Epigenetic Silencing of miR-218-5p Modulates BIRC5 and DDX21 Expression to Promote Colorectal Cancer Progression
Abstract
1. Introduction
2. Results
2.1. MiRNA Expression Profiling in Healthy Controls, Adenomatous Polyps Presenting, and CRC
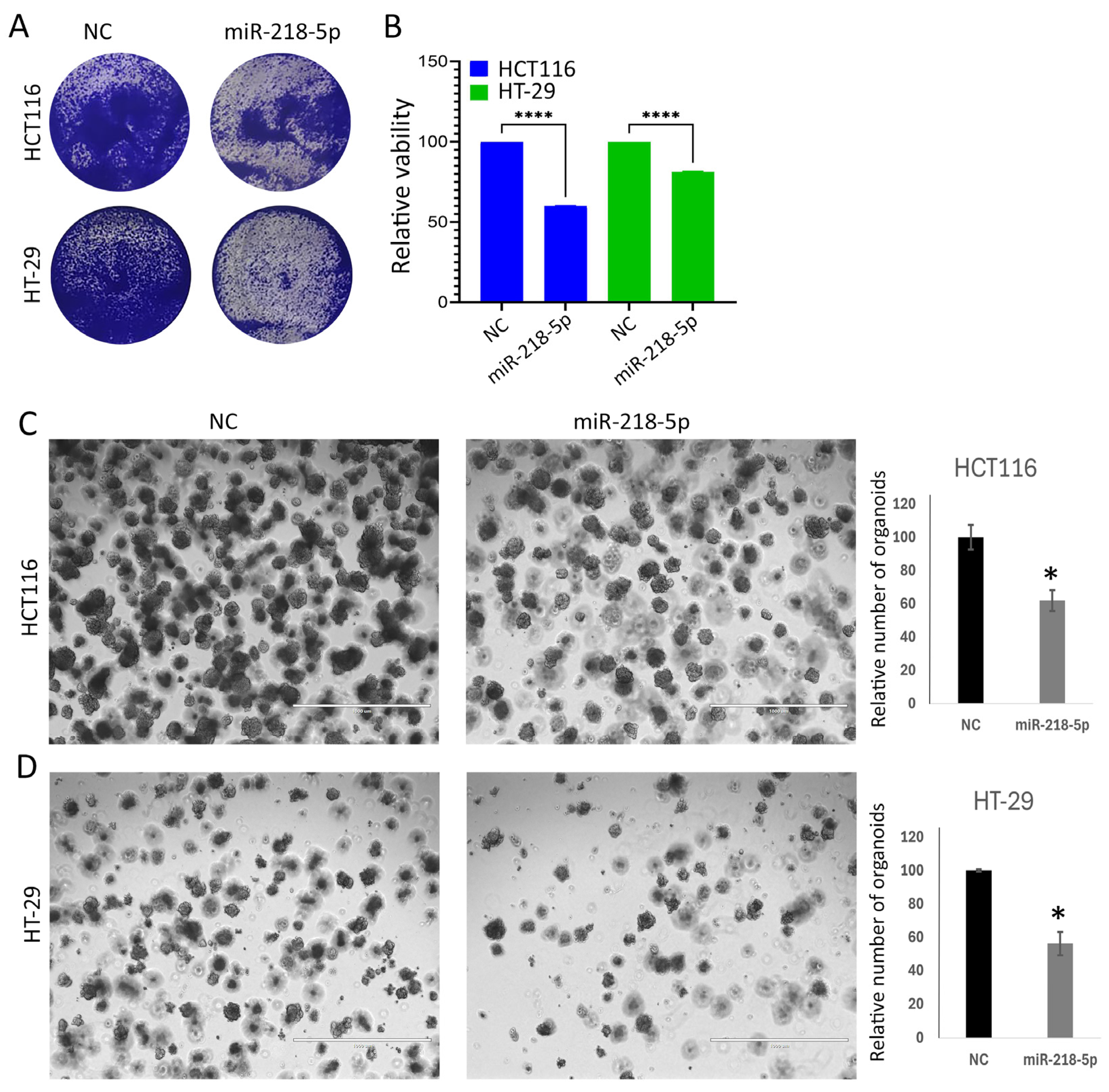
2.2. miR-218-5p Suppresses Tumorigenicity of CRC Models
2.3. Dead–Live Staining of HCT116 and HT-29 CRC Models in Response to miR-218-5p Overexpression
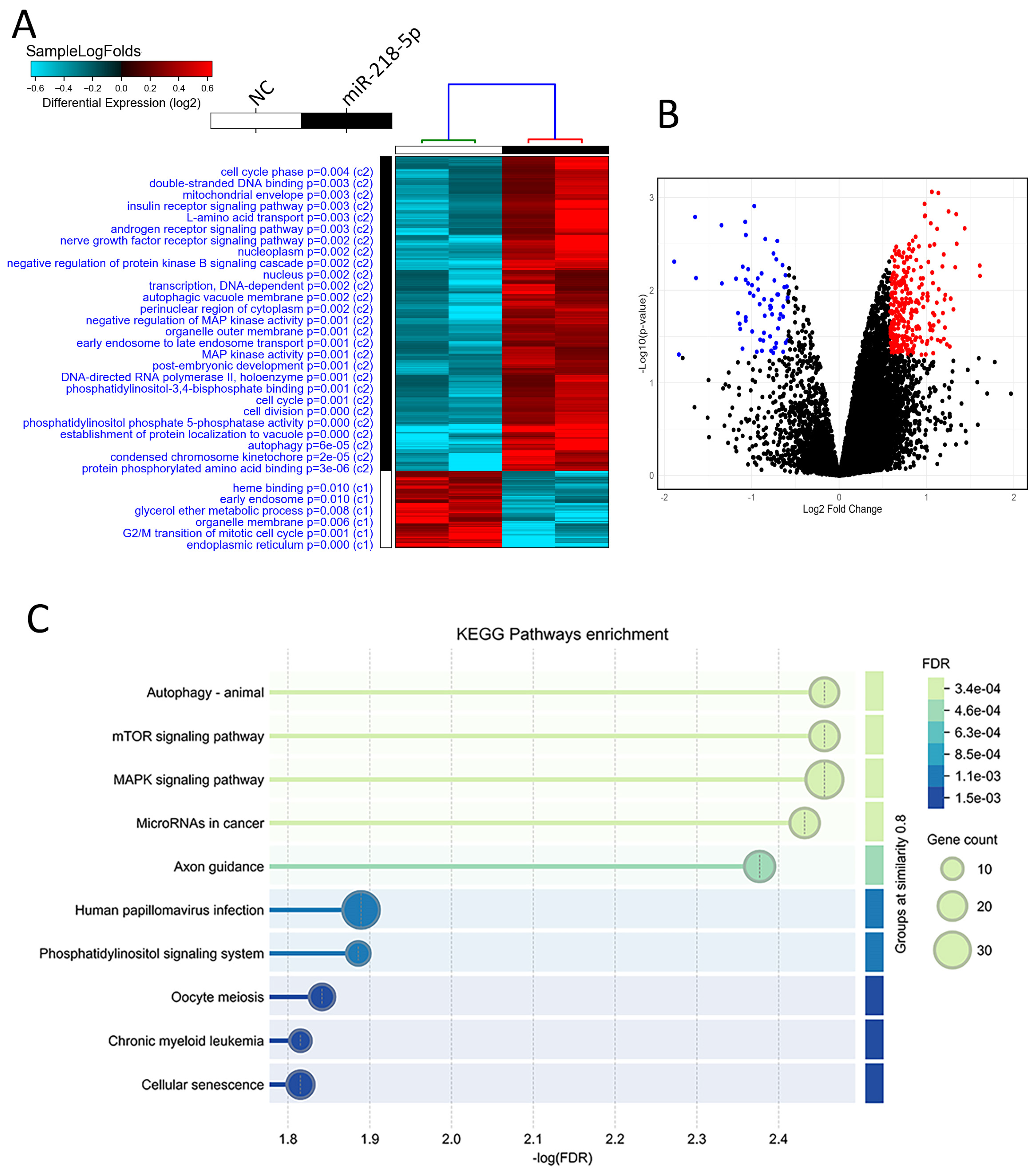
2.4. Transcriptomic Profiling and Pathway Enrichment Analysis in miR-218 Overexpressing CRC Cells
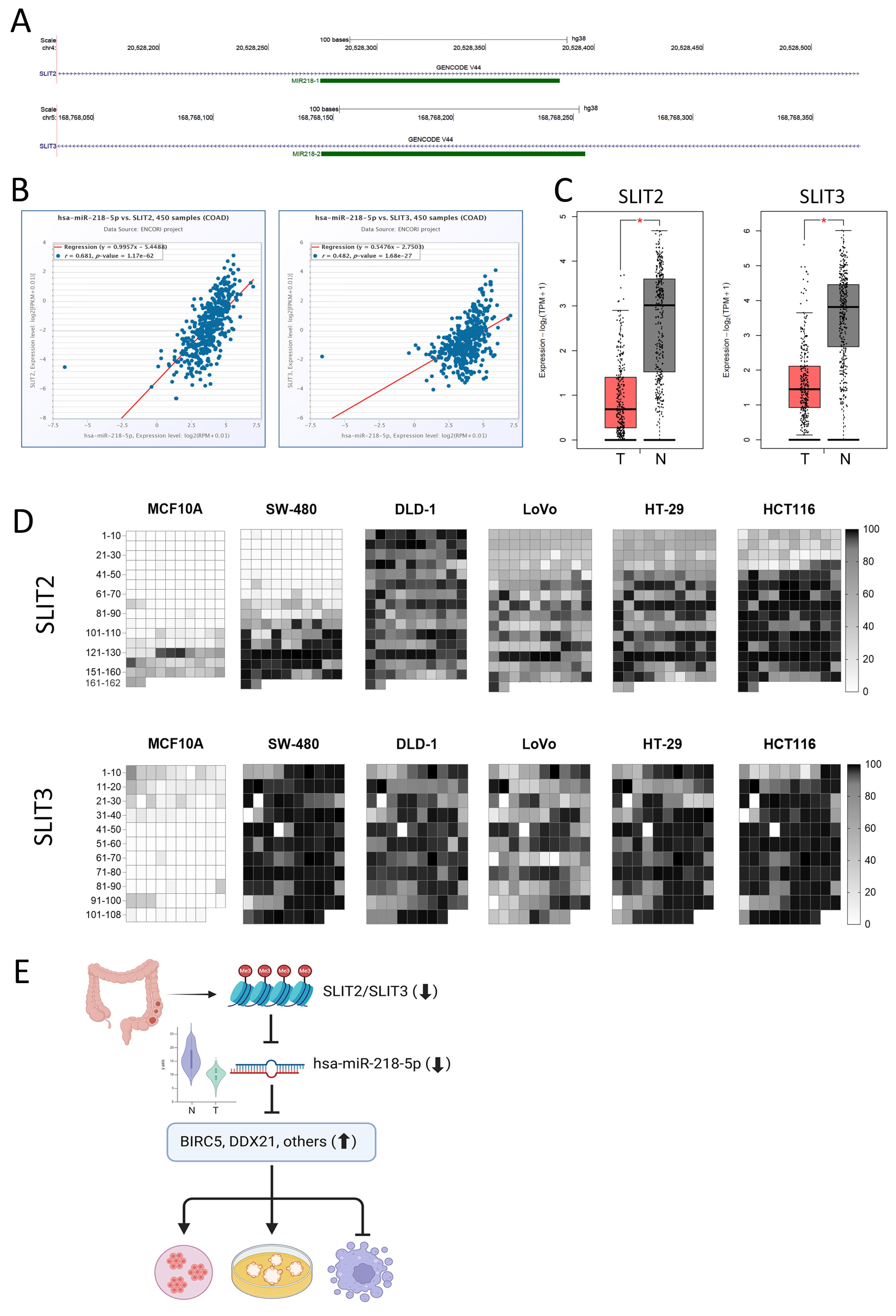
2.5. Identification of miR-218-5p Bona Fide Gene Targets
2.6. Suppression of miR-218-5p Expression Is Mediated via Promoter Hypermethylation of SLIT2 and SLIT3 Host Genes
3. Discussion
4. Materials and Methods
4.1. miRNA Quantification and Bioinformatics Analysis
4.2. Maintenance of Cancer Cell Lines
4.3. mirVana miRNA Mimic Transfection in CRC Cells
4.4. Cell Proliferation Assay
4.5. Detection of Cell Death Using Fluorescence Microscopy
4.6. 3D Organoid Culture
4.7. Total RNA Isolation and Next-Generation Sequencing (NGS) in miR-218-5p Overexpressing CRC Cells
4.8. Identification of miR-218-5p Targets
4.9. Reverse Transcriptase Quantitative Polymerase Chain Reaction (RT-qPCR)
4.10. Western Blot
4.11. Methylation Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Shaath, H.; Vishnubalaji, R.; Elango, R.; Velayutham, D.; Jithesh, P.V.; Alajez, N.M. Therapeutic targeting of the TPX2/TTK network in colorectal cancer. Cell Commun. Signal. 2023, 21, 265. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Yu, R.; Liu, Y.; Geng, X.; Liu, Q.; Liu, S.; Zhu, Y.; Li, G.; Guo, Y.; Xi, X.; et al. LncRNA H19 Participates in Leukemia Inhibitory Factor Mediated Stemness Promotion in Colorectal Cancer Cells. Biochem. Genet. 2024, 62, 3695–3708. [Google Scholar] [CrossRef] [PubMed]
- Legge, D.N.; Collard, T.J.; Stanko, E.; Hoskin, A.J.; Holt, A.K.; Bull, C.J.; Kollareddy, M.; Bellamy, J.; Groves, S.; Ma, E.H.; et al. Identifying targetable metabolic dependencies across colorectal cancer progression. Mol. Metab. 2024, 90, 102037. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jiang, D.; Liu, H.; Guo, C.; Zhang, Q.; Li, X.; Chen, X.; Chen, Z.; Feng, J.; Tan, S.; et al. Comprehensive Proteogenomic Profiling Reveals the Molecular Characteristics of Colorectal Cancer at Distinct Stages of Progression. Cancer Res. 2024, 84, 2888–2910. [Google Scholar] [CrossRef]
- Zhao, Y.; Guo, X.; Zhang, L.; Wang, D.; Li, Y. Mitochondria: A crucial factor in the progression and drug resistance of colorectal cancer. Front. Immunol. 2024, 15, 1512469. [Google Scholar] [CrossRef]
- Bahrami, A.; Khalaji, A.; Bahri Najafi, M.; Sadati, S.; Raisi, A.; Abolhassani, A.; Eshraghi, R.; Khaksary Mahabady, M.; Rahimian, N.; Mirzaei, H. NF-kappaB pathway and angiogenesis: Insights into colorectal cancer development and therapeutic targets. Eur. J. Med. Res. 2024, 29, 610. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef]
- Nemeth, K.; Bayraktar, R.; Ferracin, M.; Calin, G.A. Non-coding RNAs in disease: From mechanisms to therapeutics. Nat. Rev. Genet. 2024, 25, 211–232. [Google Scholar] [CrossRef]
- Vishnubalaji, R.; Shaath, H.; Al-Alwan, M.; Abdelalim, E.M.; Alajez, N.M. Reciprocal interplays between MicroRNAs and pluripotency transcription factors in dictating stemness features in human cancers. Semin. Cancer Biol. 2022, 87, 1–16. [Google Scholar] [CrossRef]
- Shaath, H.; Toor, S.M.; Nada, M.A.; Elkord, E.; Alajez, N.M. Integrated whole transcriptome and small RNA analysis revealed multiple regulatory networks in colorectal cancer. Sci. Rep. 2021, 11, 14456. [Google Scholar] [CrossRef]
- Abohalawa, B.Y.; Shaath, H.; Elango, R.; Vishnubalaji, R.; Rashid, S.; Al-Sarraf, R.; Akhtar, M.; Alajez, N.M. MicroRNAome profiling of breast cancer unveils hsa-miR-5683 as a tumor suppressor microRNA predicting favorable clinical outcome. Cancer Cell Int. 2024, 24, 377. [Google Scholar] [CrossRef]
- Chen, B.; Xia, Z.; Deng, Y.N.; Yang, Y.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. Emerging microRNA biomarkers for colorectal cancer diagnosis and prognosis. Open Biol. 2019, 9, 180212. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- Farasati Far, B.; Vakili, K.; Fathi, M.; Yaghoobpoor, S.; Bhia, M.; Naimi-Jamal, M.R. The role of microRNA-21 (miR-21) in pathogenesis, diagnosis, and prognosis of gastrointestinal cancers: A review. Life Sci. 2023, 316, 121340. [Google Scholar] [CrossRef]
- Li, J.; Chen, H.; Sun, G.; Zhang, X.; Ye, H.; Wang, P. Role of miR-21 in the diagnosis of colorectal cancer: Meta-analysis and bioinformatics. Pathol. Res. Pract. 2023, 248, 154670. [Google Scholar] [CrossRef]
- Huang, Z.; Kaller, M.; Hermeking, H. CRISPR/Cas9-mediated inactivation of miR-34a and miR-34b/c in HCT116 colorectal cancer cells: Comprehensive characterization after exposure to 5-FU reveals EMT and autophagy as key processes regulated by miR-34. Cell Death Differ. 2023, 30, 2017–2034. [Google Scholar] [CrossRef]
- Moravcik, R.; Olejarova, S.; Zlacka, J.; Herichova, I. Effect of miR-34a on the expression of clock and clock-controlled genes in DLD1 and Lovo human cancer cells with different backgrounds with respect to p53 functionality and 17beta-estradiol-mediated regulation. PLoS ONE 2023, 18, e0292880. [Google Scholar] [CrossRef]
- Hosseini, S.S.; Nazifi, P.; Amini, M.; Zargari, F.; Yari, A.H.; Baradaran, B.; Mahboob, S.; Mokhtarzadeh, A. Crocin Suppresses Colorectal Cancer Cell Proliferation by Regulating miR-143/145 and KRAS/RREB1 Pathways. Anticancer Agents Med. Chem. 2023, 23, 1916–1923. [Google Scholar] [CrossRef]
- Jepsen, R.K.; Novotny, G.W.; Klarskov, L.L.; Bang-Berthelsen, C.H.; Haakansson, I.T.; Hansen, A.; Christensen, I.J.; Riis, L.B.; Hogdall, E. Early metastatic colorectal cancers show increased tissue expression of miR-17/92 cluster members in the invasive tumor front. Hum. Pathol. 2018, 80, 231–238. [Google Scholar] [CrossRef]
- Bostanghadiri, N.; Razavi, S.; Shariati, A.; Talebi, M.; Mirkalantari, S.; Emami Razavi, A.; Darban-Sarokhalil, D. Exploring the interplay between Fusobacterium nucleatum with the expression of microRNA, and inflammatory mediators in colorectal cancer. Front. Microbiol. 2023, 14, 1302719. [Google Scholar] [CrossRef]
- Pavlic, A.; Urh, K.; Bostjancic, E.; Zidar, N. Analyzing the invasive front of colorectal cancer—By punching tissue block or laser capture microdissection? Pathol. Res. Prarct. 2023, 248, 154727. [Google Scholar] [CrossRef]
- Lin, Y.; Wang, W.; Wang, Y.; Fu, K. MiR-223-3p increases resistance of colorectal cancer cells to 5-fluorouracil via targeting SORBS1. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2023, 48, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Siemens, H.; Neumann, J.; Jackstadt, R.; Mansmann, U.; Horst, D.; Kirchner, T.; Hermeking, H. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and beta-catenin predicts distant metastasis of colon cancer. Clin. Cancer Res. 2013, 19, 710–720. [Google Scholar] [CrossRef]
- Shaath, H.; Toor, S.M.; Nair, V.S.; Elkord, E.; Alajez, N.M. Transcriptomic Analyses Revealed Systemic Alterations in Gene Expression in Circulation and Tumor Microenvironment of Colorectal Cancer Patients. Cancers 2019, 11, 1994. [Google Scholar] [CrossRef] [PubMed]
- Radak, M.; Fallahi, H. Unraveling molecular similarities between colorectal polyps and colorectal cancer: A systems biology approach. Intest. Res. 2024, 22, 199–207. [Google Scholar] [CrossRef]
- Li, Q.; Geng, S.; Luo, H.; Wang, W.; Mo, Y.Q.; Luo, Q.; Wang, L.; Song, G.B.; Sheng, J.P.; Xu, B. Signaling pathways involved in colorectal cancer: Pathogenesis and targeted therapy. Signal Transduct. Target. Ther. 2024, 9, 266. [Google Scholar] [CrossRef]
- Thiebes, K.P.; Nam, H.; Cambronne, X.A.; Shen, R.; Glasgow, S.M.; Cho, H.H.; Kwon, J.S.; Goodman, R.H.; Lee, J.W.; Lee, S.; et al. miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1-Lhx3. Nat. Commun. 2015, 6, 7718. [Google Scholar] [CrossRef]
- Yang, Y.; Ding, L.; Hu, Q.; Xia, J.; Sun, J.; Wang, X.; Xiong, H.; Gurbani, D.; Li, L.; Liu, Y.; et al. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol. Cancer 2017, 16, 141. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, L.; Wang, L.; Zhang, H.; Hu, X. miR-218 functions as a tumor suppressor gene in cervical cancer. Mol. Med. Rep. 2020, 21, 209–219. [Google Scholar] [CrossRef]
- Venkataraman, S.; Birks, D.K.; Balakrishnan, I.; Alimova, I.; Harris, P.S.; Patel, P.R.; Handler, M.H.; Dubuc, A.; Taylor, M.D.; Foreman, N.K.; et al. MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma. J. Biol. Chem. 2013, 288, 1918–1928. [Google Scholar] [CrossRef]
- Chen, J.; Tang, Z.; Chen, Z.; Wei, Y.; Liang, H.; Zhang, X.; Gao, Z.; Zhu, H. MicroRNA-218-5p regulates inflammation response via targeting TLR4 in atherosclerosis. BMC Cardiovasc. Disord. 2023, 23, 122. [Google Scholar] [CrossRef] [PubMed]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Hu, F.; Hu, J.; Wang, C.; Hou, J.; Yu, Z.; Wang, T.T.; Liu, X.; Huang, H. MicroRNA-218 promotes cisplatin resistance in oral cancer via the PPP2R5A/Wnt signaling pathway. Oncol. Rep. 2017, 38, 2051–2061. [Google Scholar] [CrossRef]
- Puppo, M.; Valluru, M.K.; Clezardin, P. MicroRNAs and Their Roles in Breast Cancer Bone Metastasis. Curr. Osteoporos. Rep. 2021, 19, 256–263. [Google Scholar] [CrossRef]
- Yang, J.L.; Ma, J.J.; Qu, T.Y.; Dai, Q.; Leng, J.; Fang, L.; Wu, J.; Li, Y.J.; Yu, H.F. Glycolysis-related lncRNA FTX upregulates YAP1 to facilitate colorectal cancer progression via sponging miR-215-3p. Sci. Rep. 2025, 15, 9929. [Google Scholar] [CrossRef]
- Li, J.; Han, Y.; Zhou, M.; Liu, N.; Li, H.; Huang, G.; Yu, Z.; Luo, D.; Zhang, H.; Zheng, X.; et al. Electroacupuncture ameliorates AOM/DSS-induced mice colorectal cancer by inhibiting inflammation and promoting autophagy via the SIRT1/miR-215/Atg14 axis. Aging 2023, 15, 13194–13212. [Google Scholar] [CrossRef]
- Alajez, N.M.; Lenarduzzi, M.; Ito, E.; Hui, A.B.; Shi, W.; Bruce, J.; Yue, S.; Huang, S.H.; Xu, W.; Waldron, J.; et al. MiR-218 suppresses nasopharyngeal cancer progression through downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011, 71, 2381–2391. [Google Scholar] [CrossRef]
- Lukosevicius, R.; Juzenas, S.; Salteniene, V.; Kulokiene, U.; Arstikyte, J.; Hemmrich-Stanisak, G.; Franke, A.; Link, A.; Ruzgys, P.; Satkauskas, S.; et al. miRNome Profiling and Functional Analysis Reveal Involvement of hsa-miR-1246 in Colon Adenoma-Carcinoma Transition by Targeting AXIN2 and CFTR. Int. J. Mol. Sci. 2022, 23, 2107. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, K.; Zhou, E.; Xue, X.; Yan, S.; Chen, Y.; Qiao, P.; Yang, L.; Chen, X. hsa_circ_0023305 Enhances Laryngeal Squamous Cell Carcinoma Progression and Modulates TRPM7 via miR-218-5p Sponging. Biomed. Res. Int. 2021, 2021, 9968499. [Google Scholar] [CrossRef]
- Dickinson, R.E.; Dallol, A.; Bieche, I.; Krex, D.; Morton, D.; Maher, E.R.; Latif, F. Epigenetic inactivation of SLIT3 and SLIT1 genes in human cancers. Br. J. Cancer 2004, 91, 2071–2078. [Google Scholar] [CrossRef]
- Narayan, G.; Goparaju, C.; Arias-Pulido, H.; Kaufmann, A.M.; Schneider, A.; Durst, M.; Mansukhani, M.; Pothuri, B.; Murty, V.V. Promoter hypermethylation-mediated inactivation of multiple Slit-Robo pathway genes in cervical cancer progression. Mol. Cancer 2006, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.L.; Wang, Y.; Zhang, T.J.; Chu, M.Q.; Xu, Z.J.; Yuan, Q.; Ma, J.C.; Lin, J.; Qian, J.; Zhou, J.D. SLIT2 promoter hypermethylation predicts disease progression in chronic myeloid leukemia. Eur. J. Med. Res. 2022, 27, 259. [Google Scholar] [CrossRef]
- Zhang, T.J.; Xu, Z.J.; Wen, X.M.; Gu, Y.; Ma, J.C.; Yuan, Q.; Lin, J.; Zhou, J.D.; Qian, J. SLIT2 promoter hypermethylation-mediated SLIT2-IT1/miR-218 repression drives leukemogenesis and predicts adverse prognosis in myelodysplastic neoplasm. Leukemia 2022, 36, 2488–2498. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yin, K.; Guo, X.; Feng, H.; Yuan, M.; Liu, Y.; Zhang, J.; Guo, B.; Wang, C.; Zhou, G.; et al. Diphthamide Biosynthesis 1 is a Novel Oncogene in Colorectal Cancer Cells and is Regulated by MiR-218-5p. Cell. Physiol. Biochem. 2017, 44, 505–514. [Google Scholar] [CrossRef]
- Meng, Q.; Chen, Y.; Lian, B.; Shang, Y.; Yang, H. miR-218 promotes apoptosis of SW1417 human colon cancer cells by targeting c-FLIP. Oncol. Rep. 2018, 40, 916–922. [Google Scholar] [CrossRef]
- Deng, H.; Zhang, Q.; Zhao, Z.; Wang, M.; Xu, Q. SNAI2/FTH1P3/miR-218-5p Positive Feedback Loop Promotes Colorectal Cancer Metastasis. Biochem. Genet. 2024, 62, 2210–2223. [Google Scholar] [CrossRef]
- Pu, Y.; Wei, J.; Wu, Y.; Zhao, K.; Wu, Y.; Wu, S.; Yang, X.; Xing, C. Correction: THUMPD3-AS1 facilitates cell growth and aggressiveness by the miR-218-5p/SKAP1 axis in colorectal cancer. Cell Biochem. Biophys. 2024, 82, 315. [Google Scholar] [CrossRef]
- Andrisani, O.; Liu, Q.; Kehn, P.; Leitner, W.W.; Moon, K.; Vazquez-Maldonado, N.; Fingerman, I.; Gale, M., Jr. Biological functions of DEAD/DEAH-box RNA helicases in health and disease. Nat. Immunol. 2022, 23, 354–357. [Google Scholar] [CrossRef]
- Zhang, Y.; Baysac, K.C.; Yee, L.F.; Saporita, A.J.; Weber, J.D. Elevated DDX21 regulates c-Jun activity and rRNA processing in human breast cancers. Breast Cancer Res. 2014, 16, 449. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Y.; Chen, C.; Zhu, X.; Zhang, C.; Xia, Y.; Zhao, Y.; Andrisani, O.M.; Kong, L. A double-negative feedback loop between DEAD-box protein DDX21 and Snail regulates epithelial-mesenchymal transition and metastasis in breast cancer. Cancer Lett. 2018, 437, 67–78. [Google Scholar] [CrossRef]
- Basha, S.; Jin-Smith, B.; Sun, C.; Pi, L. The SLIT/ROBO Pathway in Liver Fibrosis and Cancer. Biomolecules 2023, 13, 785. [Google Scholar] [CrossRef] [PubMed]
- Dallol, A.; Morton, D.; Maher, E.R.; Latif, F. SLIT2 axon guidance molecule is frequently inactivated in colorectal cancer and suppresses growth of colorectal carcinoma cells. Cancer Res. 2003, 63, 1054–1058. [Google Scholar]
- Qiu, H.; Zhu, J.; Yu, J.; Pu, H.; Dong, R. SLIT2 is epigenetically silenced in ovarian cancers and suppresses growth when activated. Asian Pac. J. Cancer Prev. 2011, 12, 791–795. [Google Scholar] [PubMed]
- Tseng, R.C.; Chang, J.M.; Chen, J.H.; Huang, W.R.; Tang, Y.A.; Kuo, I.Y.; Yan, J.J.; Lai, W.W.; Wang, Y.C. Deregulation of SLIT2-mediated Cdc42 activity is associated with esophageal cancer metastasis and poor prognosis. J. Thorac. Oncol. 2015, 10, 189–198. [Google Scholar] [CrossRef]
- Chen, W.F.; Gao, W.D.; Li, Q.L.; Zhou, P.H.; Xu, M.D.; Yao, L.Q. SLIT2 inhibits cell migration in colorectal cancer through the AKT-GSK3beta signaling pathway. Int. J. Color. Dis. 2013, 28, 933–940. [Google Scholar] [CrossRef]
- Li, G.J.; Yang, Y.; Yang, G.K.; Wan, J.; Cui, D.L.; Ma, Z.H.; Du, L.J.; Zhang, G.M. Slit2 suppresses endothelial cell proliferation and migration by inhibiting the VEGF-Notch signaling pathway. Mol. Med. Rep. 2017, 15, 1981–1988. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, J.H.; Baek, S.J.; Kim, S.Y.; Kim, Y.S. Specific expression and methylation of SLIT1, SLIT2, SLIT3, and miR-218 in gastric cancer subtypes. Int. J. Oncol. 2016, 48, 2497–2507. [Google Scholar] [CrossRef]
- Fadaka, A.O.; Akinsoji, T.; Klein, A.; Madiehe, A.M.; Meyer, M.; Keyster, M.; Sikhwivhilu, L.M.; Sibuyi, N.R.S. Stage-specific treatment of colorectal cancer: A microRNA-nanocomposite approach. J. Pharm. Anal. 2023, 13, 1235–1251. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M.; Collaboration, I.N.S.D. The sequence read archive. Nucleic Acids Res. 2010, 39 (Suppl. S1), D19–D21. [Google Scholar] [CrossRef]
- Emig, D.; Salomonis, N.; Baumbach, J.; Lengauer, T.; Conklin, B.R.; Albrecht, M. AltAnalyze and DomainGraph: Analyzing and visualizing exon expression data. Nucleic Acids Res. 2010, 38 (Suppl. S2), W755–W762. [Google Scholar] [CrossRef]
- Geissmann, Q. OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS ONE 2013, 8, e54072. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [PubMed]
- Vishnubalaji, R.; Alajez, N.M. Disrupted Lipid Metabolism, Cytokine Signaling, and Dormancy: Hallmarks of Doxorubicin-Resistant Triple-Negative Breast Cancer Models. Cancers 2024, 16, 4273. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaath, H.; Vishnubalaji, R.; Ouararhni, K.; Alajez, N.M. Epigenetic Silencing of miR-218-5p Modulates BIRC5 and DDX21 Expression to Promote Colorectal Cancer Progression. Int. J. Mol. Sci. 2025, 26, 4146. https://doi.org/10.3390/ijms26094146
Shaath H, Vishnubalaji R, Ouararhni K, Alajez NM. Epigenetic Silencing of miR-218-5p Modulates BIRC5 and DDX21 Expression to Promote Colorectal Cancer Progression. International Journal of Molecular Sciences. 2025; 26(9):4146. https://doi.org/10.3390/ijms26094146
Chicago/Turabian StyleShaath, Hibah, Radhakrishnan Vishnubalaji, Khalid Ouararhni, and Nehad M. Alajez. 2025. "Epigenetic Silencing of miR-218-5p Modulates BIRC5 and DDX21 Expression to Promote Colorectal Cancer Progression" International Journal of Molecular Sciences 26, no. 9: 4146. https://doi.org/10.3390/ijms26094146
APA StyleShaath, H., Vishnubalaji, R., Ouararhni, K., & Alajez, N. M. (2025). Epigenetic Silencing of miR-218-5p Modulates BIRC5 and DDX21 Expression to Promote Colorectal Cancer Progression. International Journal of Molecular Sciences, 26(9), 4146. https://doi.org/10.3390/ijms26094146






