Genome-Wide Identification and Classification of Arabinogalactan Proteins Gene Family in Gossypium Species and GhAGP50 Increases Numbers of Epidermal Hairs in Arabidopsis
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification and Sequence Analysis of AGP
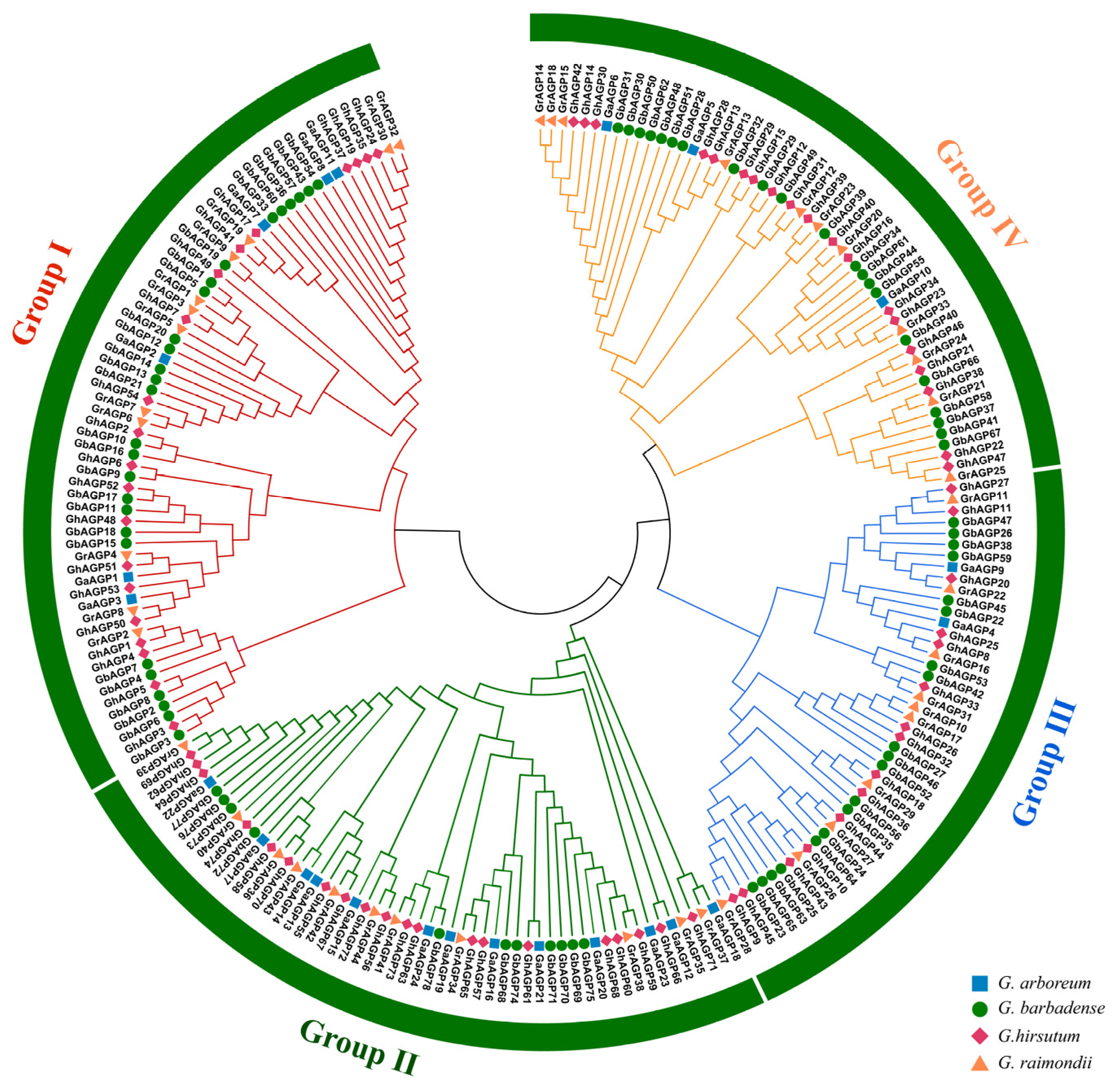
2.2. Phylogenetic Tree and Genomic Architecture of AGP Gene Family
2.3. Localization of AGP Genes on Chromosomes and Phylogenetic Analysis
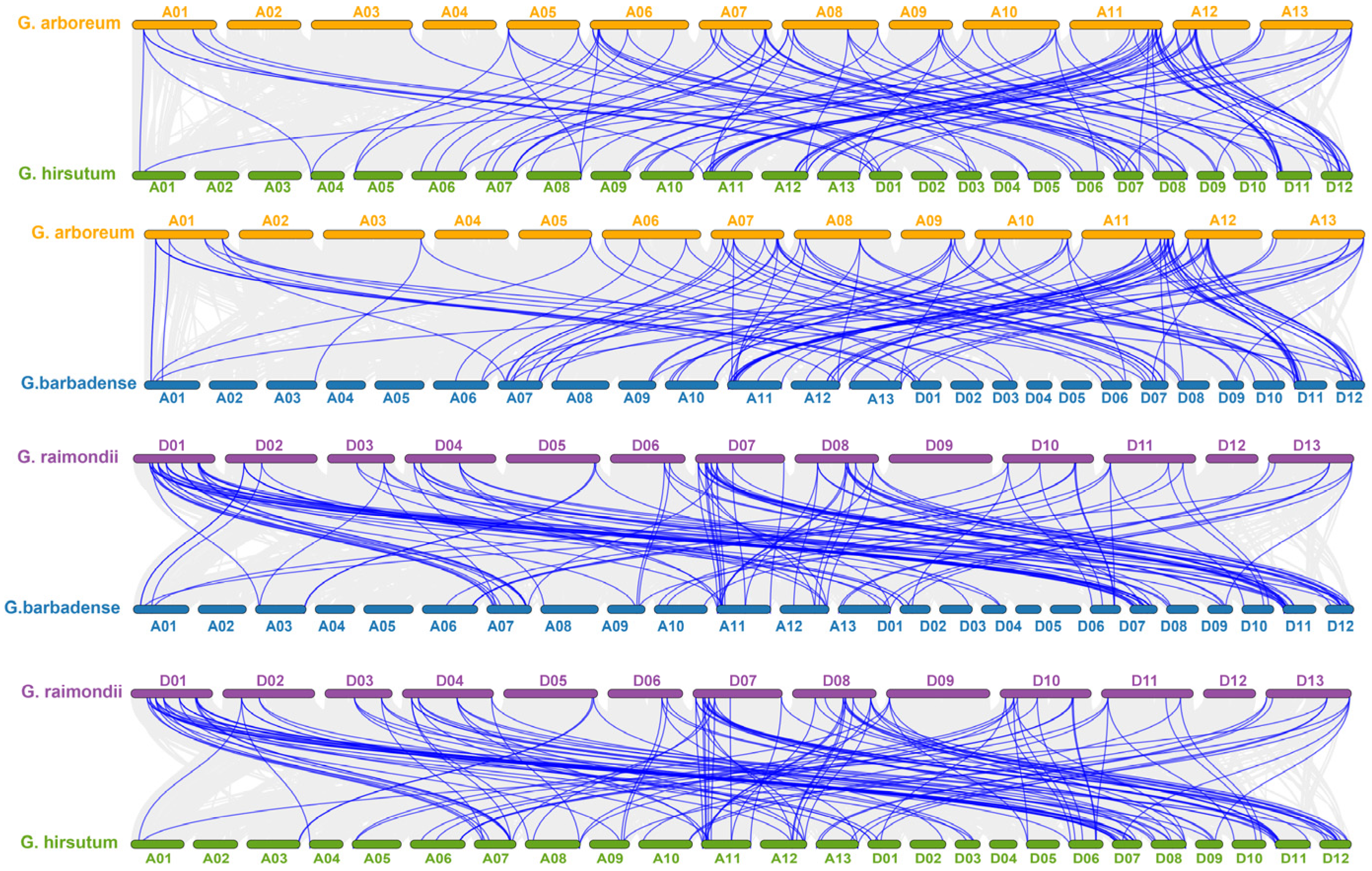
2.4. Collinearity Analysis of AGP Genes
2.5. Analysis of Orthologous Gene Clusters
2.6. cis Element Analysis of AGP Genes
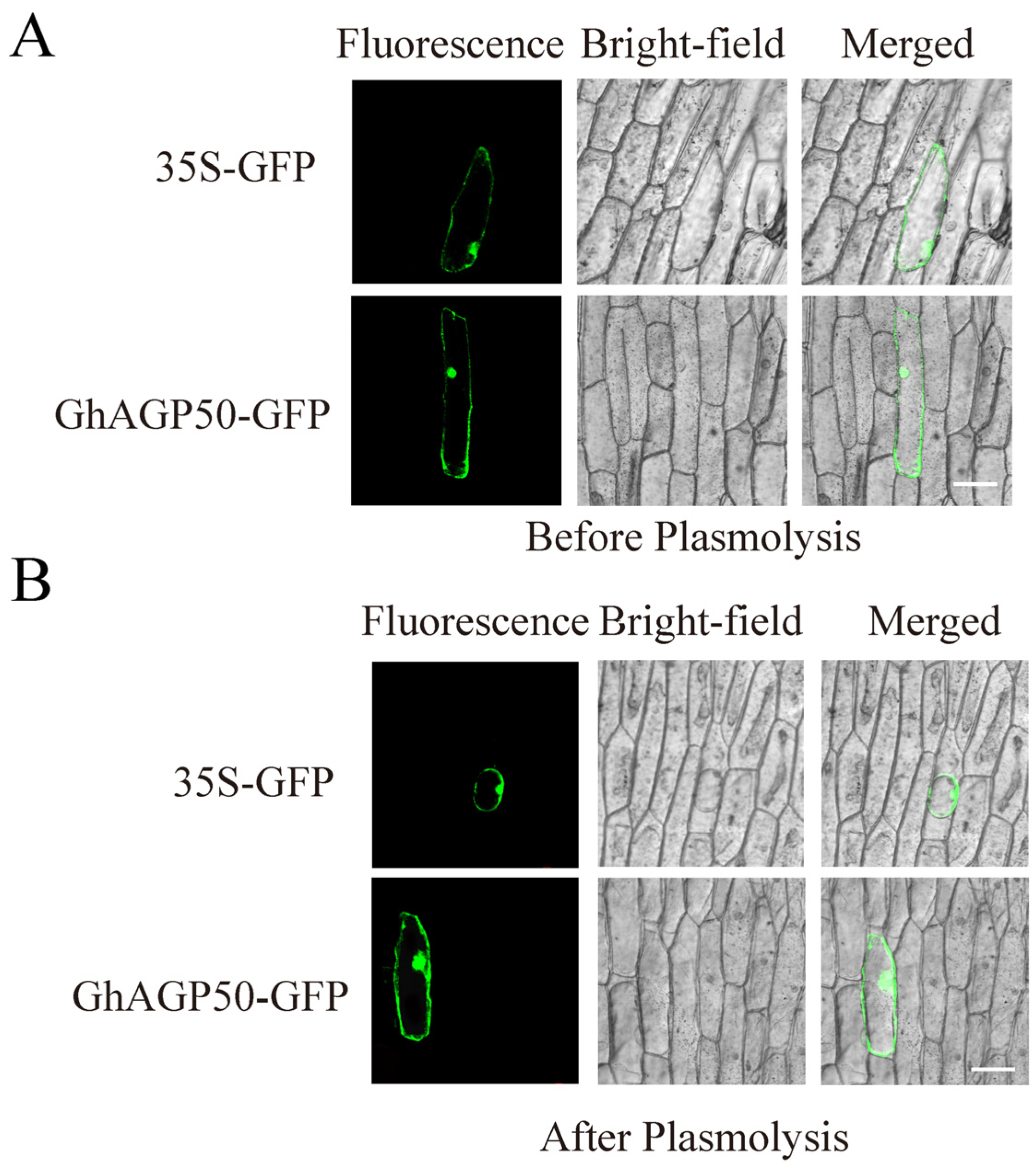
2.7. Subcellular Localization and Expression of Promoter::GUS Fusion of GhAGP50
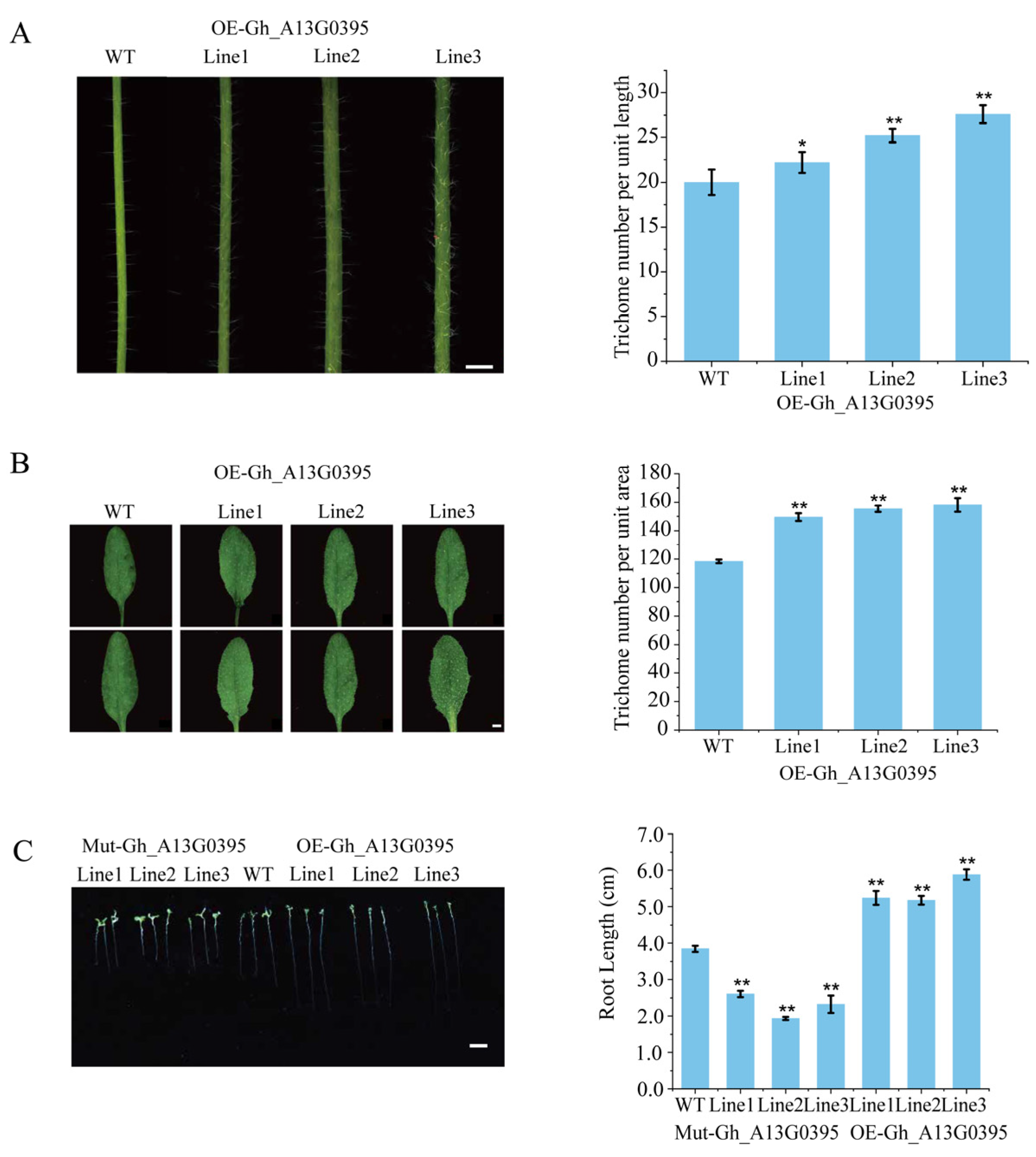
2.8. Overexpression of GhAGP50 Improved the Growth of Root Hair and Epidermal Hairs in Arabidopsis
3. Discussion
3.1. Evolution Analysis Revealed Purifying Selection
3.2. The Evolutionary Events of AGPs Influence Functional Diversification
3.3. Regulation of AGPs by Various Hormones During Plant Growth
3.4. AGPs Play Roles in the Growth and Development of Plants
3.5. AGPs Regulate Cotton Fiber Development
4. Materials and Methods
4.1. Identification and Characterization of AGP Genes
4.2. Phylogenetic Investigation and Gene Structure of AGP Genes
4.3. Chromosomal Locations and Gene Collinearity Analysis of AGP Genes
4.4. Identifying of cis Regulatory Element
4.5. Subcellular Localization
4.6. GhAGP50 Promoter Expression in Transgenic Arabidopsis Plants
4.7. Transformation in Arabidopsis
4.8. Exogenous Hormone Treatment
4.9. RNA Extraction and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sarwar, M.; Saleem, M.F.; Ali, B.; Saleem, M.H.; Rizwan, M.; Usman, K.; Keblawy, A.E.; Ali, A.; Afzal, M.; Sheteiwy, M.S. Application of Potassium, Zinc and Boron as Potential Plant Growth Modulators in Gossypium hirsutum L. under Heat Stress. Turk. J. Agric. For. 2022, 46, 567–584. [Google Scholar] [CrossRef]
- Qin, Y.-M.; Zhu, Y.-X. How Cotton Fibers Elongate: A Tale of Linear Cell-Growth Mode. Curr. Opin. Plant Biol. 2011, 14, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Haigler, C.H.; Betancur, L.; Stiff, M.R.; Tuttle, J.R. Cotton Fiber: A Powerful Single-Cell Model for Cell Wall and Cellulose Research. Front. Plant Sci. 2012, 3, 104. [Google Scholar] [CrossRef] [PubMed]
- Jareczek, J.J.; Grover, C.E.; Wendel, J.F. Cotton Fiber as a Model for Understanding Shifts in Cell Development under Domestication. Front. Plant Sci. 2023, 14, 1146802. [Google Scholar] [CrossRef]
- Cao, J.-F.; Zhao, B.; Huang, C.-C.; Chen, Z.-W.; Zhao, T.; Liu, H.-R.; Hu, G.-J.; Shangguan, X.-X.; Shan, C.-M.; Wang, L.-J.; et al. The miR319-Targeted GhTCP4 Promotes the Transition from Cell Elongation to Wall Thickening in Cotton Fiber. Mol. Plant 2020, 13, 1063–1077. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, J.-B.; Wei, N.; Liu, Z.-H.; Li, Y.; Zheng, Y.; Li, X.-B. A Type-2C Protein Phosphatase (GhDRP1) Participates in Cotton (Gossypium hirsutum) Response to Drought Stress. Plant Mol. Biol. 2021, 107, 499–517. [Google Scholar] [CrossRef]
- Yang, Y.; Lai, W.; Long, L.; Gao, W.; Xu, F.; Li, P.; Zhou, S.; Ding, Y.; Hu, H. Comparative Proteomic Analysis Identified Proteins and the Phenylpropanoid Biosynthesis Pathway Involved in the Response to ABA Treatment in Cotton Fiber Development. Sci. Rep. 2023, 13, 1488. [Google Scholar] [CrossRef]
- Xiao, Y.-H.; Li, D.-M.; Yin, M.-H.; Li, X.-B.; Zhang, M.; Wang, Y.-J.; Dong, J.; Zhao, J.; Luo, M.; Luo, X.-Y. Gibberellin 20-Oxidase Promotes Initiation and Elongation of Cotton Fibers by Regulating Gibberellin Synthesis. J. Plant Physiol. 2010, 167, 829–837. [Google Scholar] [CrossRef]
- Paunović, D.M.; Ćuković, K.B.; Bogdanović, M.D.; Todorović, S.I.; Trifunović-Momčilov, M.M.; Subotić, A.R.; Simonović, A.D.; Dragićević, M.B. The Arabinogalactan Protein Family of Centaurium Erythraea Rafn. Plants 2021, 10, 1870. [Google Scholar] [CrossRef]
- Ma, Y.; Yan, C.; Li, H.; Wu, W.; Liu, Y.; Wang, Y.; Chen, Q.; Ma, H. Bioinformatics Prediction and Evolution Analysis of Arabinogalactan Proteins in the Plant Kingdom. Front. Plant Sci. 2017, 8, 66. [Google Scholar] [CrossRef]
- Lin, S.; Miao, Y.; Huang, H.; Zhang, Y.; Huang, L.; Cao, J. Arabinogalactan Proteins: Focus on the Role in Cellulose Synthesis and Deposition during Plant Cell Wall Biogenesis. Int. J. Mol. Sci. 2022, 23, 6578. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Kim, Y.; Guo, Y.; Stevenson, B.; Zhu, J.-K. The Arabidopsis SOS5 Locus Encodes a Putative Cell Surface Adhesion Protein and Is Required for Normal Cell Expansion. Plant Cell 2003, 15, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Johnson, K. Arabinogalactan Proteins—Multifunctional Glycoproteins of the Plant Cell Wall. Cell Surf. 2023, 9, 100102. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, Y.; Carneros, E.; Berenguer, E.; Solís, M.-T.; Bárány, I.; Pintos, B.; Gómez-Garay, A.; Risueño, M.C.; Testillano, P.S. Pectin De-Methylesterification and AGP Increase Promote Cell Wall Remodeling and Are Required During Somatic Embryogenesis of Quercus Suber. Front. Plant Sci. 2019, 9, 1915. [Google Scholar] [CrossRef]
- Huang, G.-Q.; Xu, W.-L.; Gong, S.-Y.; Li, B.; Wang, X.-L.; Xu, D.; Li, X.-B. Characterization of 19 Novel Cotton FLA Genes and Their Expression Profiling in Fiber Development and in Response to Phytohormones and Salt Stress. Physiol. Plant. 2008, 134, 348–359. [Google Scholar] [CrossRef]
- Yao, K.; Yao, Y.; Ding, Z.; Pan, X.; Zheng, Y.; Huang, Y.; Zhang, Z.; Li, A.; Wang, C.; Li, C.; et al. Characterization of the FLA Gene Family in Tomato (Solanum lycopersicum L.) and the Expression Analysis of SlFLAs in Response to Hormone and Abiotic Stresses. Int. J. Mol. Sci. 2023, 24, 16063. [Google Scholar] [CrossRef]
- Wang, H.; Jin, Y.; Wang, C.; Li, B.; Jiang, C.; Sun, Z.; Zhang, Z.; Kong, F.; Zhang, H. Fasciclin-like Arabinogalactan Proteins, PtFLAs, Play Important Roles in GA-Mediated Tension Wood Formation in Populus. Sci. Rep. 2017, 7, 6182. [Google Scholar] [CrossRef]
- Gong, S.-Y.; Huang, G.-Q.; Sun, X.; Li, P.; Zhao, L.-L.; Zhang, D.-J.; Li, X.-B. GhAGP31, a Cotton Non-classical Arabinogalactan Protein, Is Involved in Response to Cold Stress during Early Seedling Development. Plant Biol. 2012, 14, 447–457. [Google Scholar] [CrossRef]
- Xue, H.; Seifert, G.J. FASCICLIN LIKE ARABINOGALACTAN PROTEIN 4 and RESPIRATORY BURST OXIDASE HOMOLOG D and F Independently Modulate Abscisic Acid Signaling. Plant Signal. Behav. 2015, 10, e989064. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome Sequence of Cultivated Upland Cotton (Gossypium hirsutum TM-1) Provides Insights into Genome Evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef]
- Li, W.-J.; Wu, N.; Chen, C.; Zhao, Y.-P.; Hou, Y.-X. Identification and Expression Analysis of Arabinogalactan Protein Genes in Cotton Reveal the Function of GhAGP15 in Verticillium Dahliae Resistance. Gene 2022, 822, 146336. [Google Scholar] [CrossRef] [PubMed]
- Borassi, C.; Gloazzo Dorosz, J.; Ricardi, M.M.; Carignani Sardoy, M.; Pol Fachin, L.; Marzol, E.; Mangano, S.; Rodríguez Garcia, D.R.; Martínez Pacheco, J.; Rondón Guerrero, Y.D.C.; et al. A Cell Surface Arabinogalactan-peptide Influences Root Hair Cell Fate. New Phytol. 2020, 227, 732–743. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Showalter, A.M. Immunolocalization of LeAGP-1, a Modular Arabinogalactan-Protein, Reveals Its Developmentally Regulated Expression in Tomato. Planta 2000, 210, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Showalter, A.M. Expression and Localization of AtAGP18, a Lysine-Rich Arabinogalactan-Protein in Arabidopsis. Planta 2007, 226, 169–179. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y.; Liang, Y.; Showalter, A.M. Expression Analyses of AtAGP17 and AtAGP19, Two Lysine-rich Arabinogalactan Proteins, in Arabidopsis. Plant Biol. 2011, 13, 431–438. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, J.; Showalter, A.M. AtAGP18 Is Localized at the Plasma Membrane and Functions in Plant Growth and Development. Planta 2011, 233, 675–683. [Google Scholar] [CrossRef]
- Kaur, D.; Held, M.A.; Smith, M.R.; Showalter, A.M. Functional Characterization of Hydroxyproline-O-Galactosyltransferases for Arabidopsis Arabinogalactan-Protein Synthesis. BMC Plant Biol. 2021, 21, 590. [Google Scholar] [CrossRef]
- Ito, S.; Suzuki, Y.; Miyamoto, K.; Ueda, J.; Yamaguchi, I. AtFLA11, a Fasciclin-Like Arabinogalactan-Protein, Specifically Localized in Screlenchyma Cells. Biosci. Biotechnol. Biochem. 2005, 69, 1963–1969. [Google Scholar] [CrossRef]
- MacMillan, C.P.; Mansfield, S.D.; Stachurski, Z.H.; Evans, R.; Southerton, S.G. Fasciclin-like Arabinogalactan Proteins: Specialization for Stem Biomechanics and Cell Wall Architecture in Arabidopsis and Eucalyptus: FLAs Specialized for Stem Biomechanics and Cell Walls. Plant J. 2010, 62, 689–703. [Google Scholar] [CrossRef]
- Huang, G.-Q.; Gong, S.-Y.; Xu, W.-L.; Li, W.; Li, P.; Zhang, C.-J.; Li, D.-D.; Zheng, Y.; Li, F.-G.; Li, X.-B. A Fasciclin-Like Arabinogalactan Protein, GhFLA1, Is Involved in Fiber Initiation and Elongation of Cotton. Plant Physiol. 2013, 161, 1278–1290. [Google Scholar] [CrossRef]
- Li, Y.; Si, Z.; Wang, G.; Shi, Z.; Chen, J.; Qi, G.; Jin, S.; Han, Z.; Gao, W.; Tian, Y. Genomic Insights into the Genetic Basis of Cotton Breeding in China. Mol. Plant 2023, 16, 662–677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Jamshed, M.; Shi, Y.; Liu, A.; Gong, J.; Wang, S.; Zhang, J.; Sun, F.; Jia, F.; et al. Genome-wide Quantitative Trait Loci Reveal the Genetic Basis of Cotton Fibre Quality and Yield-related Traits in a Gossypium hirsutum Recombinant Inbred Line Population. Plant Biotechnol. J. 2020, 18, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M. Sequencing of Allotetraploid Cotton (Gossypium hirsutum L. Acc. TM-1) Provides a Resource for Fiber Improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J. Gossypium Barbadense and Gossypium hirsutum Genomes Provide Insights into the Origin and Evolution of Allotetraploid Cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef]
- Huang, G.; Wu, Z.; Percy, R.G.; Bai, M.; Li, Y.; Frelichowski, J.E.; Hu, J.; Wang, K.; Yu, J.Z.; Zhu, Y. Genome Sequence of Gossypium herbaceum and Genome Updates of Gossypium arboreum and Gossypium hirsutum Provide Insights into Cotton A-Genome Evolution. Nat. Genet. 2020, 52, 516–524. [Google Scholar] [CrossRef]
- Cvijović, I.; Good, B.H.; Desai, M.M. The Effect of Strong Purifying Selection on Genetic Diversity. Genetics 2018, 209, 1235–1278. [Google Scholar] [CrossRef]
- Ke, S.; Jiang, Y.; Zhou, M.; Li, Y. Genome-Wide Identification, Evolution, and Expression Analysis of the WD40 Subfamily in Oryza Genus. Int. J. Mol. Sci. 2023, 24, 15776. [Google Scholar] [CrossRef]
- Jia, Q.; Song, J.; Zheng, C.; Fu, J.; Qin, B.; Zhang, Y.; Liu, Z.; Jia, K.; Liang, K.; Lin, W. Genome-Wide Analysis of Cation/Proton Antiporter Family in Soybean (Glycine max) and Functional Analysis of GmCHX20a on Salt Response. Int. J. Mol. Sci. 2023, 24, 16560. [Google Scholar] [CrossRef]
- Zhang, S.; Yan, C.; Lu, T.; Fan, Y.; Ren, Y.; Zhao, J.; Shan, X.; Guan, Y.; Song, P.; Li, D. New Insights into Molecular Features of the Genome-Wide AOX Family and Their Responses to Various Stresses in Common Wheat (Triticum aestivum L.). Gene 2023, 888, 147756. [Google Scholar] [CrossRef]
- Akram, J.; Siddique, R.; Shafiq, M.; Tabassum, B.; Manzoor, M.T.; Javed, M.A.; Anwar, S.; Nisa, B.U.; Saleem, M.H.; Javed, B. Genome-Wide Identification of CCO Gene Family in Cucumber (Cucumis sativus) and Its Comparative Analysis with A. thaliana. BMC Plant Biol. 2023, 23, 640. [Google Scholar] [CrossRef]
- Pacheco, D.D.R.; Santana, B.C.G.; Pirovani, C.P.; De Almeida, A.-A.F. Zinc/Iron-Regulated Transporter-like Protein Gene Family in Theobroma cacao L: Characteristics, Evolution, Function and 3D Structure Analysis. Front. Plant Sci. 2023, 14, 1098401. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Liu, Z.; Qi, Z.; Ma, Y.; Sun, M.; Su, L.; Niu, H.; Peng, Y.; Luo, X.; Zhu, M.; et al. Regulatory Controls of Duplicated Gene Expression during Fiber Development in Allotetraploid Cotton. Nat. Genet. 2023, 55, 1987–1997. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tu, L.; Lin, M.; Lin, Z.; Wang, P.; Yang, Q.; Ye, Z.; Shen, C.; Li, J.; Zhang, L.; et al. Asymmetric Subgenome Selection and Cis-Regulatory Divergence during Cotton Domestication. Nat. Genet. 2017, 49, 579–587. [Google Scholar] [CrossRef]
- He, J.; Zhao, H.; Cheng, Z.; Ke, Y.; Liu, J.; Ma, H. Evolution Analysis of the Fasciclin-Like Arabinogalactan Proteins in Plants Shows Variable Fasciclin-AGP Domain Constitutions. Int. J. Mol. Sci. 2019, 20, 1945. [Google Scholar] [CrossRef]
- Mao, Y.; Dai, F.; Si, Z.; Fang, L.; Zhang, T. Duplicate Mutations of GhCYP450 Lead to the Production of Ms5m6 Male Sterile Line in Cotton. Theor. Appl. Genet. 2023, 136, 2. [Google Scholar] [CrossRef]
- Lamport, D.T.A.; Tan, L.; Held, M.A.; Kieliszewski, M.J. Pollen Tube Growth and Guidance: Occam’s Razor Sharpened on a Molecular Arabinogalactan Glycoprotein Rosetta Stone. New Phytol. 2018, 217, 491–500. [Google Scholar] [CrossRef]
- Van Hengel, A.J.; Barber, C.; Roberts, K. The Expression Patterns of Arabinogalactan-protein AtAGP30 and GLABRA2 Reveal a Role for Abscisic Acid in the Early Stages of Root Epidermal Patterning. Plant J. 2004, 39, 70–83. [Google Scholar] [CrossRef]
- Suzuki, Y.; Kitagawa, M.; Knox, J.P.; Yamaguchi, I. A Role for Arabinogalactan Proteins in Gibberellin-induced A-amylase Production in Barley Aleurone Cells. Plant J. 2002, 29, 733–741. [Google Scholar] [CrossRef]
- Han, T.; Dong, H.; Cui, J.; Li, M.; Lin, S.; Cao, J.; Huang, L. Genomic, Molecular Evolution, and Expression Analysis of Genes Encoding Putative Classical AGPs, Lysine-Rich AGPs, and AG Peptides in Brassica rapa. Front. Plant Sci. 2017, 8, 397. [Google Scholar] [CrossRef]
- Shi, H.-D.; Zhang, W.-Q.; Lu, H.-Y.; Zhang, W.-Q.; Ye, H.; Liu, D.-D. Functional Characterization of a Starch Synthesis-Related Gene AmAGP in Amorphophallus muelleri. Plant Signal. Behav. 2020, 15, 1805903. [Google Scholar] [CrossRef]
- Shibata, M.; Sugimoto, K. A Gene Regulatory Network for Root Hair Development. J. Plant Res. 2019, 132, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Szarejko, I.; Melzer, M. Arabinogalactan Proteins Are Involved in Root Hair Development in Barley. J. Exp. Bot. 2015, 66, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Higashiyama, T. Arabinogalactan Proteins and Their Sugar Chains: Functions in Plant Reproduction, Research Methods, and Biosynthesis. Plant Reprod. 2018, 31, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Villa-Rivera, M.G.; Cano-Camacho, H.; López-Romero, E.; Zavala-Páramo, M.G. The Role of Arabinogalactan Type II Degradation in Plant-Microbe Interactions. Front. Microbiol. 2021, 12, 730543. [Google Scholar] [CrossRef]
- Yang, J.; Sardar, H.S.; McGovern, K.R.; Zhang, Y.; Showalter, A.M. A Lysine-rich Arabinogalactan Protein in Arabidopsis Is Essential for Plant Growth and Development, Including Cell Division and Expansion. Plant J. 2007, 49, 629–640. [Google Scholar] [CrossRef]
- Leszczuk, A.; Zając, A.; Kurzyna-Szklarek, M.; Cybulska, J.; Zdunek, A. Investigations of Changes in the Arabinogalactan Proteins (AGPs) Structure, Size and Composition during the Fruit Ripening Process. Sci. Rep. 2020, 10, 20621. [Google Scholar] [CrossRef]
- Demesa-Arévalo, E.; Vielle-Calzada, J.-P. The Classical Arabinogalactan Protein AGP18 Mediates Megaspore Selection in Arabidopsis. Plant Cell 2013, 25, 1274–1287. [Google Scholar] [CrossRef]
- Zhou, X.; Hu, W.; Li, B.; Yang, Y.; Zhang, Y.; Thow, K.; Fan, L.; Qu, Y. Proteomic Profiling of Cotton Fiber Developmental Transition from Cell Elongation to Secondary Wall Deposition. ABBS 2019, 51, 1168–1177. [Google Scholar] [CrossRef]
- Hossain, M.S.; Ahmed, B.; Ullah, M.W.; Aktar, N.; Haque, M.S.; Islam, M.S. Genome-Wide Identification of Fasciclin-like Arabinogalactan Proteins in Jute and Their Expression Pattern during Fiber Formation. Mol. Biol. Rep. 2020, 47, 7815–7829. [Google Scholar] [CrossRef]
- Qin, L.-X.; Chen, Y.; Zeng, W.; Li, Y.; Gao, L.; Li, D.-D.; Bacic, A.; Xu, W.-L.; Li, X.-B. The Cotton Beta-Galactosyltransferase 1 (GalT1) That Galactosylates Arabinogalactan Proteins Participates in Controlling Fiber Development. Plant J. 2017, 89, 957–971. [Google Scholar] [CrossRef]
- Yu, J.; Jung, S.; Cheng, C.-H.; Lee, T.; Zheng, P.; Buble, K.; Crabb, J.; Humann, J.; Hough, H.; Jones, D.; et al. CottonGen: The Community Database for Cotton Genomics, Genetics, and Breeding Research. Plants 2021, 10, 2805. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as Designed by Its Users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An Integrative Web Server to Predict Subcellular Localization of Proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, Scalable Generation of High-quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Bailey, T.L.; Gribskov, M. Concerning the Accuracy of MAST E-Values. Bioinformatics 2000, 16, 488–489. [Google Scholar] [CrossRef][Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Afzal Malik, W.; Afzal, M.; Chen, X.; Cui, R.; Lu, X.; Wang, S.; Wang, J.; Mahmood, I.; Ye, W. Systematic Analysis and Comparison of ABC Proteins Superfamily Confer Structural, Functional and Evolutionary Insights into Four Cotton Species. Ind. Crops Prod. 2022, 177, 114433. [Google Scholar] [CrossRef]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The Draft Genome of a Diploid Cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Tu, L.; Zhang, X.; Liu, D.; Jin, S.; Cao, J.; Zhu, L.; Deng, F.; Tan, J.; Zhang, C. Suitable Internal Control Genes for qRT-PCR Normalization in Cotton Fiber Development and Somatic Embryogenesis. Chin. Sci. Bull. 2007, 52, 3110–3117. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, R.; Guo, Z.; Yang, Z.; Zhao, Y.; Yan, H.; Azhar, M.T.; Zhang, Y.; Li, G.; Pan, J.; Liu, A.; et al. Genome-Wide Identification and Classification of Arabinogalactan Proteins Gene Family in Gossypium Species and GhAGP50 Increases Numbers of Epidermal Hairs in Arabidopsis. Int. J. Mol. Sci. 2025, 26, 4159. https://doi.org/10.3390/ijms26094159
Wei R, Guo Z, Yang Z, Zhao Y, Yan H, Azhar MT, Zhang Y, Li G, Pan J, Liu A, et al. Genome-Wide Identification and Classification of Arabinogalactan Proteins Gene Family in Gossypium Species and GhAGP50 Increases Numbers of Epidermal Hairs in Arabidopsis. International Journal of Molecular Sciences. 2025; 26(9):4159. https://doi.org/10.3390/ijms26094159
Chicago/Turabian StyleWei, Renhui, Ziru Guo, Zheng Yang, Yanpeng Zhao, Haoliang Yan, Muhammad Tehseen Azhar, Yamin Zhang, Gangling Li, Jingtao Pan, Aiying Liu, and et al. 2025. "Genome-Wide Identification and Classification of Arabinogalactan Proteins Gene Family in Gossypium Species and GhAGP50 Increases Numbers of Epidermal Hairs in Arabidopsis" International Journal of Molecular Sciences 26, no. 9: 4159. https://doi.org/10.3390/ijms26094159
APA StyleWei, R., Guo, Z., Yang, Z., Zhao, Y., Yan, H., Azhar, M. T., Zhang, Y., Li, G., Pan, J., Liu, A., Gong, W., Ge, Q., Gong, J., Yuan, Y., & Shang, H. (2025). Genome-Wide Identification and Classification of Arabinogalactan Proteins Gene Family in Gossypium Species and GhAGP50 Increases Numbers of Epidermal Hairs in Arabidopsis. International Journal of Molecular Sciences, 26(9), 4159. https://doi.org/10.3390/ijms26094159






