Dissect Gender-Dependent Susceptibility SNPs in Progressive Osteoarthritis Using Regulator Motif Candidate of Genetic Association Strategy (RMCGA)
Abstract
1. Introduction
2. Results
2.1. Demonstration of Potential NF-κB Binding Motif
2.2. Candidate SNPs on NF-κB Binding Sites
2.3. Characteristics of OA Patients and Control Subjects
2.4. Association Between Binding Site Gene Polymorphisms and OA Susceptibility
2.5. Messenger RNA Expression of Putative Genes Among Genotypes of SNPs
3. Discussion
4. Materials and Methods
4.1. Study Participants
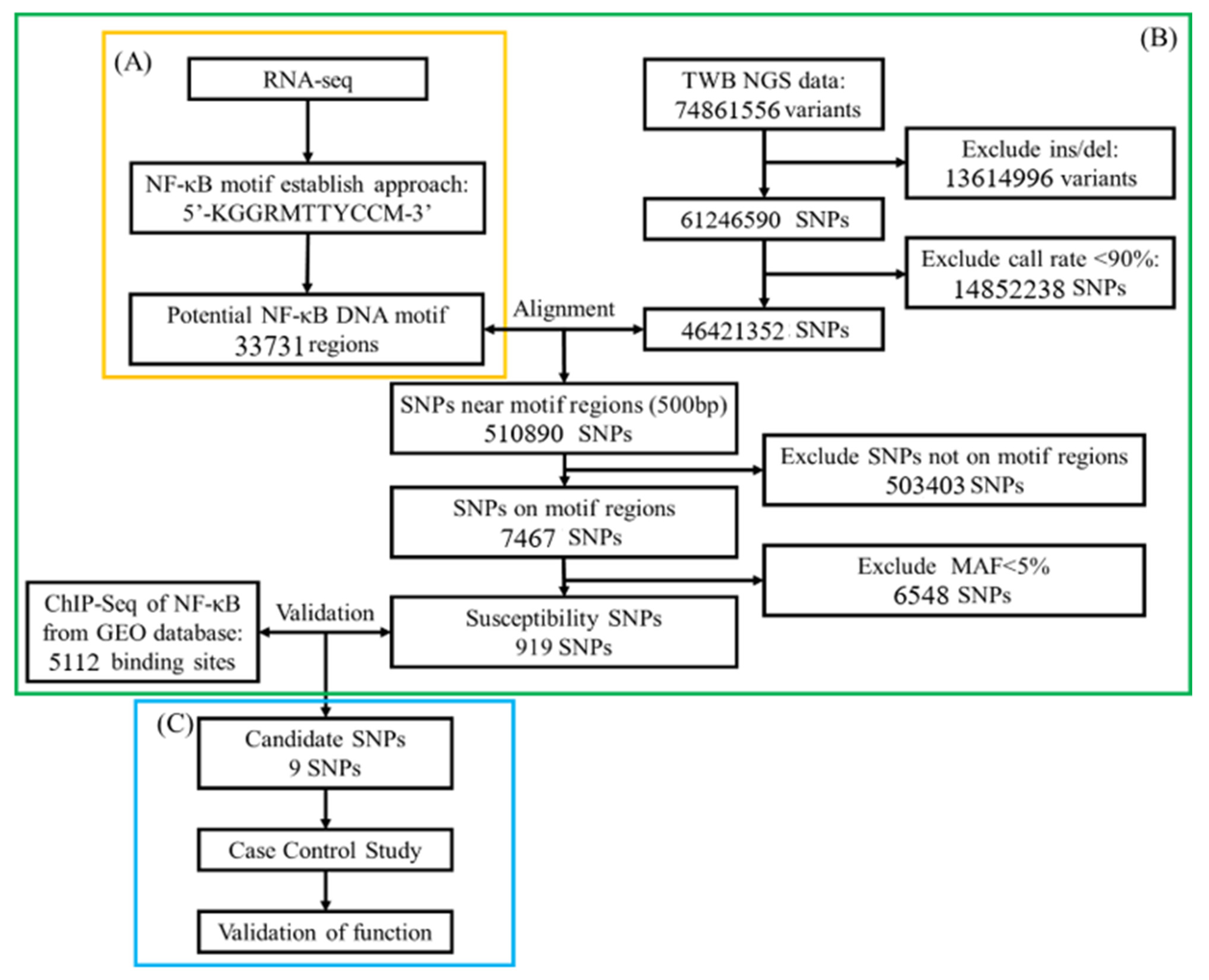
4.2. Bioinformatic Analysis in Gene Screening
Study Flowchart
4.3. Genomic DNA Extraction and SNP Genotyping
4.4. Ethics
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Safiri, S.; Kolahi, A.-A.; Smith, E.; Hill, C.; Bettampadi, D.; Mansournia, M.A.; Hoy, D.; Ashrafi-Asgarabad, A.; Sepidarkish, M.; Almasi-Hashiani, A.; et al. Global, regional and national burden of osteoarthritis 1990-2017: A systematic analysis of the Global Burden of Disease Study 2017. Ann. Rheum. Dis. 2020, 79, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Neogi, T.; Felson, D.T.; Sarno, R.; Booth, S.L. Vitamin K in hand osteoarthritis: Results from a randomised clinical trial. Ann. Rheum. Dis. 2008, 67, 1570–1573. [Google Scholar] [CrossRef]
- Glyn-Jones, S.; Palmer, A.J.; Agricola, R.; Price, A.J.; Vincent, T.L.; Weinans, H.; Carr, A.J. Osteoarthritis. Lancet 2015, 386, 376–387. [Google Scholar] [CrossRef] [PubMed]
- Loughlin, J. The genetic epidemiology of human primary osteoarthritis: Current status. Expert Rev. Mol. Med. 2005, 7, 9. [Google Scholar] [CrossRef]
- Cicuttini, F.M.; Spector, T.D. Genetics of osteoarthritis. Ann. Rheum. Dis. 1996, 55, 665–667. [Google Scholar] [CrossRef]
- Spector, T.D.; MacGregor, A.J. Risk factors for osteoarthritis: Genetics. Osteoarthr. Cartil. 2004, 12 (Suppl. A), S39–S44. [Google Scholar] [CrossRef]
- Tschon, M.; Contartese, D.; Pagani, S.; Borsari, V.; Fini, M. Gender and Sex Are Key Determinants in Osteoarthritis Not Only Confounding Variables. A Systematic Review of Clinical Data. J. Clin. Med. 2021, 10, 3178. [Google Scholar] [CrossRef]
- Sabik, O.L.; Farber, C.R. Using GWAS to identify novel therapeutic targets for osteoporosis. Transl. Res. J. Lab. Clin. Med. 2017, 181, 15–26. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef]
- Aubourg, G.; Rice, S.J.; Bruce-Wootton, P.; Loughlin, J. Genetics of osteoarthritis. Osteoarthr. Cartil. 2022, 30, 636–649. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, M.D.; Chen-Plotkin, A.S. The Post-GWAS Era: From Association to Function. Am. J. Hum. Genet. 2018, 102, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef]
- Marcu, K.B.; Otero, M.; Olivotto, E.; Borzi, R.M.; Goldring, M.B. NF-kappaB signaling: Multiple angles to target OA. Curr. Drug Targets 2010, 11, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Roman-Blas, J.A.; Jimenez, S.A. NF-kappaB as a potential therapeutic target in osteoarthritis and rheumatoid arthritis. Osteoarthr. Cartil. 2006, 14, 839–848. [Google Scholar] [CrossRef]
- Choi, M.C.; Jo, J.; Park, J.; Kang, H.K.; Park, Y. NF-κB Signaling Pathways in Osteoarthritic Cartilage Destruction. Cells 2019, 8, 734. [Google Scholar] [CrossRef]
- Fonseca, C.; Lindahl, G.E.; Ponticos, M.; Sestini, P.; Renzoni, E.A.; Holmes, A.M.; Spagnolo, P.; Pantelidis, P.; Leoni, P.; McHugh, N.; et al. A polymorphism in the CTGF promoter region associated with systemic sclerosis. N. Engl. J. Med. 2007, 357, 1210–1220. [Google Scholar] [CrossRef]
- Bansal, M. DNA structure: Revisiting the Watson–Crick double helix. Curr. Sci. 2003, 85, 1556–1563. [Google Scholar]
- Fisch, K.M.; Gamini, R.; Alvarez-Garcia, O.; Akagi, R.; Saito, M.; Muramatsu, Y.; Sasho, T.; Koziol, J.A.; Su, A.I.; Lotz, M.K. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthr. Cartil. 2018, 26, 1531–1538. [Google Scholar] [CrossRef]
- Xu, L.; Sun, C.; Zhang, S.; Xu, X.; Zhai, L.; Wang, Y.; Wang, S.; Liu, Z.; Cheng, H.; Xiao, M.; et al. Sam68 Promotes NF-κB Activation and Apoptosis Signaling in Articular Chondrocytes during Osteoarthritis. Inflamm. Res. 2015, 64, 895–902. [Google Scholar] [CrossRef]
- Hong, M.J.; Yoo, S.S.; Choi, J.E.; Kang, H.G.; Do, S.K.; Lee, J.H.; Lee, W.K.; Lee, J.; Lee, S.Y.; Cha, S.I.; et al. Functional intronic variant of SLC5A10 affects DRG2 expression and survival outcomes of early-stage non-small-cell lung cancer. Cancer Sci. 2018, 109, 3902–3909. [Google Scholar] [CrossRef]
- Hsiung, C.N.; Chu, H.W.; Huang, Y.L.; Chou, W.C.; Hu, L.Y.; Hsu, H.M.; Wu, P.E.; Hou, M.F.; Yu, J.C.; Shen, C.Y. Functional variants at the 21q22.3 locus involved in breast cancer progression identified by screening of genome-wide estrogen response elements. Breast Cancer Res. 2014, 16, 455. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.H.; Huang, G.S.; Chang, H.F.; Chen, C.Y.; Kang, C.Y.; Wang, C.C.; Lin, C.; Yang, J.H.; Su, W.; Kao, S.; et al. Gender differences between WOMAC index scores, health-related quality of life and physical performance in an elderly Taiwanese population with knee osteoarthritis. BMJ Open 2015, 5, e008542. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhang, Z.; Sheng, P.; Mobasheri, A. The role of metabolism in chondrocyte dysfunction and the progression of osteoarthritis. Ageing Res. Rev. 2021, 66, 101249. [Google Scholar] [CrossRef]
- Rumsey, S.C.; Kwon, O.; Xu, G.W.; Burant, C.F.; Simpson, I.; Levine, M. Glucose transporter isoforms GLUT1 and GLUT3 transport dehydroascorbic acid. J. Biol. Chem. 1997, 272, 18982–18989. [Google Scholar] [CrossRef]
- Kawakami, Y.; Tsuda, M.; Takahashi, S.; Taniguchi, N.; Esteban, C.R.; Zemmyo, M.; Furumatsu, T.; Lotz, M.; Belmonte, J.C.I.; Asahara, H. Transcriptional coactivator PGC-1α regulates chondrogenesis via association with Sox9. Proc. Natl. Acad. Sci. USA 2005, 102, 2414–2419. [Google Scholar] [CrossRef] [PubMed]
- Jain, L.; Jardim, C.A.; Yulo, R.; Bolam, S.M.; Monk, A.P.; Munro, J.T.; Pitto, R.; Tamatea, J.; Dalbeth, N.; Poulsen, R.C. Phenotype and energy metabolism differ between osteoarthritic chondrocytes from male compared to female patients: Implications for sexual dimorphism in osteoarthritis development? Osteoarthr. Cartil. 2024, 32, 1084–1096. [Google Scholar] [CrossRef]
- Otterness, I.G.; Eckstein, F. Women have thinner cartilage and smaller joint surfaces than men after adjustment for body height and weight. Osteoarthr. Cartil. 2007, 15, 666–672. [Google Scholar] [CrossRef]
- Li, C.; Zheng, Z. Males and Females Have Distinct Molecular Events in the Articular Cartilage during Knee Osteoarthritis. Int. J. Mol. Sci. 2021, 22, 7876. [Google Scholar] [CrossRef]
- Vina, E.R.; Kwoh, C.K. Epidemiology of osteoarthritis: Literature update. Curr. Opin. Rheumatol. 2018, 30, 160–167. [Google Scholar] [CrossRef]
- Yan, Y.S.; Qu, Z.; Yu, D.Q.; Wang, W.; Yan, S.; Huang, H.F. Sex Steroids and Osteoarthritis: A Mendelian Randomization Study. Front. Endocrinol. 2021, 12, 683226. [Google Scholar] [CrossRef]
- Peshkova, M.; Lychagin, A.; Lipina, M.; Di Matteo, B.; Anzillotti, G.; Ronzoni, F.; Kosheleva, N.; Shpichka, A.; Royuk, V.; Fomin, V.; et al. Gender-Related Aspects in Osteoarthritis Development and Progression: A Review. Int. J. Mol. Sci. 2022, 23, 2767. [Google Scholar] [CrossRef] [PubMed]
- Baldassarre, M.; Alvisi, S.; Berra, M.; Martelli, V.; Farina, A.; Righi, A.; Meriggiola, M.C. Changes in vaginal physiology of menopausal women with type 2 diabetes. J. Sex. Med. 2015, 12, 1346–1355. [Google Scholar] [CrossRef] [PubMed]
- Scordalakes, E.M.; Shetty, S.J.; Rissman, E.F. Roles of estrogen receptor alpha and androgen receptor in the regulation of neuronal nitric oxide synthase. J. Comp. Neurol. 2002, 453, 336–344. [Google Scholar] [CrossRef]
- Yabe-Nishimura, C. Aldose reductase in glucose toxicity: A potential target for the prevention of diabetic complications. Pharmacol. Rev. 1998, 50, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Xie, J.; Liu, Z.; Wang, J. Aldose reductase deficiency inhibits LPS-induced M1 response in macrophages by activating autophagy. Cell Biosci. 2021, 11, 61. [Google Scholar]
- Ulett, G.C.; Adderson, E.E. Nitric oxide is a key determinant of group B streptococcus-induced murine macrophage apoptosis. J. Infect. Dis. 2005, 191, 1761–1770. [Google Scholar] [CrossRef]
- Tinker, A.C.; Wallace, A.V. Selective inhibitors of inducible nitric oxide synthase: Potential agents for the treatment of inflammatory diseases? Curr. Top. Med. Chem. 2006, 6, 77–92. [Google Scholar] [CrossRef]
- Ulivi, V.; Giannoni, P.; Gentili, C.; Cancedda, R.; Descalzi, F. p38/NF-kB-dependent expression of COX-2 during differentiation and inflammatory response of chondrocytes. J. Cell Biochem. 2008, 104, 1393–1406. [Google Scholar] [CrossRef]
- Moncada, S.; Higgs, A. The L-arginine-nitric oxide pathway. N. Engl. J. Med. 1993, 329, 2002–2012. [Google Scholar]
- Nathan, C. Nitric oxide as a secretory product of mammalian cells. FASEB J. 1992, 6, 3051–3064. [Google Scholar] [CrossRef] [PubMed]
- Bredt, D.S.; Snyder, S.H. Nitric oxide: A physiologic messenger molecule. Annu. Rev. Biochem. 1994, 63, 175–195. [Google Scholar] [CrossRef] [PubMed]
- Baig, M.S.; Zaichick, S.V.; Mao, M.; de Abreu, A.L.; Bakhshi, F.R.; Hart, P.C.; Saqib, U.; Deng, J.; Chatterjee, S.; Block, M.L.; et al. NOS1-derived nitric oxide promotes NF-κB transcriptional activity through inhibition of suppressor of cytokine signaling-1. J. Exp. Med. 2015, 212, 1725–1738. [Google Scholar] [CrossRef] [PubMed]
- Weiner, C.P.; Lizasoain, I.; Baylis, S.A.; Knowles, R.G.; Charles, I.G.; Moncada, S. Induction of calcium-dependent nitric oxide synthases by sex hormones. Proc. Natl. Acad. Sci. USA 1994, 91, 5212–5216. [Google Scholar] [CrossRef]
- Angelini, C.; Tasca, E.; Nascimbeni, A.C.; Fanin, M. Muscle fatigue, nNOS and muscle fiber atrophy in limb girdle muscular dystrophy. Acta Myol. 2014, 33, 119–126. [Google Scholar]
- Sanchez-Lopez, E.; Coras, R.; Torres, A.; Lane, N.E.; Guma, M. Synovial inflammation in osteoarthritis progression. Nat. Rev. Rheumatol. 2022, 18, 258–275. [Google Scholar] [CrossRef]
- Ahmad, N.; Ansari, M.Y.; Haqqi, T.M. Role of iNOS in osteoarthritis: Pathological and therapeutic aspects. J. Cell Physiol. 2020, 235, 6366–6376. [Google Scholar] [CrossRef]
- Sedaghati-Khayat, B.; Boer, C.G.; Runhaar, J.; Bierma-Zeinstra, S.M.A.; Broer, L.; Ikram, M.A.; Zeggini, E.; Uitterlinden, A.G.; van Rooij, J.G.J.; van Meurs, J.B.J. Risk Assessment for Hip and Knee Osteoarthritis Using Polygenic Risk Scores. Arthritis Rheumatol. 2022, 74, 1488–1496. [Google Scholar] [CrossRef]
- Morita, Y.; Kamatani, Y.; Ito, H.; Ikegawa, S.; Kawaguchi, T.; Kawaguchi, S.; Takahashi, M.; Terao, C.; Ito, S.; Nishitani, K.; et al. Improved genetic prediction of the risk of knee osteoarthritis using the risk factor-based polygenic score. Arthritis Res. Ther. 2023, 25, 103. [Google Scholar] [CrossRef]
- Kerkhof, H.J.; Bierma-Zeinstra, S.M.; Arden, N.K.; Metrustry, S.; Castano-Betancourt, M.; Hart, D.J.; Hofman, A.; Rivadeneira, F.; Oei, E.H.; Spector, T.D.; et al. Prediction model for knee osteoarthritis incidence, including clinical, genetic and biochemical risk factors. Ann. Rheum. Dis. 2014, 73, 2116–2121. [Google Scholar] [CrossRef]
- Lacaze, P.; Wang, Y.; Polekhina, G.; Bakshi, A.; Riaz, M.; Owen, A.; Franks, A.; Abidi, J.; Tiller, J.; McNeil, J.; et al. Genomic Risk Score for Advanced Osteoarthritis in Older Adults. Arthritis Rheumatol. 2022, 74, 1480–1487. [Google Scholar] [CrossRef] [PubMed]
- Gill, R.; Liu, M.; Sun, G.; Furey, A.; Spector, T.; Rahman, P.; Zhai, G. Genomic heterozygosity is associated with a lower risk of osteoarthritis. BMC Genom. 2024, 25, 85. [Google Scholar] [CrossRef] [PubMed]
- Dean, A.G.; Sullivan, K.M.; Soe, M.M. OpenEpi: Open Source Epidemiologic Statistics for Public Health. 2013. Available online: www.OpenEpi.com (accessed on 17 April 2025).
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef] [PubMed]
- The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [CrossRef]
- Perkel, J. SNP genotyping: Six technologies that keyed a revolution. Nat. Methods 2008, 5, 447–453. [Google Scholar] [CrossRef]
| Independent Variables | Control (n = 614) | OA (n = 533) | p-Value | Severe OA(n = 134) | p-Value |
|---|---|---|---|---|---|
| Gender | 0.015 * | 0.014 * | |||
| Male | 268 (43.6%) | 195 (36.6%) | 43 (32.1%) | ||
| Female | 346 (56.4%) | 338 (63.4%) | 91 (67.9%) | ||
| Age | 72.48 ± 6.80 | 73.31 ± 6.94 | 0.043 * | 75.85 ± 7.08 | <0.001 * |
| BMI | 23.94 ± 3.41 | 24.38 ± 3.48 | 0.042 * | 25.19 ± 3.20 | <0.001 * |
| SNPs | Control | OA | HWE | Crude-OR (95% CI) | Adj-OR (95% CI) |
|---|---|---|---|---|---|
| rs11826681 C/G | 0.961 | ||||
| CC | 217 (36.0%) | 206 (38.9%) | 1.00 | 1.00 | |
| CG | 300 (49.8%) | 241 (45.6%) | 0.85 (0.66–1.09) | 0.85 (0.65–1.12) | |
| GG | 85 (14.1%) | 82 (15.5%) | 1.02 (0.71–1.45) | 0.98 (0.67–1.43) | |
| rs2257609 G/A | 0.630 | ||||
| GG | 205 (33.9%) | 193 (36.8%) | 1.00 | 1.00 | |
| GA | 294 (48.6%) | 238 (45.3%) | 0.86 (0.66–1.12) | 0.82 (0.63–1.08) | |
| AA | 106 (17.5%) | 94 (17.9%) | 0.94 (0.67–1.32) | 0.90 (0.63–1.28) | |
| rs3749606 C/T | 0.957 | ||||
| CC | 431 (71.4%) | 372 (71.3%) | 1.00 | 1.00 | |
| CT | 163 (27.0%) | 141 (27.0%) | 1.00 (0.77–1.31) | 1.03 (0.78–1.37) | |
| TT | 10 (1.7%) | 9 (1.7%) | 1.04 (0.42–2.59) | 0.96 (0.37–2.53) | |
| rs4702701 A/G | 0.911 | ||||
| AA | 528 (87.0%) | 462 (87.5%) | 1.00 | 1.00 | |
| AG | 77 (12.7%) | 64 (12.1%) | 0.95 (0.67–1.35) | 0.90 (0.62–1.31) | |
| GG | 2 (0.3%) | 2 (0.4%) | 1.14 (0.16–8.15) | 1.06 (0.15–7.63) | |
| rs545654 C/T | 0.951 | ||||
| CC | 283 (46.6%) | 238 (45.2%) | 1.00 | 1.00 | |
| CT | 260 (42.8%) | 238 (45.2%) | 1.09 (0.85–1.39) | 1.10 (0.85–1.43) | |
| TT | 64 (10.5%) | 50 (9.5%) | 0.93 (0.62–1.40) | 0.94 (0.62–1.43) | |
| rs7256865 T/G | 0.298 | ||||
| TT | 451 (74.5%) | 396 (75.0%) | 1.00 | 1.00 | |
| TG | 137 (22.6%) | 121 (22.9%) | 1.01 (0.76–1.33) | 1.03 (0.77–1.39) | |
| GG | 17 (2.8%) | 11 (2.1%) | 0.74 (0.34–1.59) | 0.77 (0.35–1.68) | |
| rs73164856 C/T | 1.000 | ||||
| CC | 453 (75.1%) | 428 (81.8%) | 1.00 | 1.00 | |
| CT | 141 (23.4%) | 89 (17.0%) | 0.67 (0.50–0.90) * | 0.68 (0.50–0.93) * | |
| TT | 9 (1.5%) | 6 (1.1%) | 0.71 (0.25–2.00) | 0.52 (0.17–1.57) | |
| rs77836284 C/T | 0.986 | ||||
| CC | 504 (83.2%) | 450 (85.9%) | 1.00 | 1.00 | |
| CT | 98 (16.2%) | 71 (13.5%) | 0.81 (0.58–1.13) | 0.75 (0.53–1.07) | |
| TT | 4 (0.7%) | 3 (0.6%) | 0.84 (0.19–3.77) | 0.81 (0.18–3.69) | |
| rs79975923 A/T | 0.720 | ||||
| AA | 420 (69.5%) | 365 (69.3%) | 1.00 | 1.00 | |
| AT | 170 (28.1%) | 149 (28.3%) | 1.01 (0.78–1.31) | 1.02 (0.78–1.35) | |
| TT | 14 (2.3%) | 13 (2.5%) | 1.07 (0.50–2.30) | 1.31 (0.59–2.94) |
| SNPs | Control | Severe OA | Crude-OR (95% CI) | Adj-OR (95% CI) |
|---|---|---|---|---|
| rs11826681 C/G | ||||
| CC | 217 (36.0%) | 52 (39.7%) | 1.00 | 1.00 |
| CG | 300 (49.8%) | 65 (49.6%) | 0.90 (0.60–1.35) | 0.90 (0.58–1.41) |
| GG | 85 (14.1%) | 14 (10.7%) | 0.69 (0.36–1.31) | 0.79 (0.41–1.56) |
| rs2257609 G/A | ||||
| GG | 205 (33.9%) | 44 (33.8%) | 1.00 | 1.00 |
| GA | 294 (48.6%) | 65 (50.0%) | 1.03 (0.68–1.57) | 0.91 (0.58–1.45) |
| AA | 106 (17.5%) | 21 (16.2%) | 0.92 (0.52–1.63) | 0.88 (0.48–1.61) |
| rs3749606 C/T | ||||
| CC | 431 (71.4%) | 88 (67.7%) | 1.00 | 1.00 |
| CT | 163 (27.0%) | 41 (31.5%) | 1.23 (0.82–1.86) | 1.35 (0.87–2.10) |
| TT | 10 (1.7%) | 1 (0.8%) | 0.49 (0.06–3.87) | 0.57 (0.07–4.66) |
| rs4702701 A/G | ||||
| AA | 528 (87.0%) | 111 (84.1%) | 1.00 | 1.00 |
| AG | 77 (12.7%) | 20 (15.2%) | 1.24 (0.73–2.10) | 1.11 (0.60–2.02) |
| GG | 2 (0.3%) | 1 (0.8%) | 2.38 (0.21–26.46) | 1.84 (0.16–21.36) |
| rs545654 C/T | ||||
| CC | 283 (46.6%) | 50 (38.5%) | 1.00 | 1.00 |
| CT | 260 (42.8%) | 66 (50.8%) | 1.44 (0.96–2.15) | 1.86 (1.19–2.92) * |
| TT | 64 (10.5%) | 14 (10.8%) | 1.24 (0.65–2.38) | 1.48 (0.75–2.93) |
| rs7256865 T/G | ||||
| TT | 451 (74.5%) | 97 (73.5%) | 1.00 | 1.00 |
| TG | 137 (22.6%) | 33 (25.0%) | 1.12 (0.72–1.74) | 1.11 (0.69–1.79) |
| GG | 17 (2.8%) | 2 (1.5%) | 0.55 (0.12–2.41) | 0.63 (0.14–2.88) |
| rs73164856 C/T | ||||
| CC | 453 (75.1%) | 107 (81.7%) | 1.00 | 1.00 |
| CT | 141 (23.4%) | 22 (16.8%) | 0.66 (0.40–1.08) | 0.72 (0.43–1.23) |
| TT | 9 (1.5%) | 2 (1.5%) | 0.94 (0.20–4.42) | 1.05 (0.22–5.02) |
| rs77836284 C/T | ||||
| CC | 504 (83.2%) | 113 (85.6%) | 1.00 | 1.00 |
| CT | 98 (16.2%) | 19 (14.4%) | 0.86 (0.51–1.47) | 0.80 (0.44–1.46) |
| TT | 4 (0.7%) | 0 (0.0%) | 0.00 (0.00–Inf) | 0.00 (0.00–Inf) |
| rs79975923 A/T | ||||
| AA | 420 (69.5%) | 84 (65.6%) | 1.00 | 1.00 |
| AT | 170 (28.1%) | 38 (29.7%) | 1.12 (0.73–1.71) | 1.10 (0.69–1.74) |
| TT | 14 (2.3%) | 6 (4.7%) | 2.14 (0.80–5.74) | 3.25 (1.11–9.54) * |
| SNPs | Male | Female | ||
|---|---|---|---|---|
| Crude-OR (95% CI) | Adj-OR (95% CI) | Crude-OR (95% CI) | Adj-OR (95% CI) | |
| rs11826681 C/G | ||||
| CC | 1.00 | 1.00 | 1.00 | 1.00 |
| CG | 0.65 (0.33–1.29) | 0.62 (0.30–1.27) | 1.06 (0.63–1.76) | 1.15 (0.64–2.05) |
| GG | 0.50 (0.16–1.57) | 0.57 (0.18–1.83) | 0.80 (0.36–1.76) | 1.04 (0.45–2.42) |
| rs2257609 G/A | ||||
| GG | 1.00 | 1.00 | 1.00 | 1.00 |
| GA | 1.12 (0.56–2.23) | 0.95 (0.46–1.98) | 0.96 (0.56–1.64) | 0.91 (0.50–1.65) |
| AA | 0.50 (0.16–1.58) | 0.44 (0.14–1.44) | 1.14 (0.58–2.26) | 1.17 (0.56–2.47) |
| rs3749606 C/T | ||||
| CC | 1.00 | 1.00 | 1.00 | 1.00 |
| CT | 0.89 (0.42–1.91) | 0.97 (0.44–2.13) | 1.38 (0.84–2.27) | 1.58 (0.91–2.74) |
| TT | 0.00 (0.00–Inf) | 0.00 (0.00–Inf) | 0.54 (0.07–4.40) | 0.83 (0.10–7.29) |
| rs4702701 A/G | ||||
| AA | 1.00 | 1.00 | 1.00 | 1.00 |
| AG | 0.93 (0.37–2.36) | 0.99 (0.38–2.58) | 1.53 (0.79–2.98) | 1.22 (0.56–2.69) |
| GG | 6.22 (0.38–101.72) | 3.82 (0.23–63.77) | 0.00 (0.00–Inf) | 0.00 (0.00–Inf) |
| rs545654 C/T | ||||
| CC | 1.00 | 1.00 | 1.00 | 1.00 |
| CT | 1.55 (0.78–3.07) | 1.60 (0.78–3.26) | 1.40 (0.84–2.32) | 2.07 (1.15–3.73) * |
| TT | 0.78 (0.21–2.84) | 0.70 (0.18–2.64) | 1.51 (0.70–3.26) | 2.24 (0.98–5.12) |
| rs7256865 T/G | ||||
| TT | 1.00 | 1.00 | 1.00 | 1.00 |
| TG | 0.96 (0.45–2.06) | 1.07 (0.48–2.35) | 1.22 (0.71–2.10) | 1.11 (0.61–2.04) |
| GG | 0.00 (0.00–Inf) | 0.00 (0.00–Inf) | 0.90 (0.19–4.25) | 0.97 (0.19–4.95) |
| rs73164856 C/T | ||||
| CC | 1.00 | 1.00 | 1.00 | 1.00 |
| CT | 0.16 (0.04–0.67) * | 0.17 (0.04–0.73) * | 0.97 (0.55–1.70) | 1.22 (0.66–2.25) |
| TT | 0.00 (0.00–Inf) | 0.00 (0.00–Inf) | 1.10 (0.22–5.40) | 1.75 (0.34–9.02) |
| rs77836284 C/T | ||||
| CC | 1.00 | 1.00 | 1.00 | 1.00 |
| CT | 0.90 (0.35–2.26) | 0.99 (0.38–2.58) | 0.83 (0.43–1.60) | 0.81 (0.37–1.75) |
| TT | 0.00 (0.00–Inf) | 0.00 (0.00–Inf) | 0.00 (0.00–Inf) | 0.00 (0.00–Inf) |
| rs79975923 A/T | ||||
| AA | 1.00 | 1.00 | 1.00 | 1.00 |
| AT | 0.85 (0.39–1.84) | 0.81 (0.36–1.82) | 1.29 (0.77–2.15) | 1.38 (0.78–2.47) |
| TT | 8.38 (2.12–33.16) * | 11.96 (2.48–57.64) * | 0.42 (0.05–3.34) | 0.80 (0.10–6.76) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, Y.-S.; Wang, D.-L.; Lee, M.-C.; Wang, C.-C.; Fang, W.-H.; Chuang, S.-W.; Chen, Y.-H.; Su, H.; Chen, C.-J.; Su, S.-L. Dissect Gender-Dependent Susceptibility SNPs in Progressive Osteoarthritis Using Regulator Motif Candidate of Genetic Association Strategy (RMCGA). Int. J. Mol. Sci. 2025, 26, 4117. https://doi.org/10.3390/ijms26094117
Bai Y-S, Wang D-L, Lee M-C, Wang C-C, Fang W-H, Chuang S-W, Chen Y-H, Su H, Chen C-J, Su S-L. Dissect Gender-Dependent Susceptibility SNPs in Progressive Osteoarthritis Using Regulator Motif Candidate of Genetic Association Strategy (RMCGA). International Journal of Molecular Sciences. 2025; 26(9):4117. https://doi.org/10.3390/ijms26094117
Chicago/Turabian StyleBai, Yin-Shiuan, Ding-Lian Wang, Meng-Chang Lee, Chih-Chien Wang, Wen-Hui Fang, Su-Wen Chuang, Yu-Hsuan Chen, Hao Su, Cheng-Jung Chen, and Sui-Lung Su. 2025. "Dissect Gender-Dependent Susceptibility SNPs in Progressive Osteoarthritis Using Regulator Motif Candidate of Genetic Association Strategy (RMCGA)" International Journal of Molecular Sciences 26, no. 9: 4117. https://doi.org/10.3390/ijms26094117
APA StyleBai, Y.-S., Wang, D.-L., Lee, M.-C., Wang, C.-C., Fang, W.-H., Chuang, S.-W., Chen, Y.-H., Su, H., Chen, C.-J., & Su, S.-L. (2025). Dissect Gender-Dependent Susceptibility SNPs in Progressive Osteoarthritis Using Regulator Motif Candidate of Genetic Association Strategy (RMCGA). International Journal of Molecular Sciences, 26(9), 4117. https://doi.org/10.3390/ijms26094117






