. Transcriptomic and physiological analyses reveal changes in secondary metabolite and endogenous hormone in ginger (Zingiber officinale Rosc.) in response to postharvest chilling stress. Plant Physiol. Biochem. 2023, 201, 107799. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, A.; Laufs, P. Meristem initiation and de novo stem cell formation. Front. Plant Sci. 2022, 13, 891228. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Iwase, A.; Rymen, B.; Lambolez, A.; Kojima, M.; Takebayashi, Y.; Heyman, J.; Watanabe, S.; Seo, M.; De Veylder, L.; et al. Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol. 2017, 175, 1158–1174. [Google Scholar] [CrossRef]
- Lee, K.; Seo, P.J. High-temperature promotion of callus formation requires the BIN2-ARF-LBD axis in Arabidopsis. Planta 2017, 246, 797–802. [Google Scholar] [CrossRef]
- Yin, J.; Tian, J.; Li, G.; Zhu, Y.; Zhou, X.; He, Y.; Nie, P.; Su, Y.; Zhong, Q.; Chen, Z. Carbohydrate, phytohormone, and associated transcriptome changes during storage root formation in alligatorweed (Alternanthera philoxeroides). Weed Sci. 2020, 68, 382–395. [Google Scholar] [CrossRef]
- Hong, L.; Fletcher, J.C. Stem cells: Engines of plant growth and development. Int. J. Mol. Sci. 2023, 24, 14889. [Google Scholar] [CrossRef]
- Zhao, X.; Song, J.; Zeng, Q.; Ma, Y.; Fang, H.; Yang, L.; Deng, B.; Liu, J.; Fang, J.; Zuo, L.; et al. Auxin and cytokinin mediated regulation involved in vitro organogenesis of papaya. J. Plant Physiol. 2021, 260, 153405. [Google Scholar] [CrossRef]
- Lombardi-Crestana, S.; da Silva Azevedo, M.; e Silva, G.F.; Pino, L.E.; Appezzato-da-Glória, B.; Figueira, A.; Nogueira, F.T.; Peres, L.E. The tomato (Solanum lycopersicum cv. Micro-Tom) natural genetic variation Rg1 and the DELLA mutant procera control the competence necessary to form adventitious roots and shoots. J. Exp. Bot. 2012, 63, 5689–5703. [Google Scholar] [CrossRef]
- Chen, L.; Tong, J.; Xiao, L.; Ruan, Y.; Liu, J.; Zeng, M.; Huang, H.; Wang, J.W.; Xu, L. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 4273–4284. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Jiao, Y.; Meyerowitz, E.M. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 2010, 18, 463–471. [Google Scholar] [CrossRef]
- Zhai, N.; Pan, X.; Zeng, M.; Xu, L. Developmental trajectory of pluripotent stem cell establishment in Arabidopsis callus guided by a quiescent center-related gene network. Development 2023, 150, dev200879. [Google Scholar] [CrossRef]
- Hu, X.; Xu, L. Transcription Factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 2016, 172, 2363–2373. [Google Scholar] [CrossRef] [PubMed]
- Guarneri, N.; Willig, J.J.; Willemsen, V.; Goverse, A.; Sterken, M.G.; Nibbering, P.; Lozano Torres, J.L.; Smant, G. WOX11-mediated cell size control in Arabidopsis attenuates growth and fecundity of endoparasitic cyst nematodes. Plant J. 2024, 120, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Kinoshita, A.; Koga, H.; Tsukaya, H. Expression analyses of CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS in the one-leaf plant Monophyllaea glabra reveal neoteny evolution of shoot meristem. Sci. Rep. 2024, 14, 11148. [Google Scholar] [CrossRef]
- Sang, Y.L.; Cheng, Z.J.; Zhang, X.S. Plant stem cells and de novo organogenesis. New. Phytol. 2018, 218, 1334–1339. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Favero, D.S.; Sakamoto, Y.; Iwase, A.; Coleman, D.; Rymen, B.; Sugimoto, K. Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 2019, 70, 377–406. [Google Scholar] [CrossRef]
- Negin, B.; Shemer, O.; Sorek, Y.; Eshed Williams, L. Shoot stem cell specification in roots by the WUSCHEL transcription factor. PLoS ONE 2017, 12, e0176093. [Google Scholar] [CrossRef]
- Matsuo, N.; Makino, M.; Banno, H. Arabidopsis ENHANCER OF SHOOT REGENERATION (ESR)1 and ESR2 regulate in vitro shoot regeneration and their expressions are differentially regulated. Plant Sci. 2011, 181, 39–46. [Google Scholar] [CrossRef]
- Yan, T.; Hou, Q.; Wei, X.; Qi, Y.; Pu, A.; Wu, S.; An, X.; Wan, X. Promoting genotype-independent plant transformation by manipulating developmental regulatory genes and/or using nanoparticles. Plant Cell Rep. 2023, 42, 1395–1417. [Google Scholar] [CrossRef] [PubMed]
- Kareem, A.; Durgaprasad, K.; Sugimoto, K.; Du, Y.; Pulianmackal, A.J.; Trivedi, Z.B.; Abhayadev, P.V.; Pinon, V.; Meyerowitz, E.M.; Scheres, B.; et al. PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 2015, 25, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Motte, H.; Vereecke, D.; Geelen, D.; Werbrouck, S. The molecular path to in vitro shoot regeneration. Biotechnol. Adv 2014, 32, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Wang, X.R.; Zeng, Q.Y. De novo assembly of white poplar genome and genetic diversity of white poplar population in Irtysh River basin in China. Sci. China. Life. Sci. 2019, 62, 609–618. [Google Scholar] [CrossRef]
- Zhao, H.Y.; Lu, S.F. Chao, R.T. Studies on tissue culture and gene engineering of Poplar. Chin. Bull. Bot. 2001, 18, 169–176. Available online: https://www.chinbullbotany.com/EN/Y2001/V18/I02/169 (accessed on 20 December 2021).

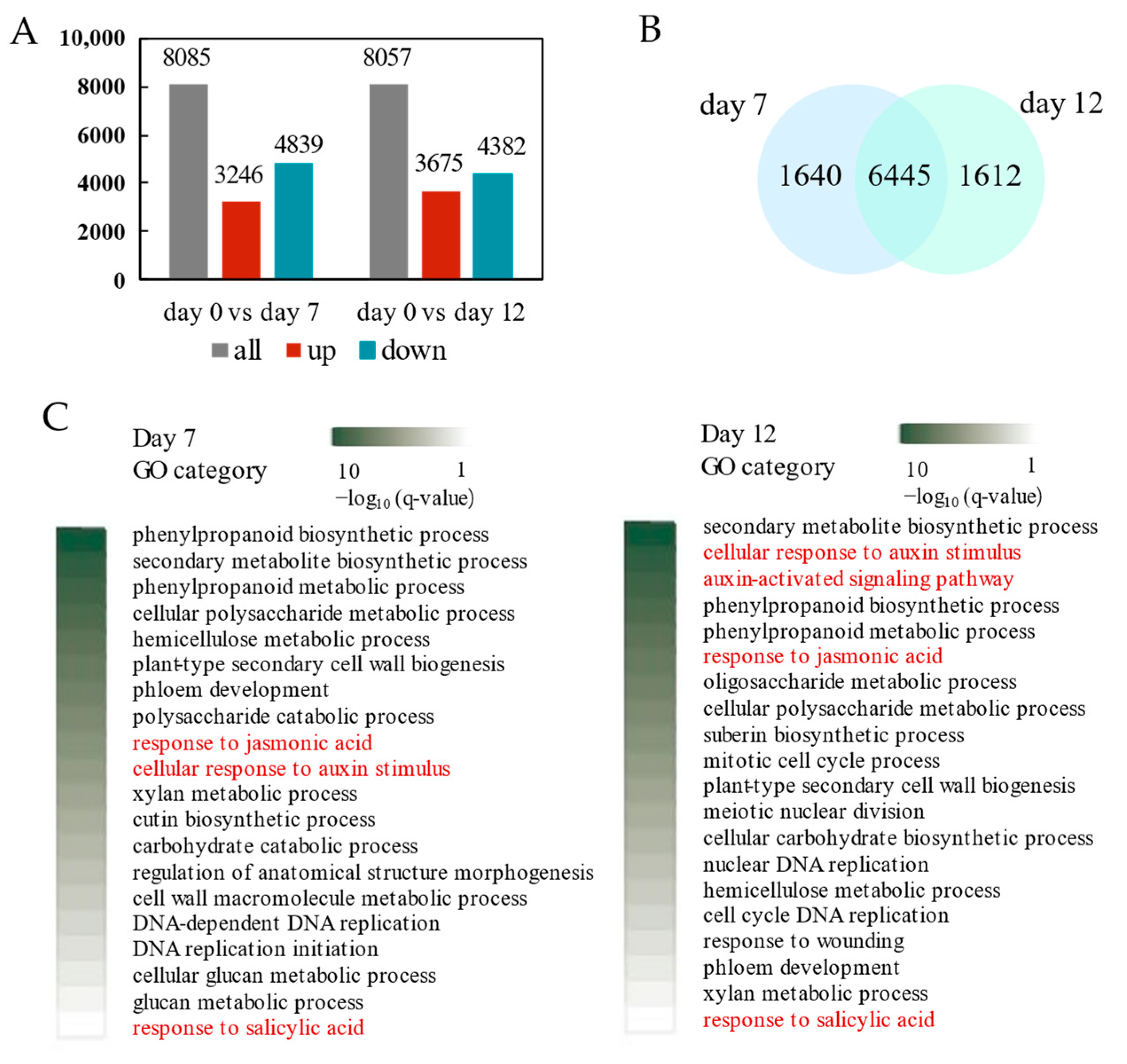
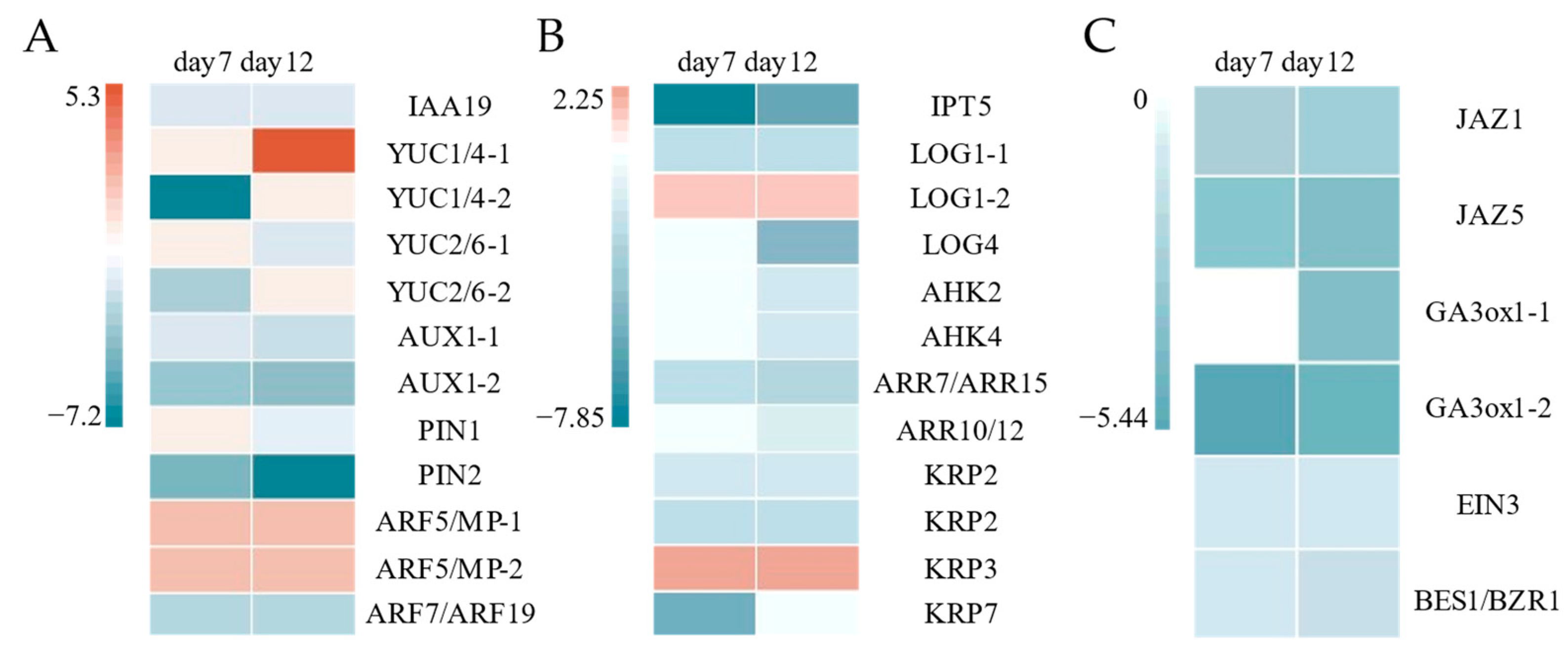
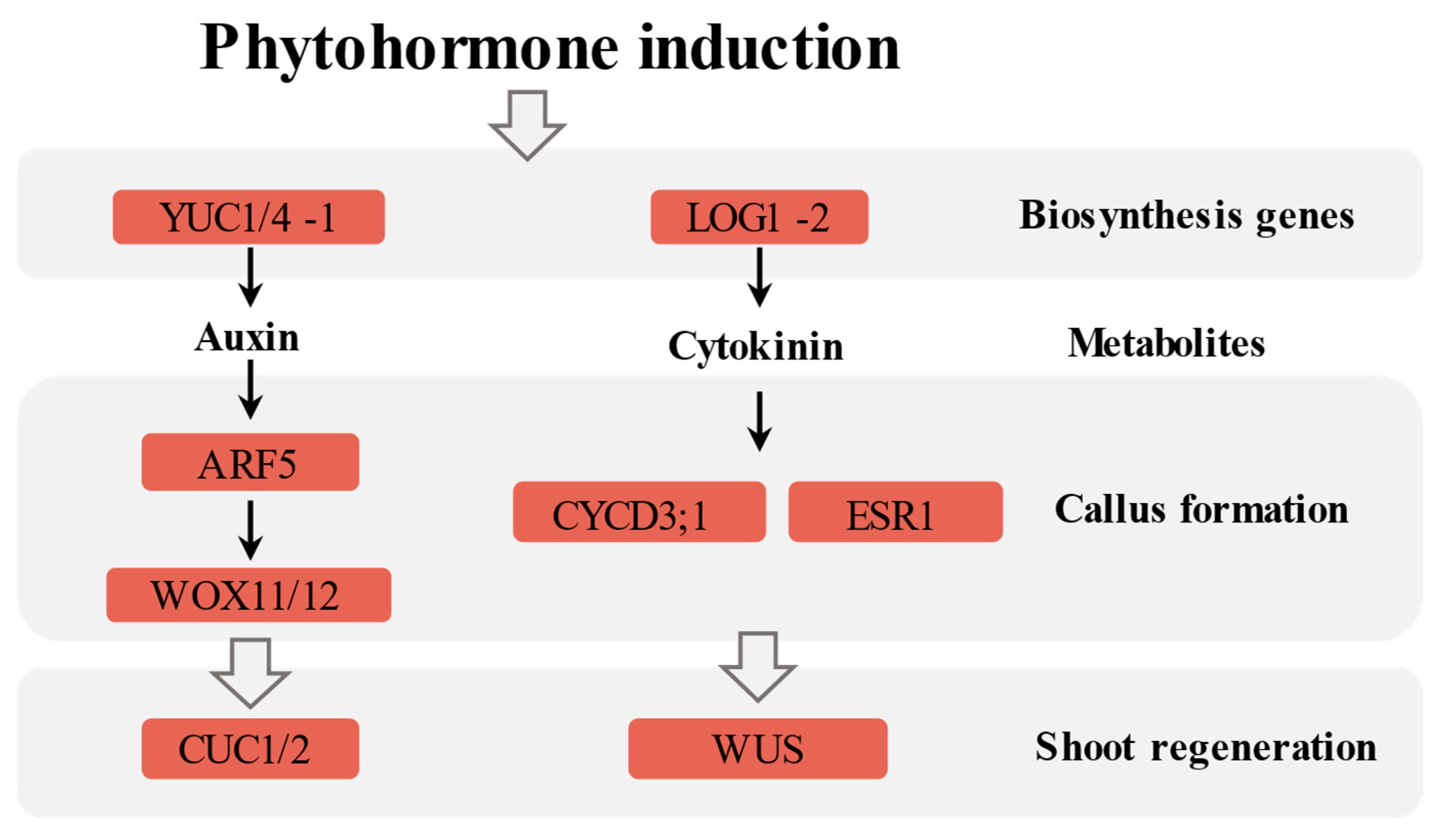
| Gene ID | Description | Log2FC | Log2FC | Process |
|---|---|---|---|---|
| (0 vs. 7) | (0 vs. 12) | |||
| Poalb02G008940 | LBD18; LOB DOMAIN-CONTAINING PROTEIN 18 | −3.3 | 0 | Auxin signaling |
| Poalb14G005550 | LBD18; LOB DOMAIN-CONTAINING PROTEIN 18 | −4.59 | −2.09 | Auxin signaling |
| Poalb01G034320 | CUC1/2; CUP-SHAPED COTYLEDON1/2 | 0 | 6.71 | Auxin/Cytokinin signaling |
| Poalb11G007910 | CUC1/2; CUP-SHAPED COTYLEDON1/2 | 0 | 10.18 | Auxin/Cytokinin signaling |
| Poalb13G011170 | WOX11/12; WUSCHEL-RELATED HOMEOBOX 11/12 | 9.87 | 10.14 | Auxin signaling |
| Poalb06G000150 | PLT5; AP2 family of transcriptional regulators | 0 | −1.2 | Auxin signaling |
| Poalb18G002670 | PLT5; AP2 family of transcriptional regulators | 0 | −1.22 | Auxin signaling |
| Poalb07G000880 | bZIP59 | 0 | −1.01 | Auxin signaling |
| Poalb05G009480 | WUSCHEL | 2.29 | 3.09 | Cytokinin signaling |
| Poalb08G016150 | ESR1; ENHANCER OF SHOOT REGENERATION 1 | 0 | 4.42 | Cytokinin signaling |
| Poalb10G003790 | ESR1; ENHANCER OF SHOOT REGENERATION 1 | 10.39 | 10.36 | Cytokinin signaling |
| Poalb08G012860 | CYCD1;1 | −2.01 | −2.06 | Cytokinin signaling |
| Poalb10G008040 | CYCD1;1 | −1.81 | −1.7 | Cytokinin signaling |
| Poalb01G026650 | CYCD3;1 | 3.13 | 2.48 | Cytokinin signaling |
| Poalb09G008730 | CYCD3;1 | 3.91 | 3.17 | Cytokinin signaling |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.-Y.; Liu, G.-F.; Zeng, Q.-Y.; Liu, Y.-J. A Comprehensive Analysis on the Regulatory Network Underlying Callus Induction and Adventitious Organogenesis Process in Stem of Populus Alba L. Int. J. Mol. Sci. 2025, 26, 4087. https://doi.org/10.3390/ijms26094087
Li X-Y, Liu G-F, Zeng Q-Y, Liu Y-J. A Comprehensive Analysis on the Regulatory Network Underlying Callus Induction and Adventitious Organogenesis Process in Stem of Populus Alba L. International Journal of Molecular Sciences. 2025; 26(9):4087. https://doi.org/10.3390/ijms26094087
Chicago/Turabian StyleLi, Xiao-Yuan, Gui-Feng Liu, Qing-Yin Zeng, and Yan-Jing Liu. 2025. "A Comprehensive Analysis on the Regulatory Network Underlying Callus Induction and Adventitious Organogenesis Process in Stem of Populus Alba L." International Journal of Molecular Sciences 26, no. 9: 4087. https://doi.org/10.3390/ijms26094087
APA StyleLi, X.-Y., Liu, G.-F., Zeng, Q.-Y., & Liu, Y.-J. (2025). A Comprehensive Analysis on the Regulatory Network Underlying Callus Induction and Adventitious Organogenesis Process in Stem of Populus Alba L. International Journal of Molecular Sciences, 26(9), 4087. https://doi.org/10.3390/ijms26094087






