Identification of eccDNA in Extracellular Vesicles Derived from Human Dermal Fibroblasts Through Nanopore Sequencing
Abstract
:1. Introduction
2. Results
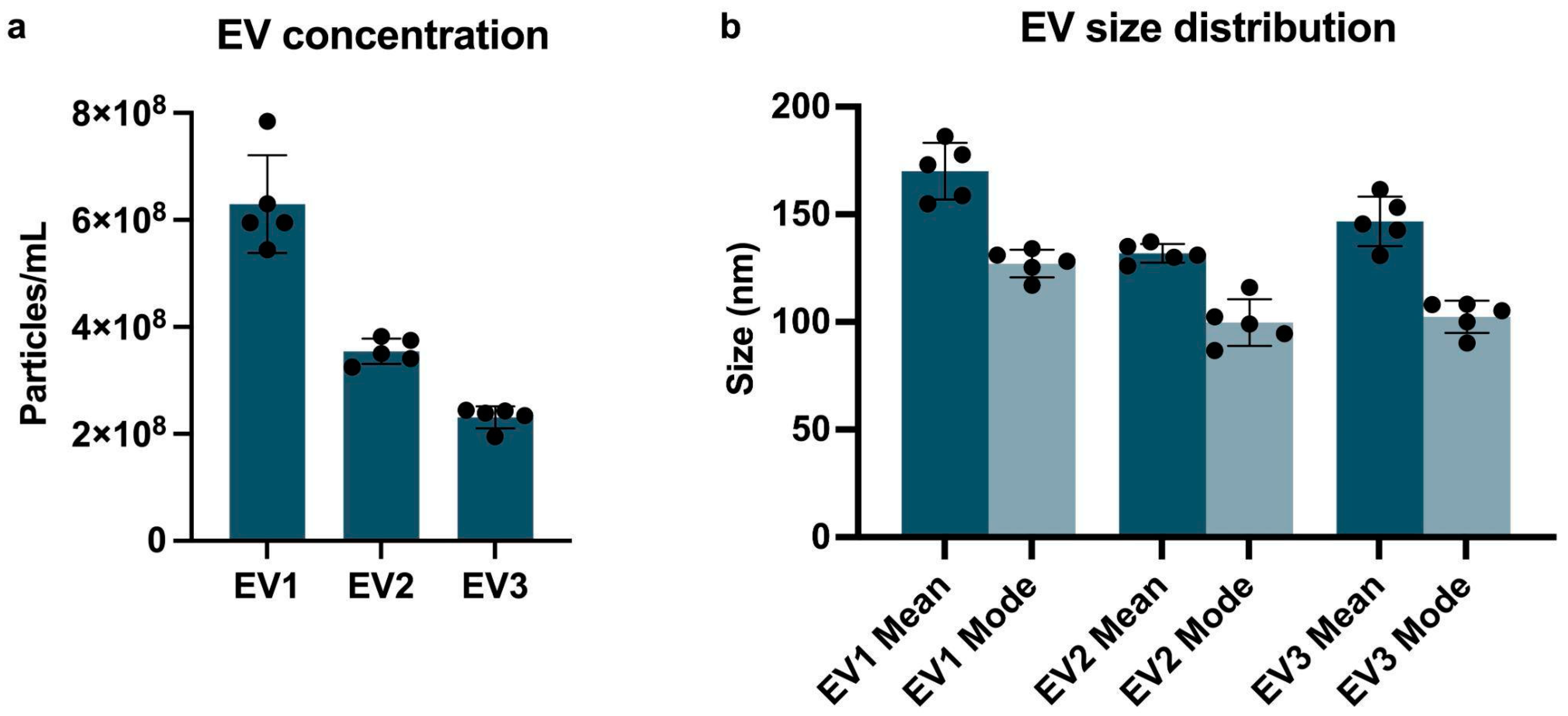
2.1. Extracellular Vesicles Characterization
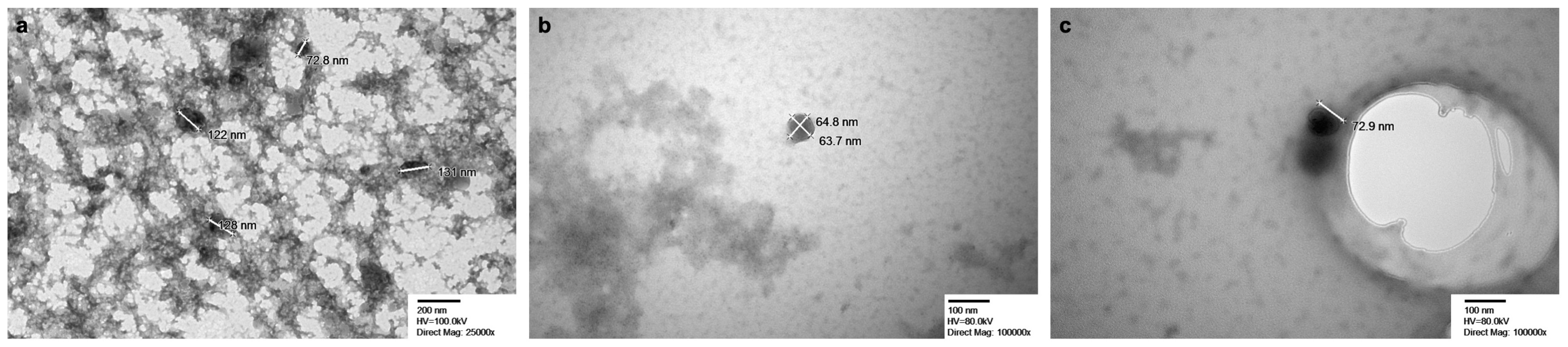
2.1.1. Nanoparticle Tracking Analysis (NTA) and Transmission Electron Microscopy (TEM)
2.1.2. Flow Cytometry
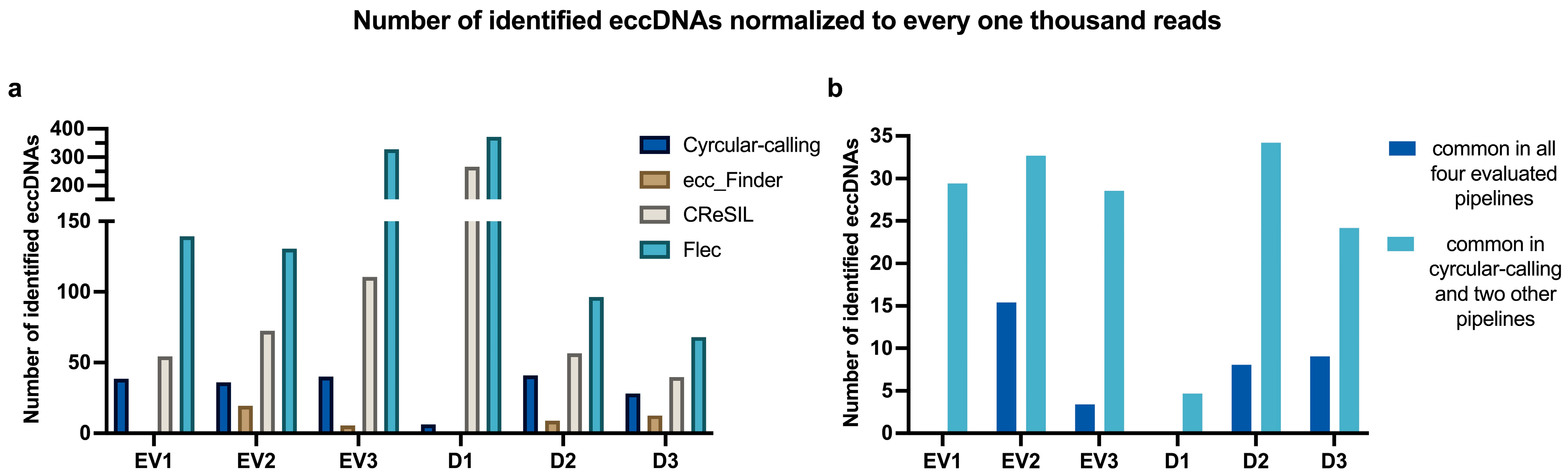
2.2. Identification of eccDNAs Isolated from the Extracellular Vesicle Fraction
2.2.1. eccDNAs Are Present in EVs, but There Is Variability Amongst Pipelines in the Identification Process
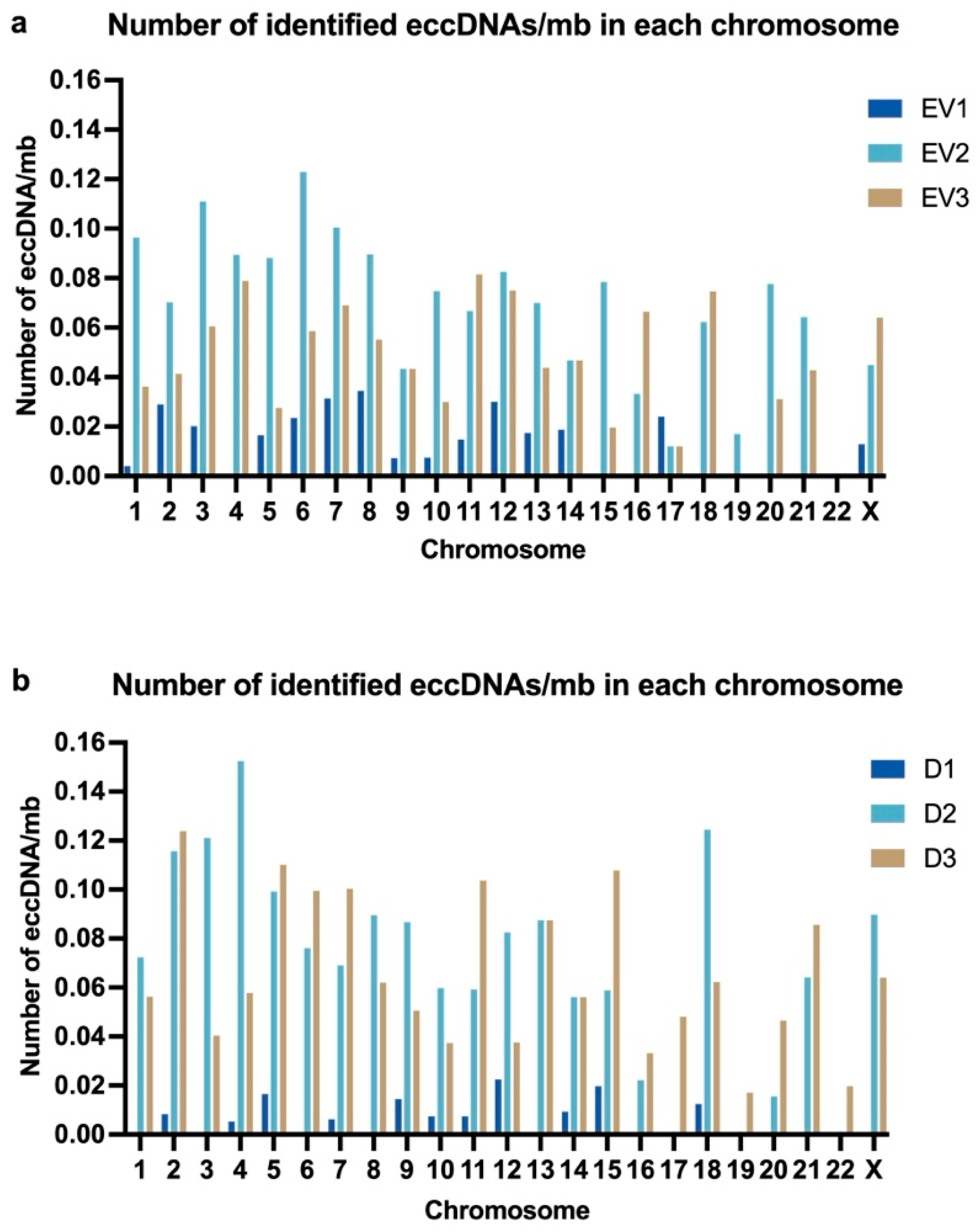
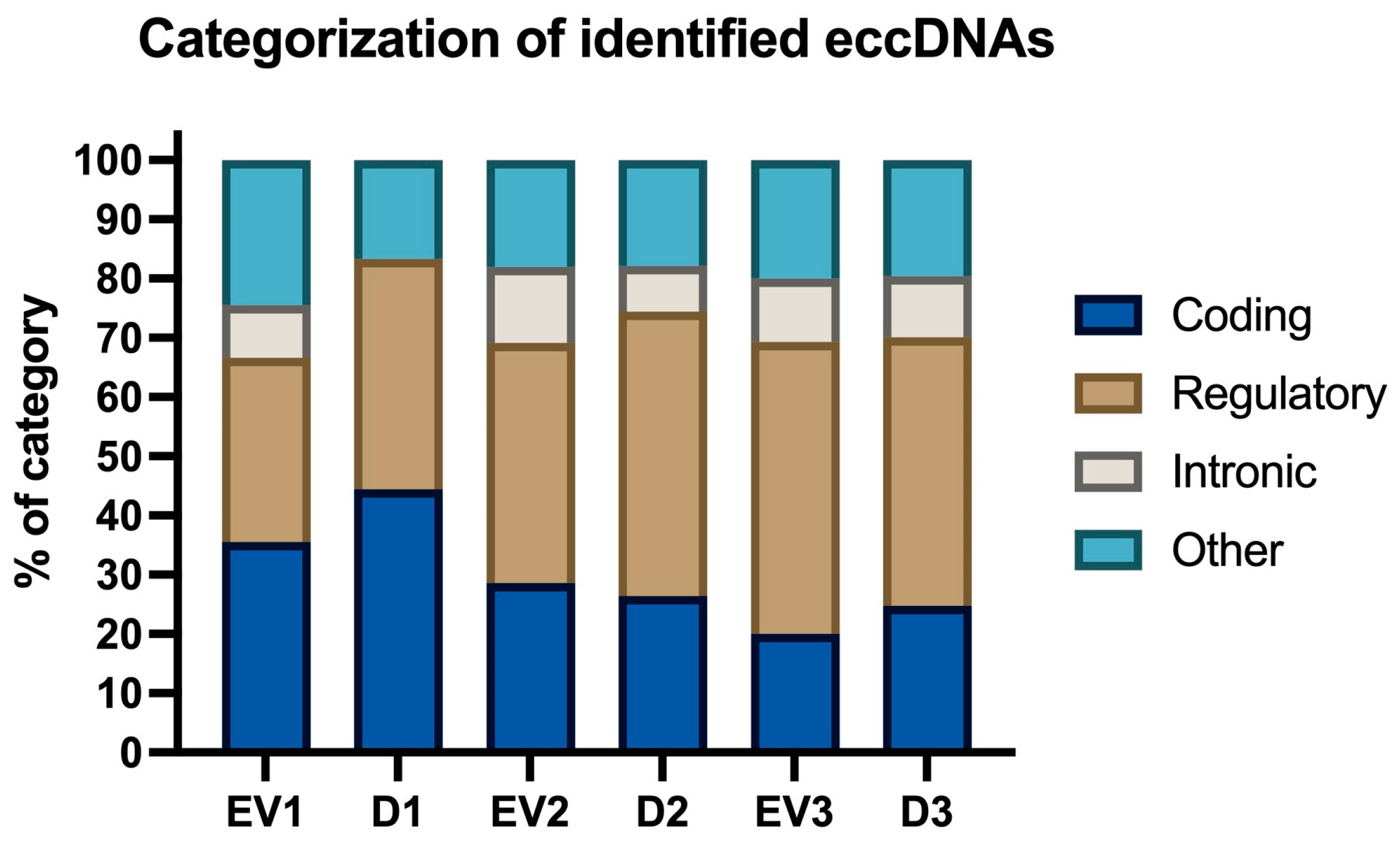
2.2.2. Characteristics of Identified eccDNAs: Size, Chromosomal Distribution, GC Content, and Categorization
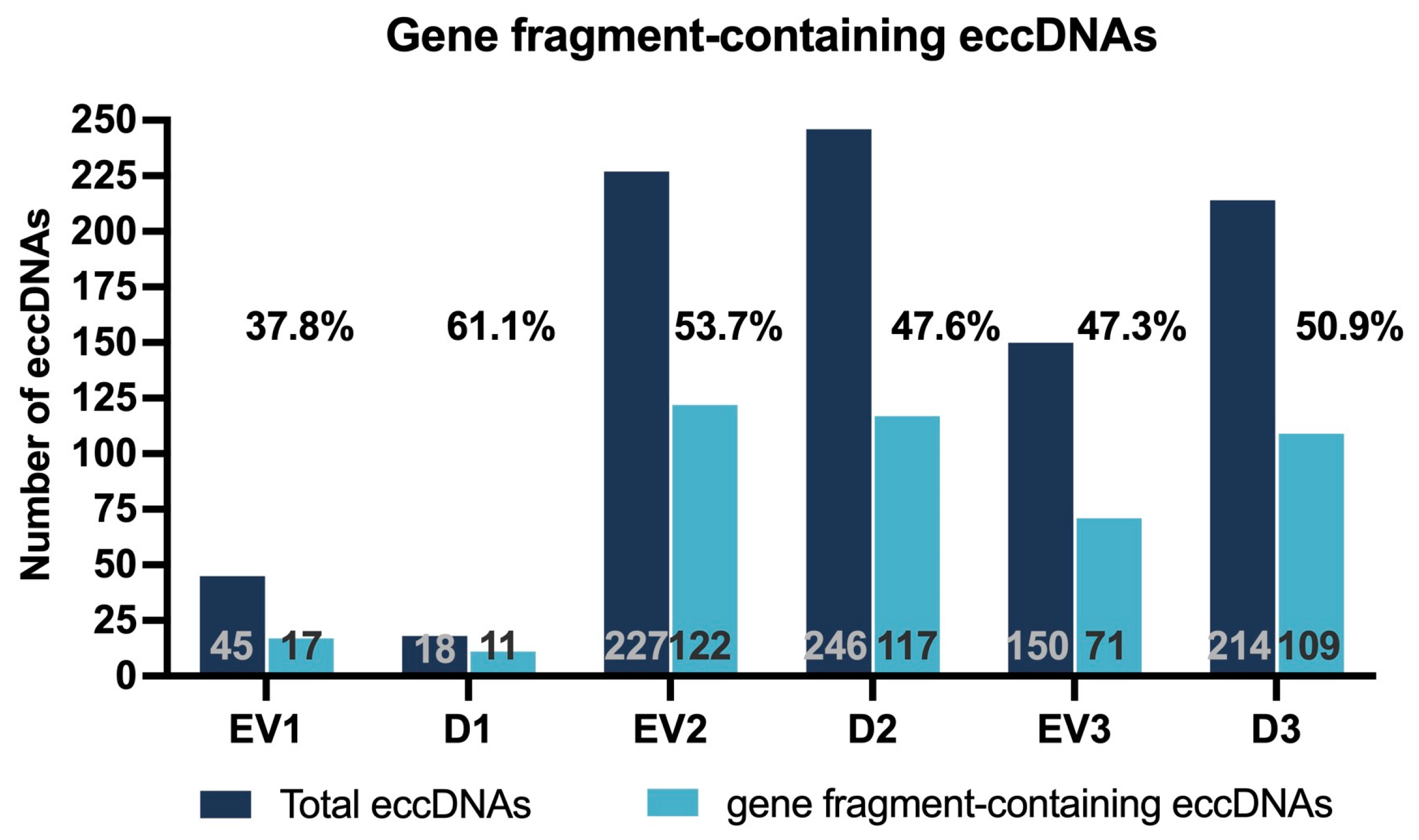
2.2.3. Gene Fragment-Containing eccDNAs
2.2.4. eccDNA Similarity in Different Samples
3. Discussion
4. Materials and Methods
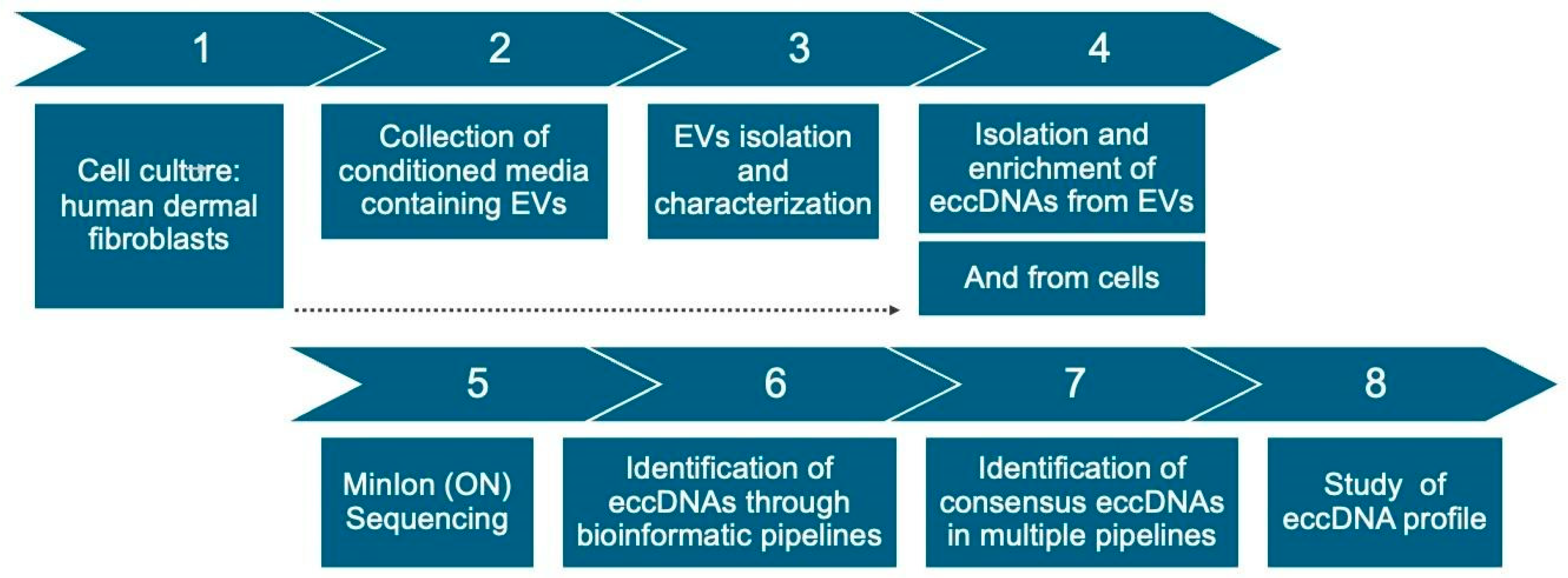
4.1. Workflow
4.2. Cell Culture
4.3. Extracellular Vesicle Isolation
4.4. Extracellular Vesicle Characterization
4.5. DNA Isolation and eccDNA Enrichment
4.6. Sequencing
4.7. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| eccDNA | Extrachromosomal Circular DNA |
| EV(s) | Extracellular Vesicles |
| NTA | Nanoparticle Tracking Analysis |
| SD | Standard Deviation |
| TEM | Transmission Electron Microscopy |
References
- Shmookler Reis, R.J.; Lumpkin, C.K., Jr.; McGill, J.R.; Riabowol, K.T.; Goldstein, S. Genome alteration during in vitro and in vivo aging: Amplification of extrachromosomal circular DNA molecules containing a chromosomal sequence of variable repeat frequency. Cold Spring Harb. Symp. Quant. Biol. 1983, 47, 1135–1139. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, T.; Kumar, P.; Koseoglu, M.M.; Dutta, A. Discoveries of Extrachromosomal Circles of DNA in Normal and Tumor Cells. Trends Genet. 2018, 34, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Dillon, L.W.; Shibata, Y.; Jazaeri, A.A.; Jones, D.R.; Dutta, A. Normal and Cancerous Tissues Release Extrachromosomal Circular DNA (eccDNA) into the Circulation. Mol. Cancer Res. 2017, 15, 1197–1205. [Google Scholar] [CrossRef]
- Xu, Z.; He, J.; Han, P.; Dai, P.; Lv, W.; Liu, N.; Liu, L.; Liu, L.; Pan, X.; Xiang, X.; et al. Plasma extrachromosomal circular DNA is a pathophysiological hallmark of short-term intensive insulin therapy for type 2 diabetes. Clin. Transl. Med. 2023, 13, e1437. [Google Scholar] [CrossRef]
- Møller, H.D.; Mohiyuddin, M.; Prada-Luengo, I.; Sailani, M.R.; Halling, J.F.; Plomgaard, P.; Maretty, L.; Hansen, A.J.; Snyder, M.P.; Pilegaard, H.; et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat. Commun. 2018, 9, 1069. [Google Scholar] [CrossRef]
- Zuo, S.; Yi, Y.; Wang, C.; Li, X.; Zhou, M.; Peng, Q.; Zhou, J.; Yang, Y.; He, Q. Extrachromosomal Circular DNA (eccDNA): From Chaos to Function. Front. Cell Dev. Biol. 2021, 9, 792555. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Wu, D.; Shen, X.; Wu, Q.; Li, C.; Xiong, H.; Xiong, Z.; Gong, R.; Liu, Z.; Wang, W.; et al. Deciphering the role of extrachromosomal circular DNA in adipose stem cells from old and young donors. Stem Cell Res. Ther. 2023, 14, 341. [Google Scholar] [CrossRef]
- Prada-Luengo, I.; Møller, H.D.; Henriksen, R.A.; Gao, Q.; Larsen, C.E.; Alizadeh, S.; Maretty, L.; Houseley, J.; Regenberg, B. Replicative aging is associated with loss of genetic heterogeneity from extrachromosomal circular DNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2020, 48, 7883–7898. [Google Scholar] [CrossRef]
- Dillon, L.W.; Kumar, P.; Shibata, Y.; Wang, Y.H.; Willcox, S.; Griffith, J.D.; Pommier, Y.; Takada, S.; Dutta, A. Production of Extrachromosomal MicroDNAs Is Linked to Mismatch Repair Pathways and Transcriptional Activity. Cell Rep. 2015, 11, 1749–1759. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, M.; Djekidel, M.N.; Chen, H.; Liu, D.; Alt, F.W.; Zhang, Y. eccDNAs are apoptotic products with high innate immunostimulatory activity. Nature 2021, 599, 308–314. [Google Scholar] [CrossRef]
- Yang, L.; Jia, R.; Ge, T.; Ge, S.; Zhuang, A.; Chai, P.; Fan, X. Extrachromosomal circular DNA: Biogenesis, structure, functions and diseases. Signal Transduct Target Ther. 2022, 7, 342. [Google Scholar] [PubMed]
- Wen, K.; Zhang, L.; Cai, Y.; Teng, H.; Liang, J.; Yue, Y.; Li, Y.; Huang, Y.; Liu, M.; Zhang, Y. Identification and characterization of extrachromosomal circular DNA in patients with high myopia and cataract. Epigenetics 2023, 18, 2192324. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Vizio, D.D.; Driedonkd, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Seo, E.C.; Lee, J.H.; Kim, H.J.; Hwangbo, C. The Multifunctional Protein Syntenin-1: Regulator of Exosome Biogenesis, Cellular Function, and Tumor Progression. Int. J. Mol. Sci. 2023, 24, 9418. [Google Scholar] [CrossRef]
- Kugeratski, F.G.; Hodge, K.; Lilla, S.; McAndrews, K.M.; Zhou, X.; Hwang, R.F.; Zanivan, S.; Kalluri, R. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat. Cell Biol. 2021, 23, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Cvjetkovic, A.; Karimi, N.; Crescitelli, R.; Thorsell, A.; Taflin, H.; Lässer, C.; Lötvall, J. Proteomic profiling of tumour tissue-derived extracellular vesicles in colon cancer. J. Extracell. Biol. 2024, 3, e127. [Google Scholar] [CrossRef]
- Wen, Y.; Chen, H.; Wang, Y.; Sun, Y.; Dou, F.; Du, X.; Liu, T.; Chen, C. Extracellular vesicle-derived TP53BP1, CD34, and PBX1 from human peripheral blood serve as potential biomarkers for the assessment and prediction of vascular aging. Hereditas 2024, 161, 3. [Google Scholar] [CrossRef]
- Lee, J.C.; Ray, R.M.; Scott, T.A. Prospects and challenges of tissue-derived extracellular vesicles. Mol. Ther. 2024, 32, 2950–2978. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Papareddy, P.; Tapken, I.; Kroh, K.; Varma Bhongir, R.K.; Rahman, M.; Baumgarten, M.; Clim, E.I.; Györffy, L.; Smeds, E.; Neumann, A.; et al. The role of extracellular vesicle fusion with target cells in triggering systemic inflammation. Nat. Commun. 2024, 15, 1150. [Google Scholar] [CrossRef]
- Ma, Q.; Beal, J.R.; Bhurke, A.; Kannan, A.; Yu, J.; Taylor, R.N.; Bagchi, I.C.; Bagchi, M.K. Extracellular vesicles secreted by human uterine stromal cells regulate decidualization, angiogenesis, and trophoblast differentiation. Proc. Natl. Acad. Sci. USA 2022, 119, e2200252119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ling, Y.; Sun, Y.; Xiao, F.; Wang, L. Extracellular Vesicles Derived from Mesenchymal Stem Cells Promote Wound Healing and Skin Regeneration by Modulating Multiple Cellular Changes: A Brief Review. Genes 2023, 14, 1516. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Liu, K.; Luo, S.; Tang, M.; Liu, N.; Jiang, C.; Fang, J.; Li, S.; Hou, Y.; Guo, C.; et al. Comparative analysis of methodologies for detecting extrachromosomal circular DNA. Nat. Commun. 2024, 15, 9208. [Google Scholar] [CrossRef] [PubMed]
- Møller, H.D.; Ramos-Madrigal, J.; Prada-Luengo, I.; Gilbert, M.T.P.; Regenberg, B. Near-Random Distribution of Chromosome-Derived Circular DNA in the Condensed Genome of Pigeons and the Larger, More Repeat-Rich Human Genome. Genome Biol. Evol. 2020, 12, 3762–3777. [Google Scholar] [CrossRef]
- Dos Santos, C.R.; Hansen, L.B.; Rojas-Triana, M.; Johansen, A.Z.; Perez-Moreno, M.; Regenberg, B. Variation of extrachromosomal circular DNA in cancer cell lines. Comput. Struct. Biotechnol. J. 2023, 21, 4207–4214. [Google Scholar] [CrossRef]
- Chitwood, D.G.; Wang, Q.; Klaubert, S.R.; Green, K.; Wu, C.H.; Harcum, S.W.; Saski, C.A. Microevolutionary dynamics of eccDNA in Chinese hamster ovary cells grown in fed-batch cultures under control and lactate-stressed conditions. Sci. Rep. 2023, 13, 1200. [Google Scholar] [CrossRef]
- Lin, M.; Chen, Y.; Xia, S.; He, Z.; Yu, X.; Huang, L.; Lin, S.; Liang, B.; Huang, Z.; Mei, S.; et al. Integrative profiling of extrachromosomal circular DNA in placenta and maternal plasma provides insights into the biology of fetal growth restriction and reveals potential biomarkers. Front. Genet. 2023, 14, 1128082. [Google Scholar] [CrossRef]
- Chen, J.P.; Diekmann, C.; Wu, H.; Chen, C.; Della Chiara, G.; Berrino, E.; Georgiadis, K.L.; Bouwman, B.A.M.; Virdi, M.; Harbers, L.; et al. scCircle-seq unveils the diversity and complexity of extrachromosomal circular DNAs in single cells. Nat. Commun. 2024, 15, 1768, Erratum in: Nat. Commun. 2024, 15, 3390. [Google Scholar] [CrossRef]
- Nodin, B.; Fridberg, M.; Uhlén, M.; Jirström, K. Discovery of dachshund 2 protein as a novel biomarker of poor prognosis in epithelial ovarian cancer. J. Ovarian Res. 2012, 5, 6. [Google Scholar] [CrossRef]
- Muraoka, S.; DeLeo, A.M.; Sethi, M.K.; Yukawa-Takamatsu, K.; Yang, Z.; Ko, J.; Hogan, J.D.; Ruan, Z.; You, Y.; Wang, Y.; et al. Proteomic and biological profiling of extracellular vesicles from Alzheimer’s disease human brain tissues. Alzheimers Dement. 2020, 16, 896–907. [Google Scholar] [CrossRef]
- Zhang, C.; Du, Q.; Zhou, X.; Qu, T.; Liu, Y.; Ma, K.; Shen, Z.; Wang, Q.; Zhang, Z.; Zhang, R. Differential expression and analysis of extrachromosomal circular DNAs as serum biomarkers in pulmonary arterial hypertension. Respir. Res. 2024, 25, 181. [Google Scholar] [CrossRef] [PubMed]
- Scull, C.E.; Zhang, Y.; Tower, N.; Rasmussen, L.; Padmalayam, I.; Hunter, R.; Zhai, L.; Bostwick, R.; Schneider, D.A. Discovery of novel inhibitors of ribosome biogenesis by innovative high throughput screening strategies. Biochem. J. 2019, 476, 2209–2219. [Google Scholar] [CrossRef] [PubMed]
- Elbahoty, M.H.; Papineni, B.; Samant, R.S. Multiple myeloma: Clinical characteristics, current therapies and emerging innovative treatments targeting ribosome biogenesis dynamics. Clin. Exp. Metastasis 2024, 41, 829–842. [Google Scholar] [CrossRef]
- Yilmaz, Ş.G.; Erdal, M.E.; Özge, A.A.; Sungur, M.A. SNP variation in microRNA biogenesis pathway genes as a new innovation strategy for alzheimer disease diagnostics: A study of 10 candidate genes in an understudied population from the Eastern Mediterranean. Alzheimer Dis. Assoc. Disord. 2016, 30, 203–209. [Google Scholar] [CrossRef]
- Hariharan, H.; Kesavan, Y.; Raja, N.S. Impact of native and external factors on exosome release: Understanding reactive exosome secretion and its biogenesis. Mol. Biol. Rep. 2021, 48, 7559–7573. [Google Scholar] [CrossRef]
- Li, S.R.; Man, Q.W.; Gao, X.; Lin, H.; Wang, J.; Su, F.C.; Wang, H.Q.; Bu, L.L.; Liu, B.; Chen, G. Tissue-derived extracellular vesicles in cancers and non-cancer diseases: Present and future. J. Extracell. Vesicles 2021, 10, e12175. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Han, Y.; Meng, J.; Zhong, B.; Chen, J.; Zhang, H.; Qin, J.; Pang, J.; Liu, L. Small extrachromosomal circular DNA (eccDNA): Major functions in evolution and cancer. Mol. Cancer 2021, 20, 113. [Google Scholar] [CrossRef]
- Jin, W.; Xu, Z.; Song, Y.; Chen, F. Extrachromosomal circular DNA promotes prostate cancer progression through the FAM84B/CDKN1B/MYC/WWP1 axis. Cell. Mol. Biol. Lett. 2024, 29, 103. [Google Scholar] [CrossRef]
- Peng, Y.; Li, Y.; Zhang, W.; ShangGuan, Y.; Xie, T.; Wang, K.; Qiu, J.; Pu, W.; Hu, B.; Zhang, X.; et al. The characteristics of extrachromosomal circular DNA in patients with end-stage renal disease. Eur. J. Med. Res. 2023, 28, 134. [Google Scholar] [CrossRef]
- Qian, Y.; Hong, X.; Yu, Y.; Du, C.; Li, J.; Yu, J.; Xiao, W.; Chen, C.; Huang, D.; Zhong, T.; et al. Characterization and functional analysis of extrachromosomal circular DNA discovered from circulating extracellular vesicles in liver failure. Clin. Transl. Med. 2024, 14, e70059. [Google Scholar] [CrossRef]
- Lane, R.E.; Korbie, D.; Trau, M.; Hill, M.M. Purification Protocols for Extracellular Vesicles. Methods Mol. Biol. 2017, 1660, 111–130. [Google Scholar]
- Barcelos, S.M.; Rosa, P.M.d.S.; Moura, A.B.B.; Villarroel, C.L.P.; Bridi, A.; Bispo, E.C.I.; Garcez, E.M.; Oliveira, G.d.S.; Almeida, M.A.; Malard, P.F.; et al. Extracellular vesicles derived from bovine adipose-derived mesenchymal stromal cells enhance in vitro embryo production from lesioned ovaries. Cytotherapy 2024, 26, 1141–1151. [Google Scholar] [CrossRef]
- Wanchai, V.; Jenjaroenpun, P.; Leangapichart, T.; Arrey, G.; Burnham, C.M.; Tümmler, M.C.; Delgado-Calle, J.; Regenberg, B.; Nookaew, I. CReSIL: Accurate identification of extrachromosomal circular DNA from long-read sequences. Brief Bioinform. 2022, 23, bbac422. [Google Scholar] [CrossRef] [PubMed]
- Tüns, A.I.; Hartmann, T.; Magin, S.; González, R.C.; Henssen, A.G.; Rahmann, S.; Schramm, A.; Köster, J. Detection and Validation of Circular DNA Fragments Using Nanopore Sequencing. Front. Genet. 2022, 13, 867018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Peng, H.; Llauro, C.; Bucher, E.; Mirouze, M. ecc_finder: A Robust and Accurate Tool for Detecting Extrachromosomal Circular DNA From Sequencing Data. Front. Plant Sci. 2021, 12, 743742. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, M.; Zhang, Y. Purification, full-length sequencing and genomic origin mapping of eccDNA. Nat. Protoc. 2023, 18, 683–699. [Google Scholar] [CrossRef]
- Khan, A.; Mathelier, A. Intervene: A tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinform. 2017, 18, 287. [Google Scholar] [CrossRef]


| Number of Reads (Top) Total Bases (Bottom) | Cyrcular-Calling | ecc_Finder | CReSIL | Flec | Common in All Pipelines | Common in Cyrcular- Calling and at Least Two Other Pipelines | |
|---|---|---|---|---|---|---|---|
| EV1 | 1529 11,394,724 | 59 | 0 | 83 | 213 | 0 | 45 |
| EV2 | 6945 57,736,046 | 251 | 136 | 504 | 907 | 107 | 227 |
| EV3 | 5259 48,300,293 | 211 | 29 | 581 | 1725 | 18 | 150 |
| D1 | 3836 30,999,272 | 24 | 0 | 1022 | 1427 | 0 | 18 |
| D2 | 7194 49,947,289 | 295 | 64 | 407 | 694 | 58 | 246 |
| D3 | 8855 66,141,179 | 250 | 113 | 352 | 603 | 80 | 214 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simonassi-Paiva, B.; Luz, J.A.; Ribeiro, J.H.; da Silveira, J.C.; de Souza, C.A.; Pappas Jr, G.J.; Carvalho, J.L.d.; Lynch, M.; Pogue, R.; Rowan, N.J. Identification of eccDNA in Extracellular Vesicles Derived from Human Dermal Fibroblasts Through Nanopore Sequencing. Int. J. Mol. Sci. 2025, 26, 4144. https://doi.org/10.3390/ijms26094144
Simonassi-Paiva B, Luz JA, Ribeiro JH, da Silveira JC, de Souza CA, Pappas Jr GJ, Carvalho JLd, Lynch M, Pogue R, Rowan NJ. Identification of eccDNA in Extracellular Vesicles Derived from Human Dermal Fibroblasts Through Nanopore Sequencing. International Journal of Molecular Sciences. 2025; 26(9):4144. https://doi.org/10.3390/ijms26094144
Chicago/Turabian StyleSimonassi-Paiva, Bianca, Julia Alves Luz, Julia Hellena Ribeiro, Juliano Coelho da Silveira, Camila Azzolin de Souza, Georgios Joannis Pappas Jr, Juliana Lott de Carvalho, Mark Lynch, Robert Pogue, and Neil J. Rowan. 2025. "Identification of eccDNA in Extracellular Vesicles Derived from Human Dermal Fibroblasts Through Nanopore Sequencing" International Journal of Molecular Sciences 26, no. 9: 4144. https://doi.org/10.3390/ijms26094144
APA StyleSimonassi-Paiva, B., Luz, J. A., Ribeiro, J. H., da Silveira, J. C., de Souza, C. A., Pappas Jr, G. J., Carvalho, J. L. d., Lynch, M., Pogue, R., & Rowan, N. J. (2025). Identification of eccDNA in Extracellular Vesicles Derived from Human Dermal Fibroblasts Through Nanopore Sequencing. International Journal of Molecular Sciences, 26(9), 4144. https://doi.org/10.3390/ijms26094144







