Abstract
The Aesculapian snake (Zamenis longissimus) is distributed in Central and Southern Europe, the Balkans, Anatolia, and Iran, but had a wider mid-Holocene distribution into Northern Europe. To investigate the genetic affinity of a Danish population that went extinct in historical times, we analysed three ethanol-preserved individuals dating back to 1810 using a silica-in-solution ancient DNA extraction method, combined with next-generation sequencing. Bioinformatic mapping of the reads against the published genome of a related colubrid snake revealed that two of the three specimens contained endogenous snake DNA (up to 8.6% of the reads), and this was evident for tooth, bone, and soft tissue samples. The DNA was highly degraded, observed by very short average sequence lengths (<50 bp) and 11–15% C to T deamination damage at the first 5′ position. This is an effect of specimen age, combined with suboptimal, and possibly damaging, molecular preservation conditions. Phylogeographic analyses of a 1638 bp mtDNA sequence securely placed the two Danish Aesculapian snakes in the Eastern (Balkan glacial refugium) clade within this species, and revealed one previously-undescribed haplotype. These results provide new information on the past distribution and postglacial re-colonization patterns of this species.
1. Introduction
Collections of biological specimens represent an essential resource for studying biodiversity on our planet. Over the past decade it has become increasingly clear that such museum collections not only offer a wealth of information on morphological biological diversity, but also serve as a record of past and present genetic diversity [1,2]. DNA obtained from historical collections can offer direct insight into the gene pools of extinct species or populations, thus providing unique phylogenetic and phylogeographic information, not possible to obtain from living individuals. The extinct Danish population of the Aesculapian snake (Zamenis longissimus) represents such a case.
The Aesculapian snake is a non-venomous colubrid snake distributed in Central and Southern Europe including the Balkans, Anatolia, and Iran, with introduced populations in Wales and London (Figure 1). The species’ northern distribution limit coincides with the 48th to 50th parallel north, but c. 10 isolated populations in Germany, Czech Republic, and Poland exist outside the main distribution, restricted to warm and humid microhabitats in river valleys [3,4,5]. A further five northern populations have gone extinct during the last 150 years [6], including the population in Denmark. These isolated populations, whether extinct or extant, are possible remnants of an earlier, more widespread distribution, evidenced by numerous fossil findings of this species [7,8] (Figure 1). In the mid-Holocene, the mean temperature in the northern hemisphere was higher than the present day which is why some thermophilous European species, including Z. longissimus, had a distribution extending further north than their current distribution [9,10]. Aesculapian snake fossils from Denmark represent the species’ Mid-Holocene northernmost distribution border [9,11,12]. The presence of this species during historical times in Denmark is evidenced by a number of direct observations and museum specimens collected throughout the 17th to 19th centuries [13,14,15]. All reliable observations of living Aesculapian snakes in Denmark derive from the southern tip of the island Zealand (Sjælland) [16,17] (Figure 1). This area was earlier characterized by coppice of hazel [18], but the management practice ceased in late 1800s and the habitat changed, likely becoming unfavourable for the snakes. Three ethanol-preserved specimens exist from the historical Danish population from Southern Zealand, collected in 1810, 1851, and 1863, respectively, but no previously-published study on Z. longissimus morphology or phylogenetics has included these specimens.
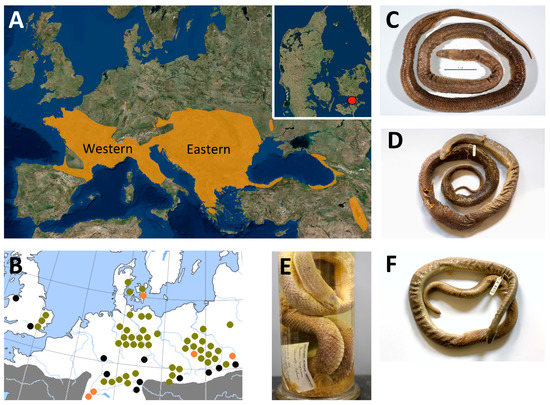
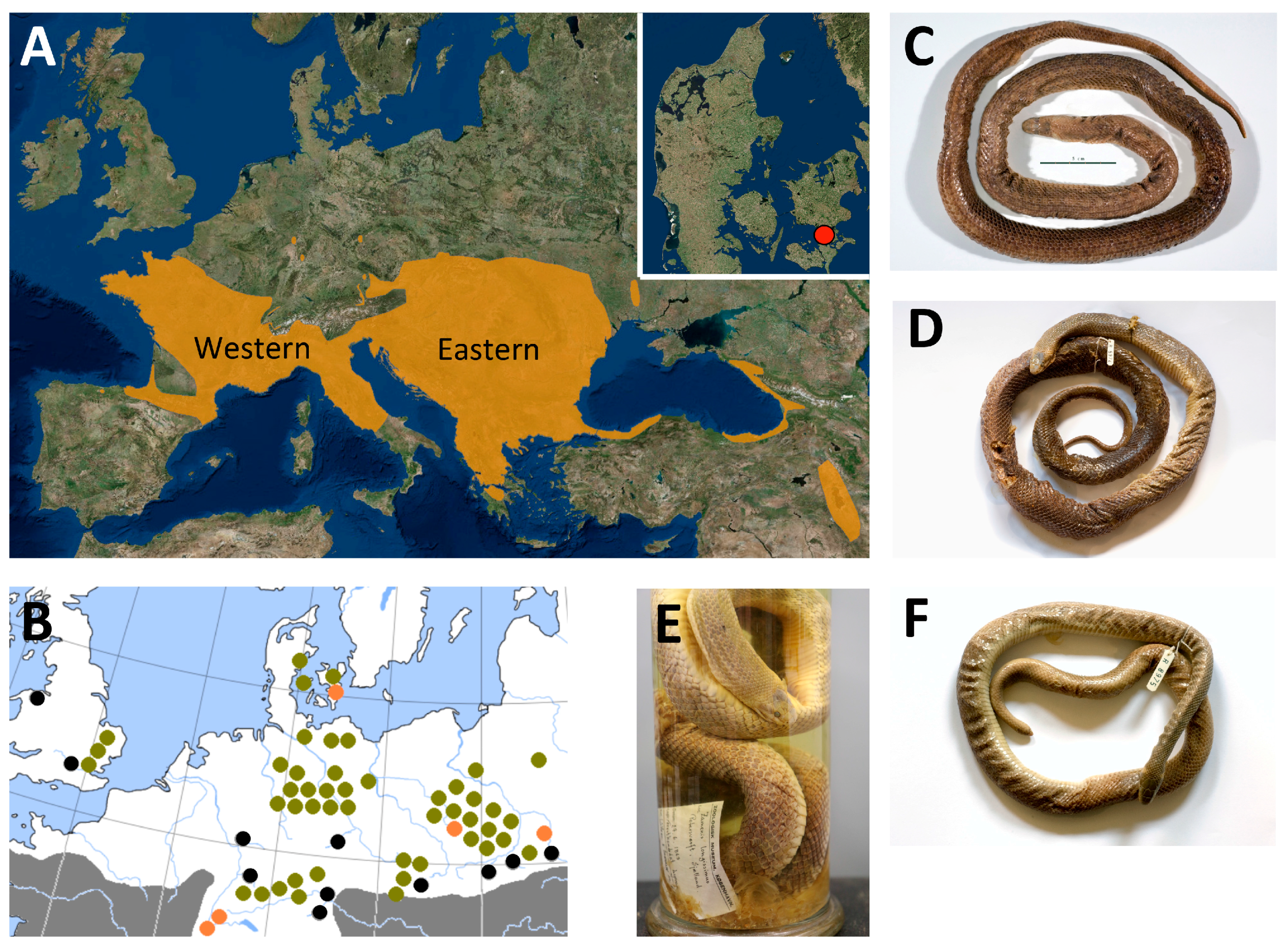
Figure 1.
A: the main distribution of Z. longissimus (downloaded and modified from iucnredlist.org), highlighting the Eastern and Western genetic clades identified in Musilová et al. [19] and the extinct Danish population from Southern Zealand (map insert). B: the current relictual (continent) or introduced (UK) populations marked with black dots. Populations that went extinct in historical times are marked with orange dots and fossil finds are marked with green dots. Modified from Gomille [6], Richter and Noe-Nygaard [11], and Kristensen [12]. Three historical Danish specimens were sampled for this study: C: HK2489, D+E: ZMUC R8974, F: ZMUC R8975. Photos: N. Ioannou and M.E. Allentoft.
The objective in the current study is two-fold. First and foremost, we aim to investigate the genetic relationship between the extinct Danish population and the genetically well-described extant Z. longissimus populations. Musilová et al. [19] demonstrated a clear population genetic structuring with four main mtDNA clades in this species. Furthermore they showed that the current small and isolated populations north of the core distribution belong to the Eastern clade, suggesting that it was snakes from a Southern Balkan glacial refugium that colonized Northern Europe during the mid-Holocene climatic optimum. We aim to investigate if the genetic profiles of the Danish Aesculapian snakes are consistent with this scenario.
A second objective is to explore the molecular potential in these old ethanol-preserved specimens. By using highly-optimized ancient DNA (aDNA) extraction protocols combined with next-generation sequencing technology it is possible to obtain data from severely degraded DNA that will fail with standard extraction methods and PCR-based technology. We test the Aesculapian snake material using a silica-in-solution extraction method that has previously been used in large-scale genomic studies on ancient human skeletal material [20]. In this context we compare DNA preservation in different types of snake tissue (tooth, bone, and soft tissue). We believe that the protocols and results presented here have high relevance to scientists that engage in museum collection-based molecular research.
2. Materials and Methods
2.1. Samples
Three historical specimens were sampled for this study (Figure 1), representing all known museum specimens of Z. longissimus from Denmark. HK2489 is a 102 cm long female collected in 1810 (noted locality: Petersgaard Skovdistrikt, Sydsjælland); ZMUC R8974 is a 128 cm long female, collected in 1863 (noted locality: Petersværft Skovridergård, Sydsjælland), and ZMUC R8975 is an 81 cm long female collected in 1851 (Noted locality: Petersgaard Skovdistrikt, Sydsjælland), received alive at the Natural History Museum of Denmark and killed and stored in ethanol. The exact preservation history of the other two specimens is unknown but today they are stored in ethanol (Figure 1) and have presumably been so since the time of collection. Small pieces of muscle and skin tissue (<0.2 g) were removed from all three, and we also obtained 3–4 teeth from each of two individuals, and a complete vertebra from one individual (Table 1).

Table 1.
Sequencing and molecular preservation of three Z. longissimus museum specimens.
2.2. DNA Extraction
A total of six DNA extracts were prepared for this study: three soft tissue samples (muscle/skin), two tooth samples, and one bone sample. The DNA was extracted using a silica-in-solution method optimized for retaining short and degraded DNA molecules [20]. The samples were incubated for 24 h at 45 °C in 5 mL digestion buffer containing 4.7 mL 0.5 M EDTA buffer, 50 μL of Proteinase K (0.14–0.22 mg/mL, Roche, Basel, Switzerland), 250 μL 10% N-laurylsarcosyl, and 50 μL TE buffer (100×). The solution was spun down and the supernatant transferred to a 50 mL tube, where it was mixed with 100 μL of silica suspension (see [20]) and 40 mL of binding buffer. The binding buffer was prepared in bulk by mixing 500 mL buffer PB (Qiagen, Hilden, Germany) with 9 mL of sodium acetate (5 M), and 2.5 mL of sodium chloride (5 M), pH adjusted to 4–5 with 37% HCL. After 1 h of incubation (room temperature) with binding buffer and silica, the supernatant was removed and the pelleted silica was re-suspended in 1 mL of new binding buffer, spun down, and washed twice with 1 mL 80% cold ethanol. Finally, the DNA was eluted in 80 μL of EB buffer (Qiagen). Extraction blanks were included with each round of extractions. The work was conducted using strict aDNA guidelines (e.g., [21]) in a dedicated clean lab at the Centre for GeoGenetics at the Natural History Museum of Denmark.
2.3. Library Preparation and Sequencing
DNA extract was built into a blunt-end library using the NEBNext DNA Sample Prep kit E6070 (New England Biolabs, Ipswich, MA, USA) and Illumina-specific adapters (Illumina, San Diego, CA, USA). The libraries were prepared as described previously [22] with a few modifications: the end-repair step was performed with 20 μL of DNA extract, 2.5 μL repair buffer, and 1.25 μL repair enzyme mix. The solution was incubated for 20 min at 12 °C, followed by 15 min at 37 °C, and purified using the same binding buffer as used for extractions (see above) with Qiagen MinElute columns, and eluted in 15 μL of EB buffer. Next, Illumina-specific adapters (prepared as in [23]) were ligated to the end-repaired DNA in 25 μL reactions (15 μL of DNA, 5 μL of ligation buffer, 0.5 μL of adapter mix, 2.5 uL of Quick T4 ligase, 2 μL of H2O). This solution was incubated for 15 min at 20 °C and purified with PB buffer on Qiagen MinElute columns, before eluted in 20 μL EB buffer. The adapter fill-in reaction was performed in a 24 μL volume (20 μL of DNA, 2.5 uL of fill-in buffer, and 1.5 μL of Bst polymerase) and incubated for 20 min at 37 °C followed by 20 min at 80 °C to inactivate the Bst enzyme. To determine the number of PCR cycles required to reach sufficient DNA concentration for sequencing, 1 μL of library was amplified with qPCR and SYBR Green detection chemistry and the CT values recorded. The entire remaining DNA library (ca. 24 μL) was then amplified and indexed in 50 μL PCR reactions containing 1X KAPA HiFi HotStart Uracil + ReadyMix (KAPA Biosystems, Woburn, MA, USA) and 200 nM of each of Illumina’s Multiplexing PCR primer in PE1.0. Thermocycling conditions were as follows: 1 min at 94 °C, followed by a sample-specific number of cycles (13–20 cycles) of 15 s at 94 °C, 20 s at 60 °C, and 20 s at 72 °C, and a final extension step of 1 min at 72 °C. The amplified library was purified with PB buffer on Qiagen MinElute columns, before being eluted in 30 μL EB buffer. Negative library controls constructed on H2O were included, as well as libraries constructed on the negative extractions controls. Finally the DNA concentration of the individual libraries was measured on an Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA) before being pooled equimolarly and ‘shotgun’ sequenced on an Illumina HiSeq 2500 platform using 81 bp single read chemistry (87 bp minus 6 bp index).
2.4. Bioinformatics
The data were base-called using the Illumina software CASAVA 1.8.2 and sequences were de-multiplexed with a requirement of a full match of the six nucleotide indices that were used. The raw reads were trimmed for adapters using AdapterRemoval2 [24], and trimmed reads shorter than 30 bp were discarded. Since no reference genome exists for Z. longissimus, we performed an initial round of mapping against the draft genome sequence of another colubrid snake species, Thamnophis sirtalis [25] allowing to test for the presence of genomic snake DNA in our samples. The trimmed reads were mapped using BWA v. 0.7.15 [26] with default settings, and Samtools v. 1.5 [27] was used to remove duplicate sequences. To investigate the level of DNA degradation in our samples, the mapped sequences were analysed with mapDamage v.2.0.5 [28].
For the phylogeographic analyses, we mapped the shotgun data (as above) against two published Z. longissimus mtDNA sequences (cytochrome c oxidase subunit I and cytochrome b, coxI and cytb) representing each of the four different phylogeographic lineages (Eastern E1, Western W1, Greek G1, Asian T1) as defined previously [19]. The individual bamfiles were then imported into Geneious v.8.1.7 [29] for manual inspection and consensus sequence construction, applying a 75% site identity threshold and a minimum site coverage of 3×. The consensus sequences are available at GenBank under accession numbers MH018690-MH018693.
2.5. Alignment and Network Analysis
The consensus sequences were then concatenated (coxI + cytb) and aligned in Geneious against 92 Z. longissimus sequences (1638 bp), representing the dataset analysed in Musilová et al. [19]. The alignment was converted to a Nexus file format and imported into POPART [30] to construct an unrooted median-joining network.
3. Results
3.1. Sequencing, Trimming, and Mapping
Roughly 500 million DNA sequences were generated for this project, ranging from 33 to 172 million per extract (Table 1). Following bioinformatic trimming and mapping against the T. sirtalis genome between 0.6% and 8.6% of the sequences could be aligned, representing an overall mapping efficiency (clonal reads, low quality reads, and <30 bp reads removed) between 0.06% and 1.3% (Table 1). Given that we had to rely on the T. sirtalis genome as reference for the genome-wide mapping, these numbers are likely lower than the true endogenous DNA content of the samples. Regardless, based on these data it is evident that two of the three specimens (HK2489 and ZMUC R8974) showed promising potential. Moreover, for ZMUC R8974 all three substrate types (muscle/skin, bone, tooth) yielded snake DNA; the vertebrae sample performed best with an overall mapping efficiency of 1.3% (Table 1).
3.2. DNA Preservation
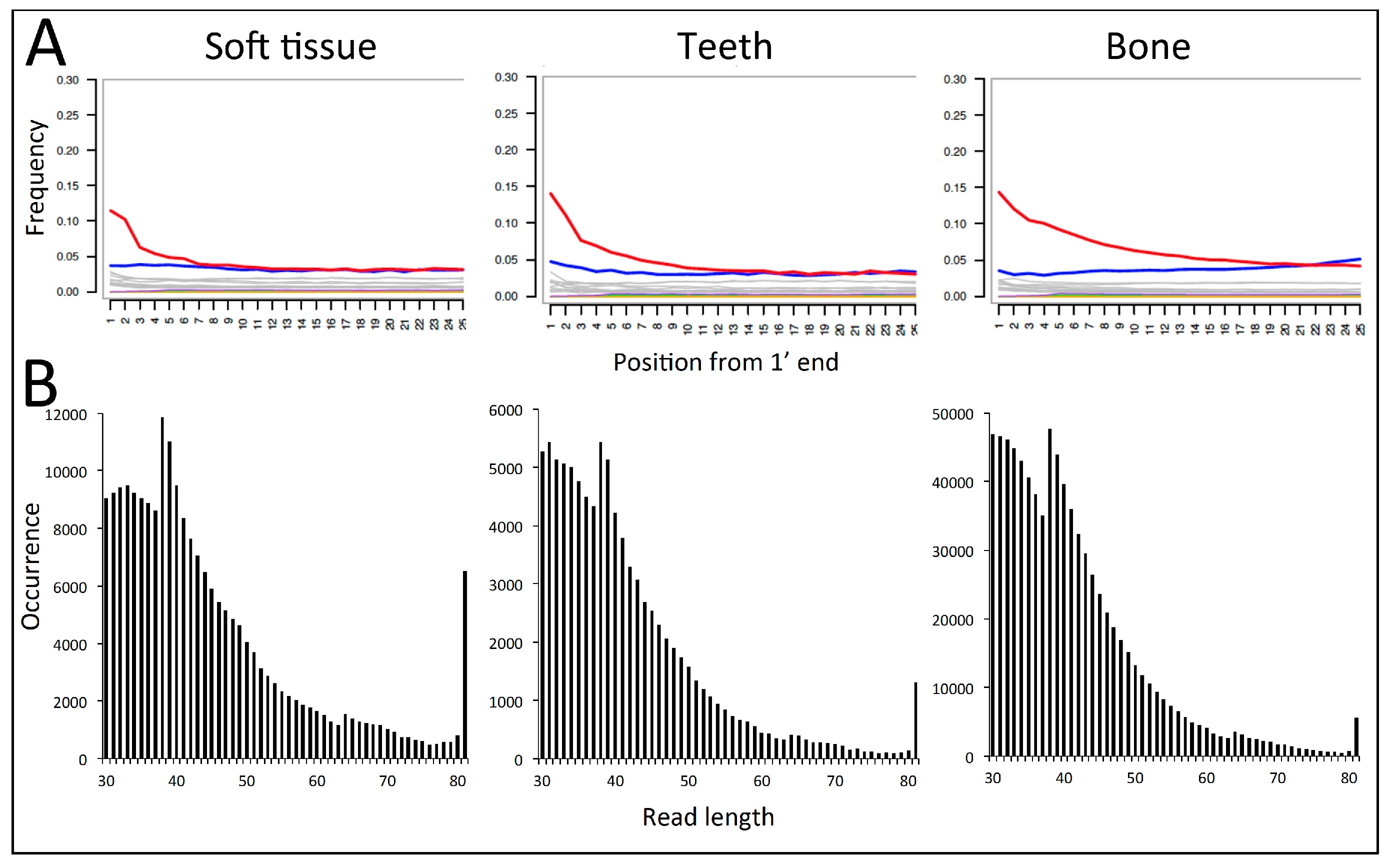
When mapped against the T. sirtalis genome and analysed in mapDamage2, the data from all DNA extracts showed a marked increase in C to T deamination damage towards the 5′-ends of the sequences, ranging from ca. 11% to 15% at position 1 (Table 1, Figure 2). The C-T damage rates appeared slightly lower in the soft tissue samples compared to bone and tooth (10% vs. 14% in ZMUC R8974 and 11% vs. 15% in ZMUC R8975). The DNA was highly degraded with length distributions of the mapped reads peaking well below 50 bp (Figure 2). There was a tendency for soft tissue samples to display slightly longer average sequence lengths than bone and tooth (Figure 2).
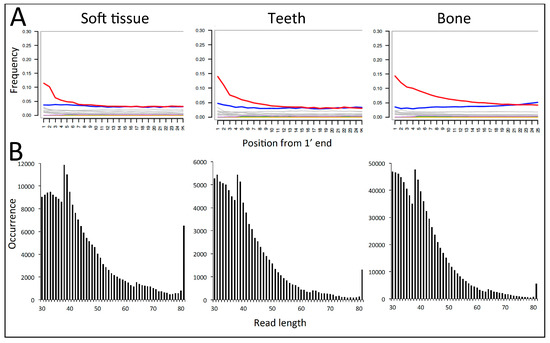
Figure 2.
DNA damage based on shotgun sequencing data of three types of tissue (soft tissue, tooth, bone) obtained from the same historical Z. longissimus individual (ZMUC R8974), and using T. sirtalis as reference genome in the mapping. A: C to T deamination damage (red line) recorded at each position in the read from the 5′ end of the molecules (output from mapDamage). B: The mapped read length distribution showing very short average read lengths and an exponentially declining distribution as typical for degraded DNA [31]. The peaks at ca. 38 and 64 bp represent bioinformatic mapping artefacts. In BWA the number of allowed mismatches when aligning each sequence to the reference genome is defined by the sequence length, increasing from 2 to 3 allowed mismatches at 38 bp, and 4 allowed mismatches at 64 bp. This relax in stringency is observed as more reads mapping. The peak observed at 81 bp represents the tail of the distribution piling up at the maximum sequencing length applied in this run.
3.3. Phylogeography
When the sequences were mapped against the coxI + cytb mitochondrial sequences from Z. longissimus obtained from GenBank, HK2489 and ZMUC R8974 again displayed far more sequences aligning than ZMUC R8975 (Table 2). With only very few sequences retained it was impossible to generate a consensus sequence for ZMUC R8975 and was, therefore, excluded in downstream analyses. For the two good samples we obtained approximately twice as many hits when mapping against Z. longissimus mitochondrial genes compared to sequences of Z. lineatus (Table 2), confirming the taxonomic identity of these two specimens. Moreover, we observed that both specimens had slightly higher affinity (more hits) to the Eastern lineage within Z. longissimus than to the other three lineages (Western, Greek, Asian) described previously [19].

Table 2.
Genetic affinity by mapping.
The merged bamfiles (duplicates removed) for HK2489 (467 reads) and ZMUC R8974 (2225 reads) were imported and re-aligned in Geneious revealing coverage values of 58× (cytb) and 70X (coxI) for ZMUC R8974, and 13× (cytB) and 15X (cox1) for HK2489. Following consensus calling and sequence concatenation, a 1636 bp sequence was generated for ZMUC R8974 and a 1624 bp sequence for HK2489, thus missing only very few sites compared to the 1638 bp concatenated sequences analysed in Musilová et al. [19].
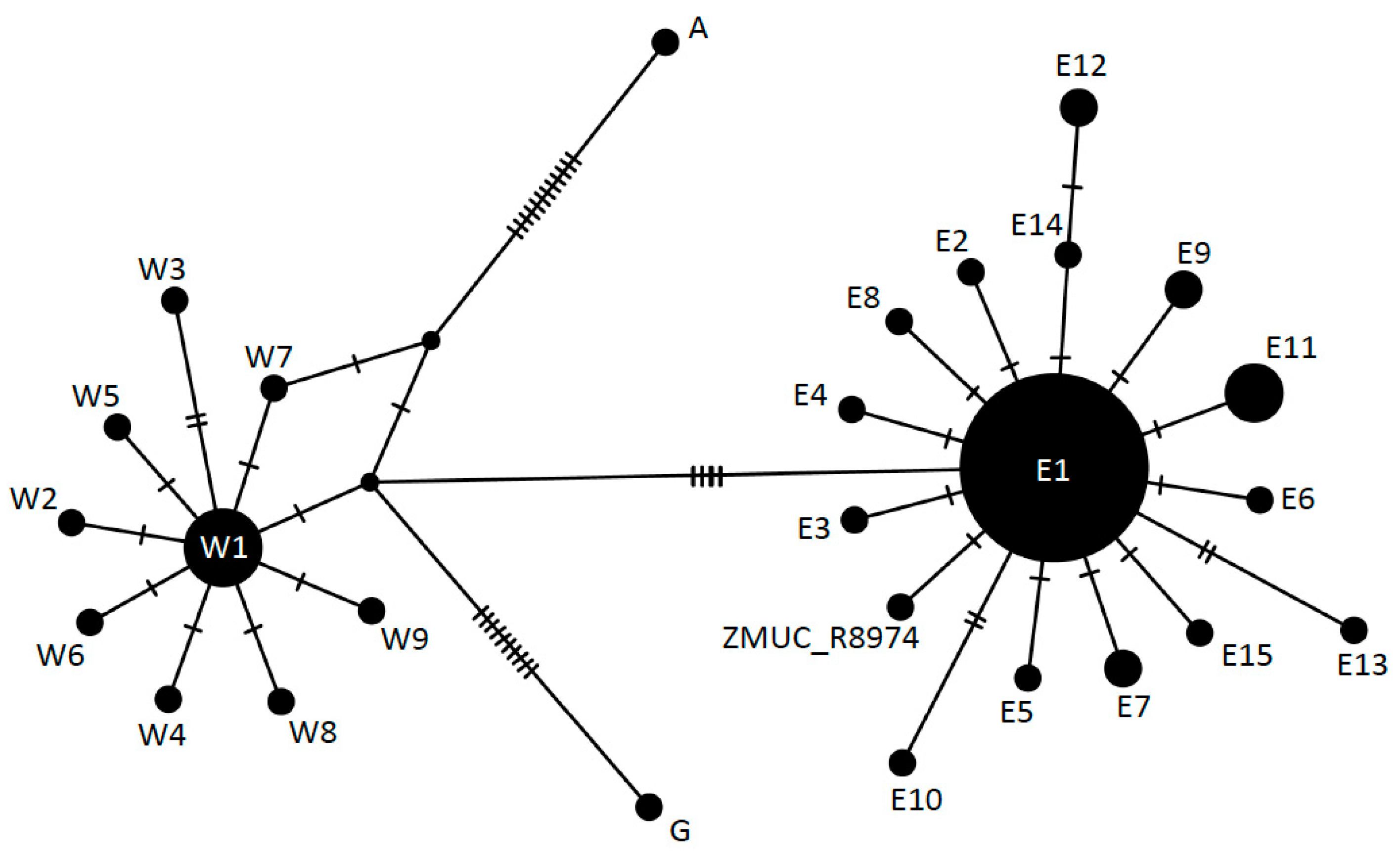
The network analysis securely placed the sequences of the two Danish individuals in the Eastern lineage (Figure 3). The specimen HK2489 displayed the previously defined haplotype E1, whereas ZMUC R8974 showed one unique mutation separating it from E1, and not previously observed in the Z. longissimus gene pool (Figure 3). This nucleotide (position 1616 in the concatenated sequence) is a T in ZMUC R8974 instead of the C observed in all previously published sequences. The position was covered 12 times with unique non-clonal reads, all showing a T nucleotide, ruling out that this variant call could be a result of deamination damage.
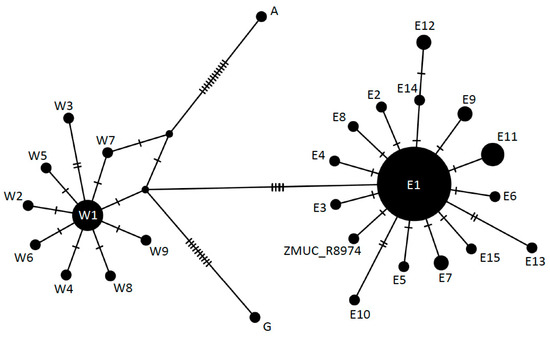
Figure 3.
Unrooted median joining network of Z. longissimus mtDNA haplotypes, based on 94 concatenated sequences (cytb + coxI, 1638 bp). Nomenclature according to Musilová et al. [19]: E = Eastern, W = Western, G = Greek, A = Asian. The two Danish individuals are found in the Eastern clade. HK2489 shows the previously described E1 haplotype and ZMUC R8974 shows a new haplotype derived from E1.
4. Discussion
4.1. DNA Preservation
By using a silica-in-solution DNA extraction method with a binding buffer optimized for recovering very short molecules [20], we succeeded in obtaining and sequencing DNA from two of the three 19th century specimens of Z. longissimus included in this study. Despite not being a large systematic study, comparing different methods on many specimens, the results are encouraging nonetheless and show that this aDNA extraction method is certainly applicable to old ethanol-preserved specimens. More systematic studies are needed in order to test the performance in comparison to other extraction methods that have worked on similar material (e.g., [32,33,34]). In terms of utilizing different parts of a specimen, we observe that soft tissue, bone, and tooth samples all yielded positive results for one of the snakes. To our knowledge this is the first study to report endogenous DNA from snake teeth. There may be situations where one substrate is preferred over another and now the molecular arguments are in place for targeting each of these substrates. The vertebra yielded the best DNA library (highest efficiency) of the three substrates, but more tests are needed before this can be confirmed as a general feature. In ZMUC R8975 both soft tissue and teeth failed to yield DNA at levels that were feasible for further sequencing. Whichever factors that have affected DNA preservation negatively in this specimen did not discriminate between the substrates.
The DNA in the snakes was highly degraded, showing short fragment lengths and C-T damage fractions that are comparable to DNA from bones thousands of years old (e.g., [20]). This explains why PCR-based experiments often fail on such material; the template molecules are too short to include both primer binding regions and the target region between them. Although ethanol preservation has ensured long-term morphological integrity of these snake specimens, the DNA molecules have clearly been highly compromised. This is perhaps not surprising considering that many such specimens are preserved in only 70% ethanol solution, likely causing DNA to degrade spontaneously in the presence of 30% water [35]. Moreover, although these particular snakes were collected prior to the use of formalin in preservation practices [36], formalin could very well have been added at a later stage, perhaps explaining the high level of DNA degradation we observe [36].
Nonetheless, the fact that we can successfully align > 4% of the reads to the genome of a distantly-related snake species (T. sirtalis) is encouraging and highlights the genomic potential in these samples. With a Z. longissimus reference genome available, our two historical specimens would likely qualify for complete genome assembly and future studies will hopefully be able to take advantage of the genome-wide shotgun data we have generated in this study.
4.2. Phylogeography
The primary objective of this investigation was to determine the genetic affinity of the extinct Danish Z. longissimus population. Four major clades have been described within this species: two small and basal clades near the Black Sea and in Greece, respectively, and two larger clades that expanded northwards from their southwestern and southeastern refugia—likely in Southern France and the Southern Balkans, respectively [19,37,38]. Our analyses clearly place the two Danish individuals in the eastern clade, likely deriving from a Balkan refugium. The south of the Balkans served as a glacial refugium for many reptiles (e.g., [39,40,41,42,43]) and our results suggest that it was an Z. longissimus expansion from this refugium that reached as far north as Denmark during the mid-Holocene warming period. This expansion mirrors the results from Musilová et al. [19] who demonstrated that the extant isolated populations in Germany and Czech Republic also belong to this Eastern clade.
The question remains, however, whether these isolated extant and recently-extinct populations are relicts of the wider mid-Holocene distribution demonstrated by fossils, or if they were introduced at a later stage. Before fossil evidence was available, it was suggested that these isolated populations could have been introduced, amongst others, by the ancient Romans, or later by Italian or French noble families or Greek merchants [6,44,45]. While the presented data (here and elsewhere) cannot exclude a later re-introduction based on individuals from the Eastern clade, they certainly exclude an origin from the Western clade (Italy, France, etc.) or Greek clade, which would have been the null hypothesis if they had been introduced by Romans or later merchants. Thus, our results suggest that the now extinct Danish population from Southern Zealand was probably a true relict population from the mid-Holocene warming period. In summary, this suggests that Z. longissimus occurred in Denmark from at least 7500 BC up to the early 1900s, when the last observations were made.
Acknowledgments
We thank the staff at the Danish National High Throughput Sequencing Centre and Jesper Stenderup for technical assistance. We are grateful to Petr Kotlik for making the published mtDNA sequences directly available to us, and we thank Kristine Vesterdorf for valuable input to the manuscript. This work was funded by The Aage V. Jensen Foundation and the Villum Foundation (Young Investigator Programme, grant no. 10120). Centre for GeoGenetics is funded by The Danish National Research Foundation (DNRF94).
Author Contributions
H.V.K., A.R.R., and M.E.A. conceived and designed the study. M.E.A. performed the experiments, analysed the data, and drafted the manuscript with input from A.R.R. and H.V.K.
Conflicts of Interest
The authors declare no conflicts of interest
References
- Guschanski, K.; Krause, J.; Sawyer, S.; Valente, L.M.; Bailey, S.; Finstermeier, K.; Sabin, R.; Gilissen, E.; Sonet, G.; Nagy, Z.T. Next-generation museomics disentangles one of the largest primate radiations. Syst. Boil. 2013, 62, 539–554. [Google Scholar] [CrossRef] [PubMed]
- Wandeler, P.; Hoeck, P.E.; Keller, L.F. Back to the future: Museum specimens in population genetics. Trends Ecol. Evol. 2007, 22, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Heimes, P. Die reptilien des rheingautaunus unter berücksichtigung der schutzproblematik der äskulapnatter, Elaphe longissima (laurenti, 1768). Naturschutz-Zentrum Hessen, Wetzlar 1988. Unpublished work. [Google Scholar]
- Musilová, R.; Zavadil, V.; Kotlík, P. Isolated populations of Zamenis longissimus (reptilia: Squamata) above the northern limit of the continuous range in europe: Origin and conservation status. Acta Soc. Zool. Bohem. 2007, 71, 197–208. [Google Scholar]
- Waitzmann, M. Zur situation der äskulapnatter Elaphe longissima (laurenti, 1768) in der bundesrepublik deutschland. Mertensiella 1993, 3, 115–133. [Google Scholar]
- Gomille, A. Die Äskulapnatter-Elaphe longissima: Verbreitung und Lebensweise in Mitteleuropa; Edition Chimaira: Frankfurt am Main, Germany, 2002; pp. 1–158. [Google Scholar]
- Szyndlar, Z. Fossil snakes from poland. Acta Zool. Cracov. 1984, 28, 1–156. [Google Scholar]
- Szyndlar, Z. A review of neogene and quaternary snakes of central and eastern europe. Part i: Scolecophidia, Boidae, Colubrinae. Estudios Geológicos 1991, 47, 103–126. [Google Scholar]
- Ljungar, L. First subfossil find of the aesculapian snake. Amphibia-Reptilia 1995, 16, 93–94. [Google Scholar] [CrossRef]
- Sommer, R.S.; Persson, A.; Wieseke, N.; Fritz, U. Holocene recolonization and extinction of the pond turtle, Emys orbicularis (l., 1758), in europe. Quat. Sci. Rev. 2007, 26, 3099–3107. [Google Scholar] [CrossRef]
- Richter, J.; Noe-Nygaard, N. A late mesolithic hunting station at agernæs, fyn, denmark. Acta Archaeol. 2003, 74, 1–64. [Google Scholar] [CrossRef]
- Kristensen, H.V. Første danske fund af knogler fra æskulapsnog (Zamenis longissimus) i bronzealdergrav. Nord. Herpetol. Foren. 2008, 51, 82–86. [Google Scholar]
- Boulenger, G.A. The Snakes of Europe; Methuen & Company, Limited: London, UK, 1913. [Google Scholar]
- Sarauw, G. En stenalders boplads i maglemose ved mullerup sammenholdt med beslaegtede. In Aarbøger for Nordisk Oldkyndighed; Lund Universitet: Lund, Sweden, 1903; Volume 1903, pp. 148–315. [Google Scholar]
- Sarauw, G.F. Rodsymbiose og Mykorrhizer Saerlig Hos Skovtraeerne; H. Hagerups Forlag: Copenhagen, Denmark, 1893; Volume 18, pp. 127–259. [Google Scholar]
- Fog, K.; Schmedes, A.; de Lasson, D.R. Nordens Padder og Krybdyr; G.E. C. Gad: Copenhagen, Denmark, 1997. [Google Scholar]
- Hvass, H. Danmarks æskulapsnoge [Danish aesculapian snake]. Naturens Verden 1942, 26, 74–86. [Google Scholar]
- Munk, H. Hassselskoven. En Skov- og Landbrugskulturhistorisk Studie fra Sydsjælland i 1600–1700; Holger Munk: Frederikssund, Denmark, 1969; p. 249. [Google Scholar]
- Musilová, R.; Zavadil, V.; Marková, S.; Kotlík, P. Relics of the europe’s warm past: Phylogeography of the aesculapian snake. Mol. Phylogenet. Evol. 2010, 57, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Allentoft, M.E.; Sikora, M.; Sjögren, K.-G.; Rasmussen, S.; Rasmussen, M.; Stenderup, J.; Damgaard, P.B.; Schroeder, H.; Ahlström, T.; Vinner, L. Population genomics of bronze age eurasia. Nature 2015, 522, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Willerslev, E.; Cooper, A. Ancient DNA. Proc. R. Soc. Lond. B Biol. Sci. 2005, 272, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Orlando, L.; Ginolhac, A.; Zhang, G.; Froese, D.; Albrechtsen, A.; Stiller, M.; Schubert, M.; Cappellini, E.; Petersen, B.; Moltke, I.; et al. Recalibrating equus evolution using the genome sequence of an early middle pleistocene horse. Nature 2013, 499, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.; Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protoc. 2010, 2010, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Lindgreen, S.; Orlando, L. Adapterremoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Warren, W.C.; Wilson, R. Thamnophis Sirtalis Isolate EDBJR-23777, Whole Genome Shotgun Sequencing Project. 2015. Available online: https://www.ncbi.nlm.nih.gov/nuccore/LFLD00000000.1. (accessed on 9 February 2018).
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and samtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Jónsson, H.; Ginolhac, A.; Schubert, M.; Johnson, P.L.F.; Orlando, L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 2013, 29, 1682–1684. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Allentoft, M.E.; Collins, M.; Harker, D.; Haile, J.; Oskam, C.L.; Hale, M.L.; Campos, P.F.; Samaniego, J.A.; Gilbert, M.T.P.; Willerslev, E. The half-life of DNA in bone: Measuring decay kinetics in 158 dated fossils. Proc. R. Soc. Lond. B Biol. Sci. 2012, 279, 4724–4733. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, A.R.; Elmberg, J.; Sanders, K.L.; Gravlund, P. Rediscovery of the rare sea snake Hydrophis parviceps smith 1935: Identification and conservation status. Copeia 2012, 2012, 276–282. [Google Scholar] [CrossRef]
- Friedman, M.; DeSalle, R. Mitochondrial DNA extraction and sequencing of formalin-fixed archival snake tissue. DNA Seq. 2008, 19, 433–437. [Google Scholar] [CrossRef]
- Ruane, S.; Austin, C.C. Phylogenomics using formalin-fixed and 100+ year-old intractable natural history specimens. Mol. Ecol. Resour. 2017, 17, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, M.; Rasmussen, A.R.; Simonsen, K.P. Enzymatic detection of formalin-fixed museum specimens for DNA analysis and enzymatic maceration of formalin-fixed specimens. Collect. Forum 2016, 30, 1–6. [Google Scholar] [CrossRef][Green Version]
- Joger, U.; Fritz, U.; Guicking, D.; Kalyabina-Hauf, S.; Nagy, Z.T.; Wink, M. Phylogeography of western palaearctic reptiles–spatial and temporal speciation patterns. Zool. Anz. 2007, 246, 293–313. [Google Scholar] [CrossRef]
- Joger, U.; Fritz, U.; Guicking, D.; Kalyabina-Hauf, S.; Nagy, Z.T.; Wink, M. Relict populations and endemic clades in palearctic reptiles: Evolutionary history and implications for conservation. In Relict Species; Springer: Berlin, Germany, 2010; pp. 119–143. [Google Scholar]
- Boehme, M.U.; Fritz, U.; Kotenko, T.; Ljubisavljević, K.; Tzankov, N.; Berendonk, T.U. Phylogeography and cryptic variation within the Lacerta viridis complex (lacertidae, reptilia). Zool. Scr. 2007, 36, 119–131. [Google Scholar] [CrossRef]
- Guicking, D.; Lawson, R.; Joger, U.; Wink, M. Evolution and phylogeny of the genus Natrix (serpentes: Colubridae). Biol. J. Linn. Soc. 2006, 87, 127–143. [Google Scholar] [CrossRef]
- Lenk, P.; Fritz, U.; Joger, U.; Wink, M. Mitochondrial phylogeography of the european pond turtle, Emys orbicularis (linnaeus 1758). Mol. Ecol. 1999, 8, 1911–1922. [Google Scholar] [CrossRef] [PubMed]
- Sommer, R.S.; Fritz, U.; Seppä, H.; Ekström, J.; Persson, A.; Liljegren, R. When the pond turtle followed the reindeer: Effect of the last extreme global warming event on the timing of faunal change in northern europe. Glob. Chang. Biol. 2011, 17, 2049–2053. [Google Scholar] [CrossRef]
- Kindler, C.; Graciá, E.; Fritz, U. Extra-mediterranean glacial refuges in barred and common grass snakes (Natrix helvetica, N. natrix). Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Böhme, W. Äskulapnatter (elaphe longissima laurenti 1768). Handbuch der Reptilien und Amphibien Europas 1993, 3, 331–372. [Google Scholar]
- Mikátová, B.; Zavadil, V. Aesculapian snake—Elaphe longissima (laurenti, 1768). In Atlas of the Distribution of Reptiles in the Czech Republic; Mikátová, B., Vlašín, M., Zavadil, V., Eds.; Agentura Ochrany Prirody: Brno-Prague, Czech Republic, 2001; pp. 113–123. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).