Genome-Wide Analysis of the MsRCI2 Gene Family in Medicago sativa and Functional Characterization of MsRCI2B in Salt Tolerance
Abstract
:1. Introduction
2. Result
2.1. MsRCI2 Gene Family Analysis
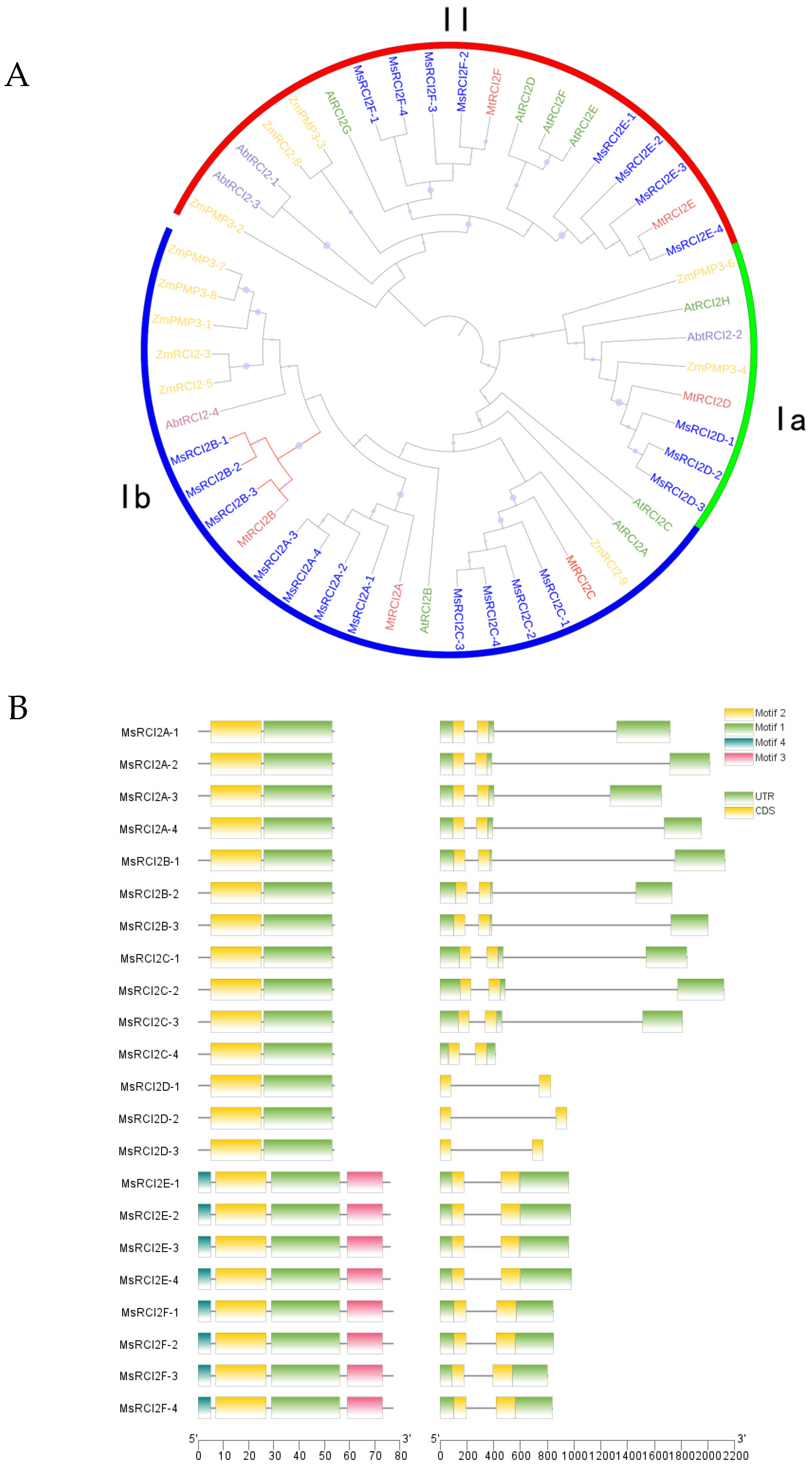
2.1.1. Sequence and Phylogenetic Analysis
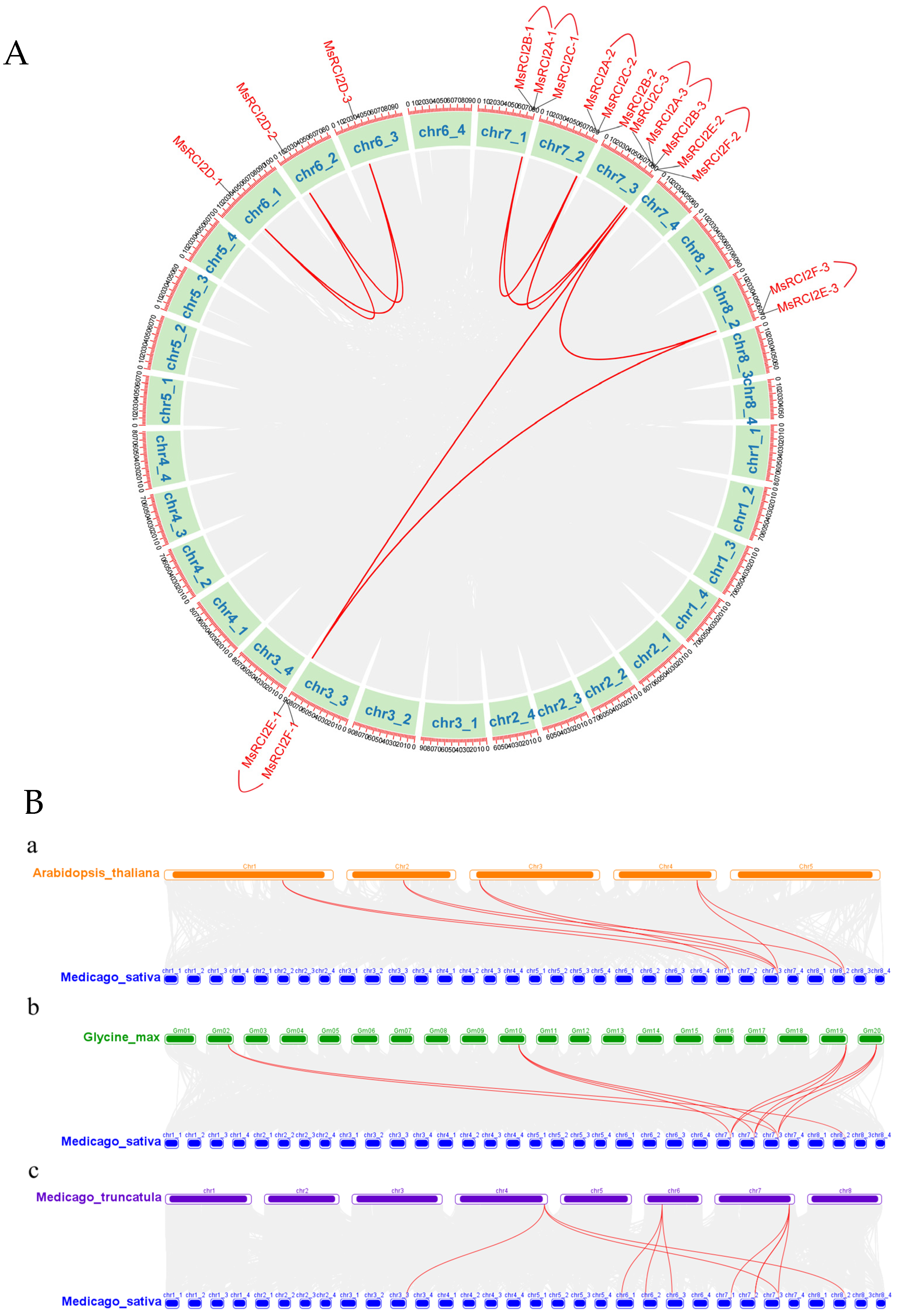
2.1.2. Chromosomal Localization and Collinearity Analysis
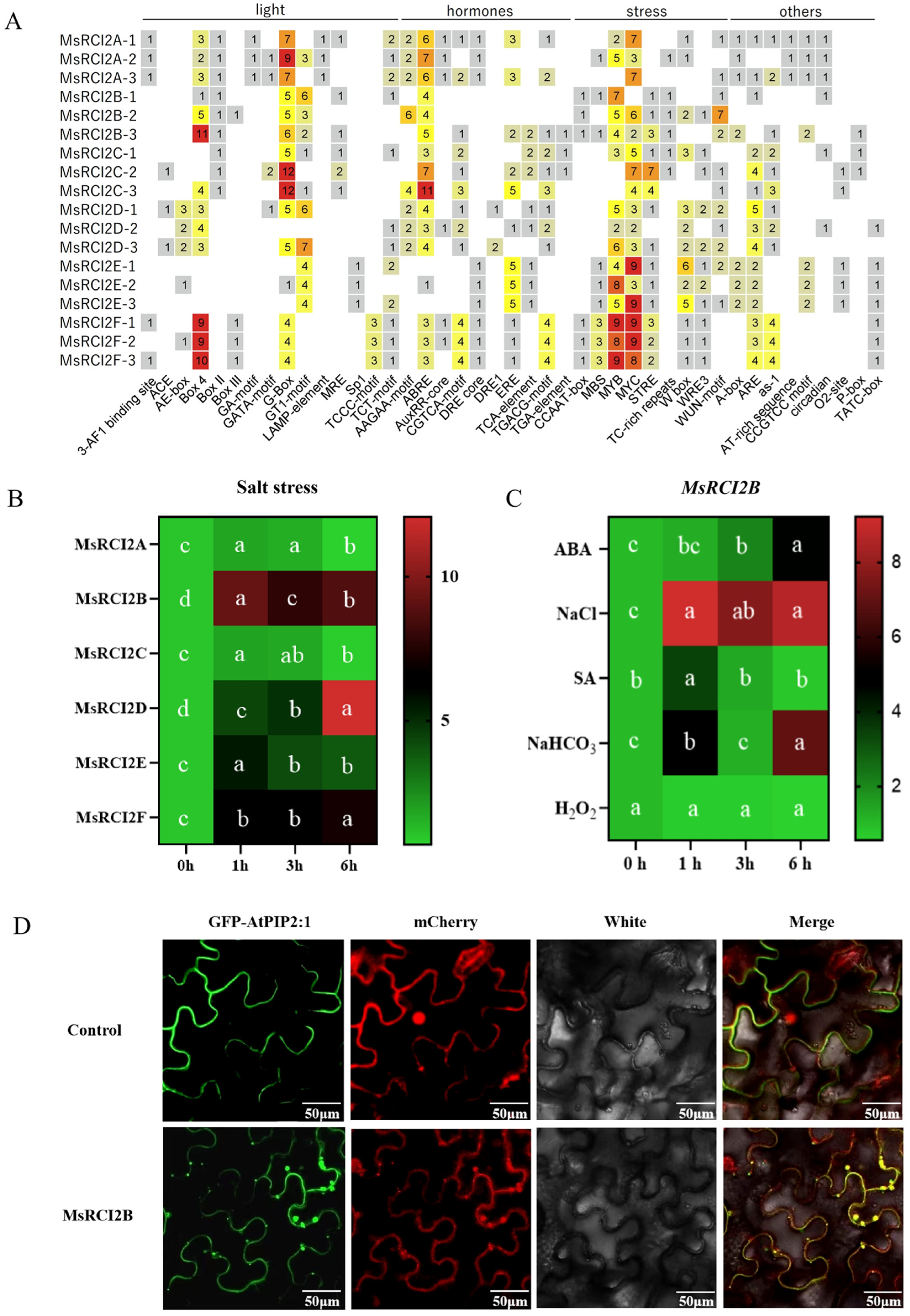
2.1.3. Role of MsRCI2B in Salt Tolerance
2.2. Overexpression of MsRCI2B Enhances Salt Tolerance and Antioxidant Capacity in Medicago sativa
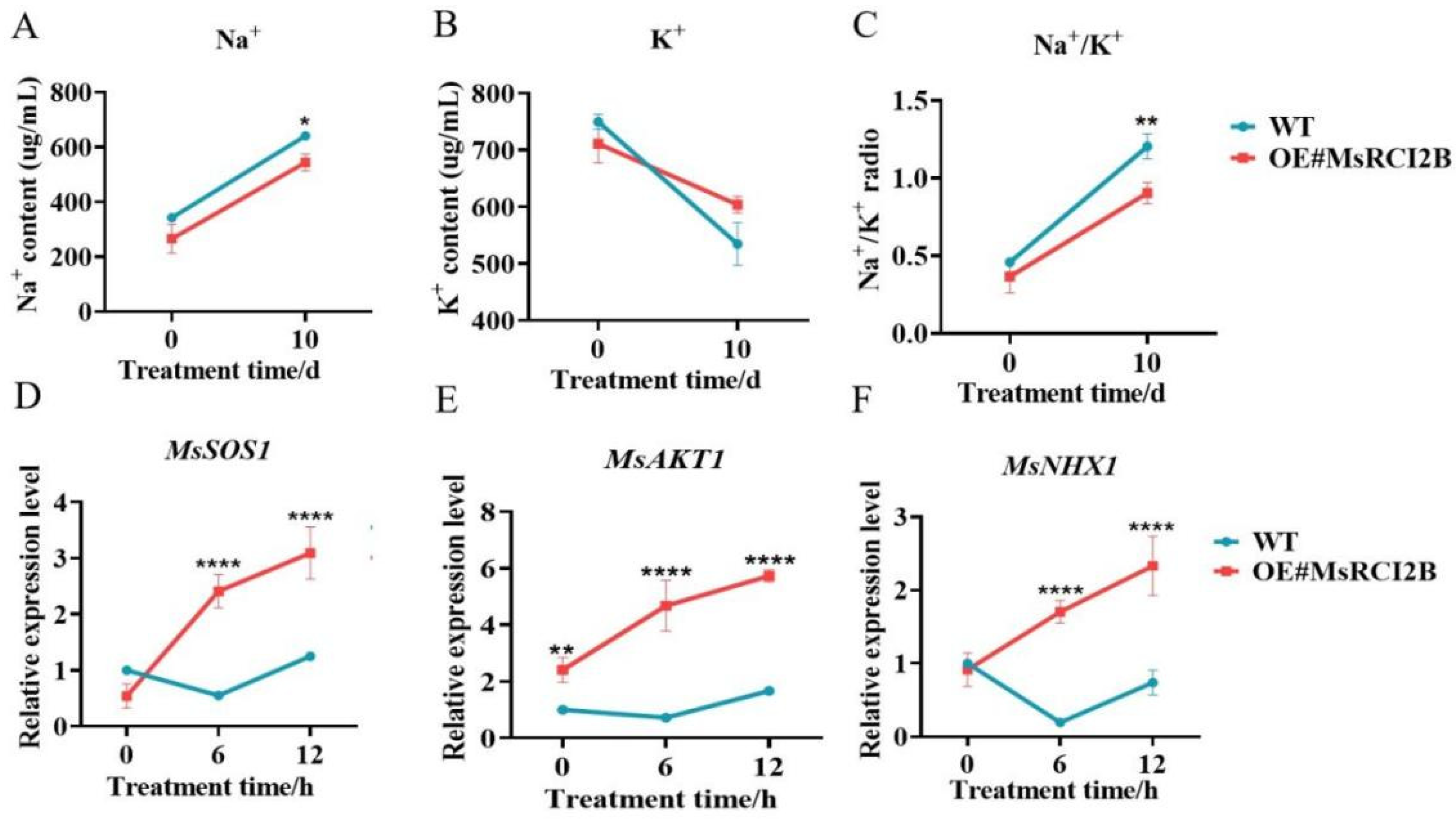
2.3. MsRCI2B Modulates Ion Homeostasis Under Salt Stress
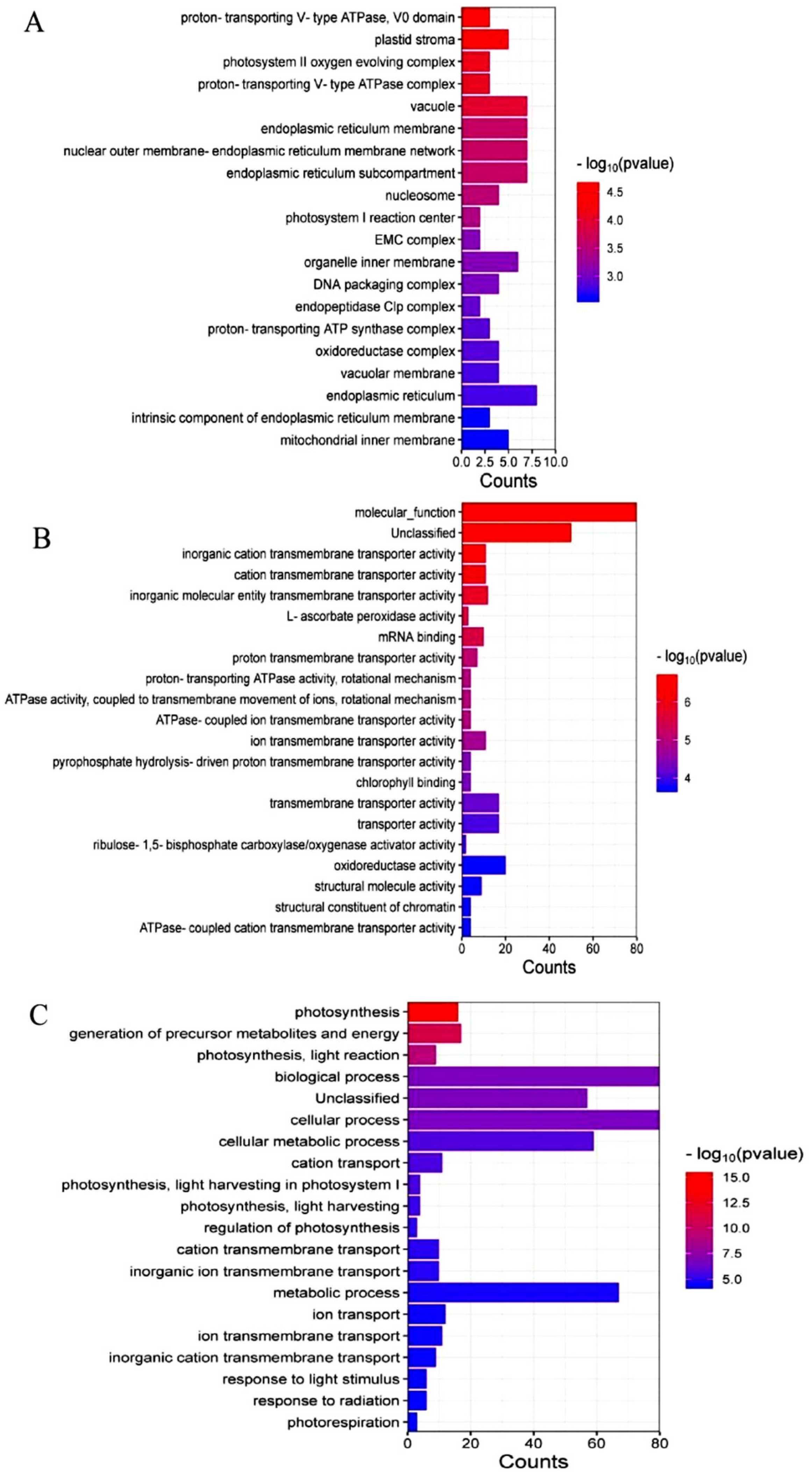
2.4. Screening and Functional Clustering (GO) Analysis of Candidate Interacting Proteins
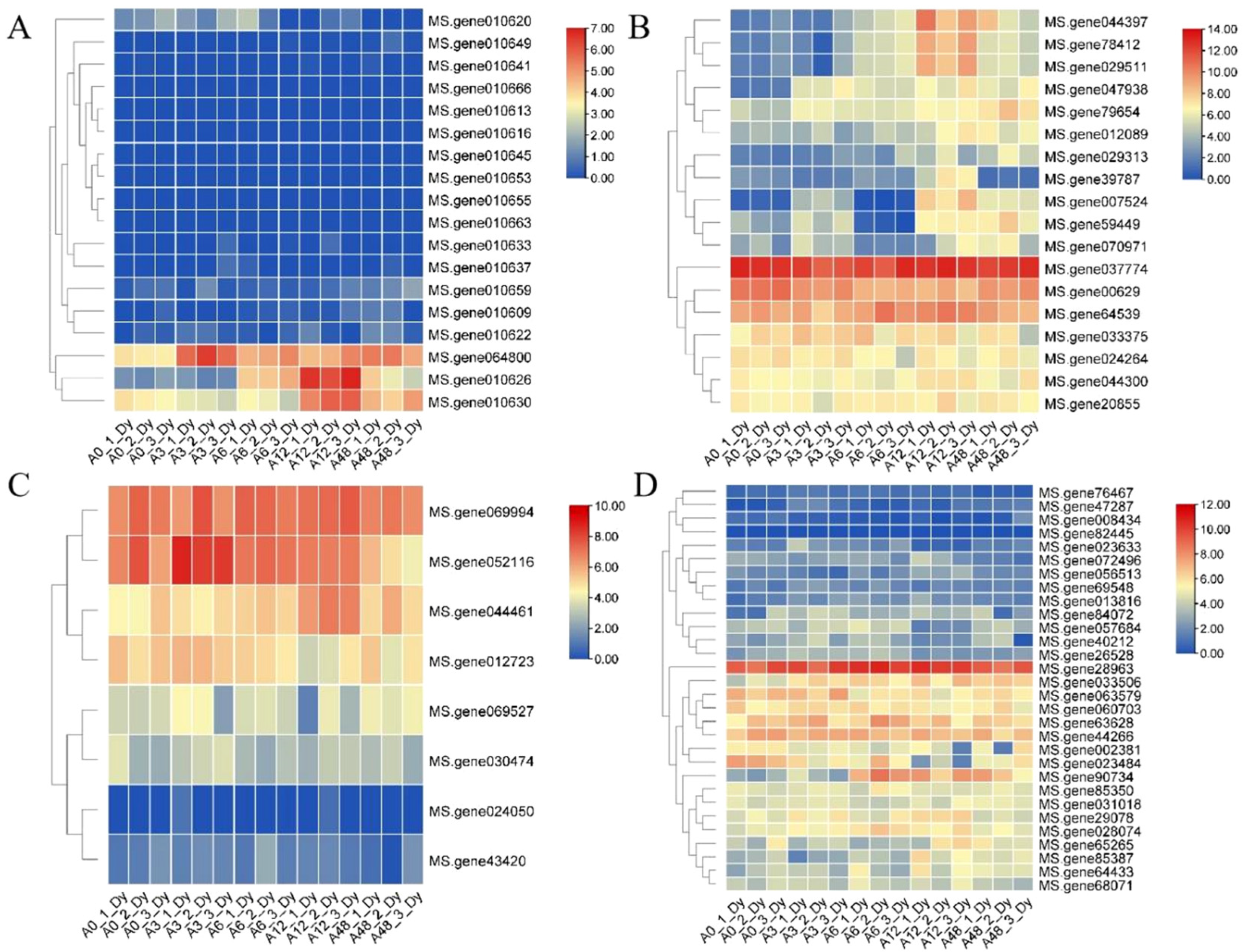
2.5. Expression Analysis of Candidate MsRCI2B-Interacting Protein Genes
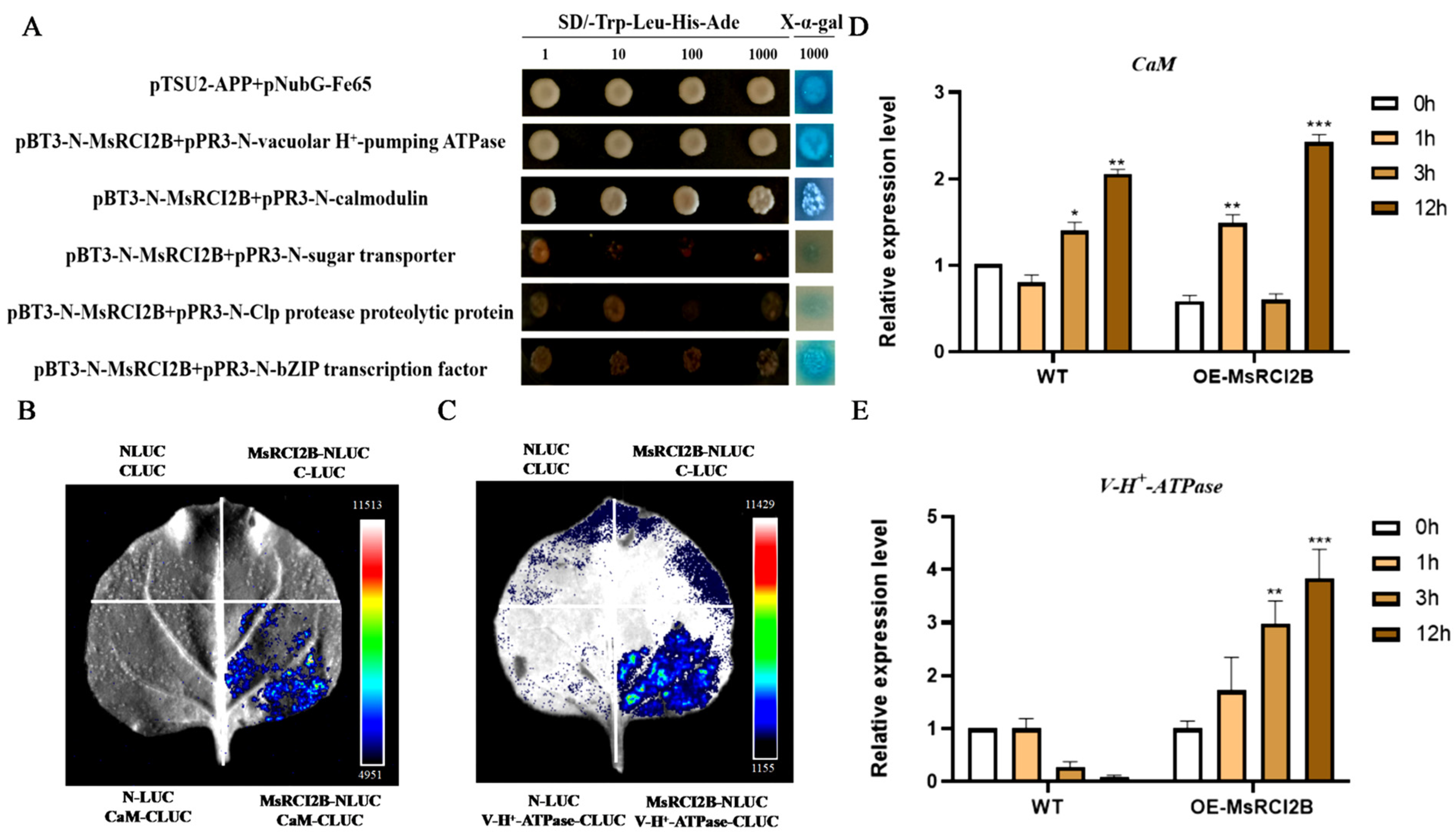
2.6. Further Verification of Candidate Interacting Proteins with MsRCI2B
3. Discussion
3.1. Identification and Evolutionary Characteristics of the MsRCI2 Gene Family
3.2. Functional Role of MsRCI2B in Salt Stress Tolerance
3.3. MsRCI2B Interacts with CaM and V-H+-ATPase to Modulate Salt Stress Response
3.4. Proposed Model of MsRCI2B Function in Salt Stress and Agricultural Implications
4. Conclusions
5. Materials and Methods
5.1. Identification of the RCI2 Family in Alfalfa
5.2. Phylogenetic Analysis, Conserved Motif, and Cis-Element Analysis of the MsRCI2 Gene Family
5.3. Construction of Alfalfa cDNA Library
5.4. Gene Cloning and Decoy Expression Vector Construction
5.5. Detection and Functional Verification of Decoy Protein Self-Activation Activity
5.6. Screening of Yeast Two-Hybrid Library with Membrane System
5.7. Positive Transformant Sequencing and Bioinformatics Analysis
5.8. Analysis of Expression of Candidate Interacting Protein Genes
5.9. Cloning of Full-Length Genes and In Vivo Interactions in Yeast
5.10. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR (RT-qPCR)
5.11. LCI (Luciferase Complementation Imaging)
5.12. Evaluation of Salt Tolerance
5.13. Determination of Na+ and K+ Content
5.14. Subcellular Localization Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.; Li, J.; Wang, P.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Ye, J.; Yang, Y.; Lin, H.; Yue, L.; Luo, J.; Long, Y.; Fu, H.; Liu, X.; Zhang, Y.; et al. The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 2019, 48, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.S.; Ren, H.L.; Wei, Z.W.; Wang, Y.W.; Ren, W.B. Effects of neutral salt and alkali on ion distributions in the roots, shoots, and leaves of two alfalfa (Medicago sativa) cultivars with differing degrees of salt tolerance. J. Integr. Agric. 2017, 16, 1800–1807. [Google Scholar] [CrossRef]
- Rahman, M.A.; Woo, J.H.; Lee, S.; Park, H.S.; Kabir, A.H.; Raza, A.; Sabagh, A.E.; Lee, K. Regulation of Na+/H+ exchangers, Na+/K+ transporters, and lignin biosynthesis genes, along with lignin accumulation, sodium extrusion, and antioxidant defense, confers salt tolerance in alfalfa (Medicago sativa). Front. Plant Sci. 2022, 13, 1041764. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Ge, G.; Sa, D.; Wang, Z.; Hou, M.; Jia, Y.S. Effects of salt stress levels on nutritional quality and microorganisms of alfalfa-influenced soil. PeerJ 2021, 9, e11729. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Li, G.; Cui, Y.; Xie, J.; Gao, X.; Teng, X.; Zhao, X.; Guan, F.; Liang, Z. Variation characteristics of root traits of different alfalfa (Medicago sativa) cultivars under saline-alkaline stress and their relationship with soil environmental factors. Phyton-Int. J. Exp. Bot. 2024, 93, 29–43. [Google Scholar]
- Jalil, S.U.; Ansari, M.I. Physiological role of Gamma-aminobutyric acid in salt stress tolerance. In Salt and Drought Stress Tolerance in Plants: Signaling Networks and Adaptive Mechanisms; Springer: Berlin/Heidelberg, Germany, 2020; pp. 337–350. [Google Scholar]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic stress and reactive oxygen species: Generation, signaling, and defense mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef]
- Tiwari, S.; Tiwari, S.; Singh, M.; Singh, A.; Prasad, S.M. Generation mechanisms of reactive oxygen species in the plant cell: An overview. In Reactive Oxygen Species in Plants: Boon Or Bane-Revisiting the Role of ROS; Wiley: Hoboken, NJ, USA, 2017; pp. 1–22. [Google Scholar]
- Ding, D. Adaptability of high quality alfalfa. Chin. Livest. Poult. Seed Ind. 2020, 16, 76–77. [Google Scholar]
- Bhattarai, S.; Biswas, D.; Fu, Y.B.; Biligetu, B. Morphological, physiological, and genetic responses to salt stress in alfalfa: A review. Agronomy 2020, 10, 577. [Google Scholar] [CrossRef]
- Cai, H.; Li, C.; Cong, C.; Zhan, L.; He, K.; Dong, L.; Xu, H. Effect of overexpression of MsRCI2A/B/C genes on drought tolerance of alfalfa (Medicago sativa L.). J. Northeast Agric. Univ.-Nat. Sci. Ed. 2022, 50, 57. [Google Scholar]
- Rocha, P.S.C.F. Plant abiotic stress-related RCI2/PMP3s: Multigenes for multiple roles. Planta 2016, 243, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Navarre, C.; Goffeau, A. Membrane hyperpolarization and salt sensitivity induced by deletion of PMP3, a highly conserved small protein of yeast plasma membrane. EMBO J. 2000, 19, 2515–2524. [Google Scholar] [CrossRef]
- Morsy, M.R.; Almutairi, A.M.; Gibbons, J.; Yun, S.J.; de los Reyes, B.G. The OsLti6 genes encoding low-molecular-weight membrane proteins are differentially expressed in rice cultivars with contrasting sensitivity to low temperature. Gene 2005, 344, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Imai, R.; Koike, M.; Sutoh, K.; Kawakami, A.; Torada, A.; Oono, K. Molecular characterization of a cold-induced plasma membrane protein gene from wheat. Mol. Genet. Genom. 2005, 274, 445–453. [Google Scholar] [CrossRef]
- Goddard, N.J.; Dunn, M.A.; Zhang, L.; White, A.J.; Jack, P.L.; Hughes, M.A. Molecular analysis and spatial expression pattern of a low-temperature-specific barley gene, blt101. Plant Mol. Biol. 1993, 23, 871–879. [Google Scholar] [CrossRef]
- Zhao, Y.; Tong, H.; Cai, R.; Peng, X.; Li, X.; Gan, D.; Zhu, S. Identification and characterization of the RCI2 gene family in maize (Zea mays). J. Genet. 2014, 93, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, J.E.; Jang, H.Y.; Kwak, K.J.; Ahn, S.J. CsRCI2A and CsRCI2E genes show opposite salt sensitivity reaction due to membrane potential control. Acta Physiol. Plant. 2016, 38, 50. [Google Scholar] [CrossRef]
- Li, C.; Song, T.; Zhan, L.; Cong, C.; Xu, H.; Dong, L.; Cai, H. Overexpression of MsRCI2A, MsRCI2B, and MsRCI2C in Alfalfa (Medicago sativa L.) Provides Different Extents of Enhanced Alkali and Salt Tolerance Due to Functional Specialization of MsRCI2s. Front. Plant Sci. 2021, 12, 702195. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Z.; Li, C.; Xing, Y.; Luo, Y.; Wang, X.; Li, D.; Ma, Z.; Cai, H. Overexpression of MsRCI2D and MsRCI2E enhances salt tolerance in alfalfa (Medicago sativa L.) by stabilizing antioxidant activity and regulating ion homeostasis. Int. J. Mol. Sci. 2022, 23, 9810. [Google Scholar] [CrossRef]
- Kim, Y.O.; Lim, H.G.; Kim, H.S.; Ahn, S.J. Overexpression of CsRCI2H enhances salt tolerance in Camelina sativa (L.). Plant Biotechnol. Rep. 2020, 14, 439–449. [Google Scholar] [CrossRef]
- Ben Romdhane, W.; Ben-Saad, R.; Meynard, D.; Verdeil, J.-L.; Azaza, J.; Zouari, N.; Fki, L.; Guiderdoni, E.; Al-Doss, A.; Hassairi, A. Ectopic expression of Aeluropus littoralis plasma membrane protein gene AlTMP1 confers abiotic stress tolerance in transgenic tobacco by improving water status and cation homeostasis. Int. J. Mol. Sci. 2017, 18, 692. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Feng, D.; Zhang, B.; Mu, P.; Zhang, Y.; He, Y.; Qi, K.; Wang, J.; Wang, H. Musa paradisica RCI complements AtRCI and confers Na+ tolerance and K+ sensitivity in Arabidopsis. Plant Sci. 2012, 184, 102–111. [Google Scholar] [CrossRef]
- Zeng, H.; Xu, L.; Singh, A.; Wang, H.; Du, L.; Poovaiah, B.W. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 2015, 6, 600. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ge, L.; Li, G.; He, P.; Yang, Y.; Liu, S. In silico identification and expression analysis of Rare Cold Inducible 2 (RCI2) gene family in cucumber. J. Plant Biochem. Biotechnol. 2020, 29, 56–66. [Google Scholar] [CrossRef]
- Capel, J.; Jarillo, J.A.; Salinas, J.; Martinez-Zapater, J.M. Two homologous low-temperature-inducible genes from Arabidopsis encode highly hydrophobic proteins. Plant Physiol. 1997, 115, 569–576. [Google Scholar] [CrossRef]
- Medina, J.; Ballesteros, M.L.; Salinas, J. Phylogenetic and functional analysis of Arabidopsis RCI2 genes. J. Exp. Bot. 2007, 58, 4333–4346. [Google Scholar] [CrossRef]
- Brunetti, S.C.; Arseneault, M.K.M.; Gulick, P.J. Characterization of the Esi3/RCI2/PMP3 gene family in the Triticeae. BMC Genom. 2018, 19, 1–16. [Google Scholar] [CrossRef]
- Zhang, D.; Shen, Z.; He, P.; Wang, J.; Li, D.; Meng, J.; Zhang, D.; You, J.; Luo, Y.; Wang, X.; et al. The synergistic roles of MsRCI2B and MsRCI2E in the regulation of ion balance and ROS homeostasis in alfalfa under salt stress. Int. J. Biol. Macromol. 2025, 300, 140093. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Melino, V.J.; Sweetman, C.; Soole, K.L. Manipulation of alternative oxidase can influence salt tolerance in Arabidopsis thaliana. Physiol. Plant. 2009, 137, 459–472. [Google Scholar] [CrossRef]
- Oh, D.-H.; Leidi, E.; Zhang, Q.; Hwang, S.-M.; Li, Y.; Quintero, F.J.; Jiang, X.; D’Urzo, M.P.; Lee, S.Y.; Zhao, Y.; et al. Loss of halophytism by interference with SOS1 expression. Plant Physiol. 2009, 151, 210–222. [Google Scholar] [CrossRef]
- Ardie, S.W.; Liu, S.; Takano, T. Expression of the AKT1-type K+ channel gene from Puccinellia tenuiflora, PutAKT1, enhances salt tolerance in Arabidopsis. Plant Cell Rep. 2010, 29, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cai, S.; Wang, G.; Wang, F.; Dong, F.; Mak, M.; Holford, P.; Ji, J.; Salih, A.; Zhou, M.; et al. Halophytic NHXs confer salt tolerance by altering cytosolic and vacuolar K+ and Na+ in Arabidopsis root cell. Plant Growth Regul. 2017, 82, 333–351. [Google Scholar] [CrossRef]
- Chin, D.; Means, A.R. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000, 10, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, R.; Singh, A.; Chattopadhyay, S. Calmodulin 7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell. 2008, 20, 1747–1759. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Li, J.; Zhang, J.; Shen, S.; Li, C.; Gao, Y.; Zhang, S. AtCaM4 interacts with a sec14-like protein, PATL1, to regulate freezing tolerance in Arabidopsis in a CBF-independent manner. J. Exp. Bot. 2018, 69, 5241–5253. [Google Scholar] [CrossRef]
- Rao, S.S.; El-Habbak, M.H.; Havens, W.M.; Singh, A.; Zheng, D.; Vaughn, L.; Haudenshield, J.S.; Hartman, G.L.; Korban, S.S.; Ghabrial, S.A. Overexpression of GmCaM4 in soybean enhances resistance to pathogens and tolerance to salt stress. Mol. Plant Pathol. 2014, 15, 145–160. [Google Scholar] [CrossRef]
- Yu, S.; Wu, J.; Sun, Y.; Zhu, H.; Sun, Q.; Zhao, P.; Huang, R.; Guo, Z. A calmodulin-like protein (CML10) interacts with cytosolic enzymes GSTU8 and FBA6 to regulate cold tolerance. Plant Physiol. 2022, 190, 1321–1333. [Google Scholar] [CrossRef]
- Poovaiah, B.W.; Du, L.; Wang, H.; Yang, T. Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol. 2013, 163, 531–542. [Google Scholar] [CrossRef]
- Chen, J.; Xu, X.; Hu, Z.; Yang, S. Recent Advances on Salt Stress Sensitivity and Related Calcium Signals in Plants. Plant Res. 2022, 42, 713–720. [Google Scholar]
- Sanders, D.; Pelloux, J.; Brownlee, C.; Harper, J.F. Calcium at the crossroads of signaling. Plant Cell 2002, 14 (Suppl. S1), S401–S417. [Google Scholar] [CrossRef]
- Li, Y.; Zeng, H.; Xu, F.; Yan, F.; Xu, W. H+-ATPases in plant growth and stress responses. Annu. Rev. Plant Biol. 2022, 73, 495–521. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Krishnakumar, V.; Bidwell, S.; Rosen, B.; Chan, A.; Zhou, S.; Gentzbittel, L.; Childs, K.L.; Yandell, M.; Gundlach, H.; et al. An improved genome release (version Mt4. 0) for the model legume Medicago truncatula. BMC Genom. 2014, 15, 312. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]


| Name | Number of Amino Acids | Theoretical pI | Molecular Weight | Instability Index | Stability | Aliphatic Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| MsRCI2As | 54 | 5.82 | 5931.38 | 35.83 | Y | 164.26 | 1.602 | vacu |
| MsRCI2Bs | 54 | 6.49 | 5946.35 | 40.45 | N | 151.67 | 1.526 | vacu, plas |
| MsRCI2Cs | 54 | 4.68 | 6020.44 | 23.67 | Y | 162.41 | 1.607 | vacu |
| MsRCI2D-1 | 54 | 4.00 | 5921.19 | 31.66 | Y | 153.15 | 1.478 | vacu |
| MsRCI2D-2/3 | 54 | 4.00 | 5903.16 | 31.66 | Y | 160.37 | 1.513 | vacu |
| MsRCI2Es | 76 | 5.59 | 8697.50 | 59.49 | N | 138.55 | 0.851 | plas |
| MsRCI2Fs | 77 | 5.05 | 8911.71 | 70.12 | N | 127.92 | 0.758 | plas, vacu |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiao, H.; Zhang, D.; Zhang, Z.; Meng, J.; He, P.; Zhang, S.; Wang, Y.; Cai, H.; Li, Y. Genome-Wide Analysis of the MsRCI2 Gene Family in Medicago sativa and Functional Characterization of MsRCI2B in Salt Tolerance. Int. J. Mol. Sci. 2025, 26, 4165. https://doi.org/10.3390/ijms26094165
Qiao H, Zhang D, Zhang Z, Meng J, He P, Zhang S, Wang Y, Cai H, Li Y. Genome-Wide Analysis of the MsRCI2 Gene Family in Medicago sativa and Functional Characterization of MsRCI2B in Salt Tolerance. International Journal of Molecular Sciences. 2025; 26(9):4165. https://doi.org/10.3390/ijms26094165
Chicago/Turabian StyleQiao, Huiru, Depeng Zhang, Zizhao Zhang, Jing Meng, Pin He, Shichao Zhang, Yan Wang, Hua Cai, and Yong Li. 2025. "Genome-Wide Analysis of the MsRCI2 Gene Family in Medicago sativa and Functional Characterization of MsRCI2B in Salt Tolerance" International Journal of Molecular Sciences 26, no. 9: 4165. https://doi.org/10.3390/ijms26094165
APA StyleQiao, H., Zhang, D., Zhang, Z., Meng, J., He, P., Zhang, S., Wang, Y., Cai, H., & Li, Y. (2025). Genome-Wide Analysis of the MsRCI2 Gene Family in Medicago sativa and Functional Characterization of MsRCI2B in Salt Tolerance. International Journal of Molecular Sciences, 26(9), 4165. https://doi.org/10.3390/ijms26094165





