No Change Detected in Culturable Fungal Assemblages on Cave Walls in Eastern Canada with the Introduction of Pseudogymnoascus destructans
Abstract
:1. Introduction
2. Materials and Methods
Statistical Analysis
3. Results
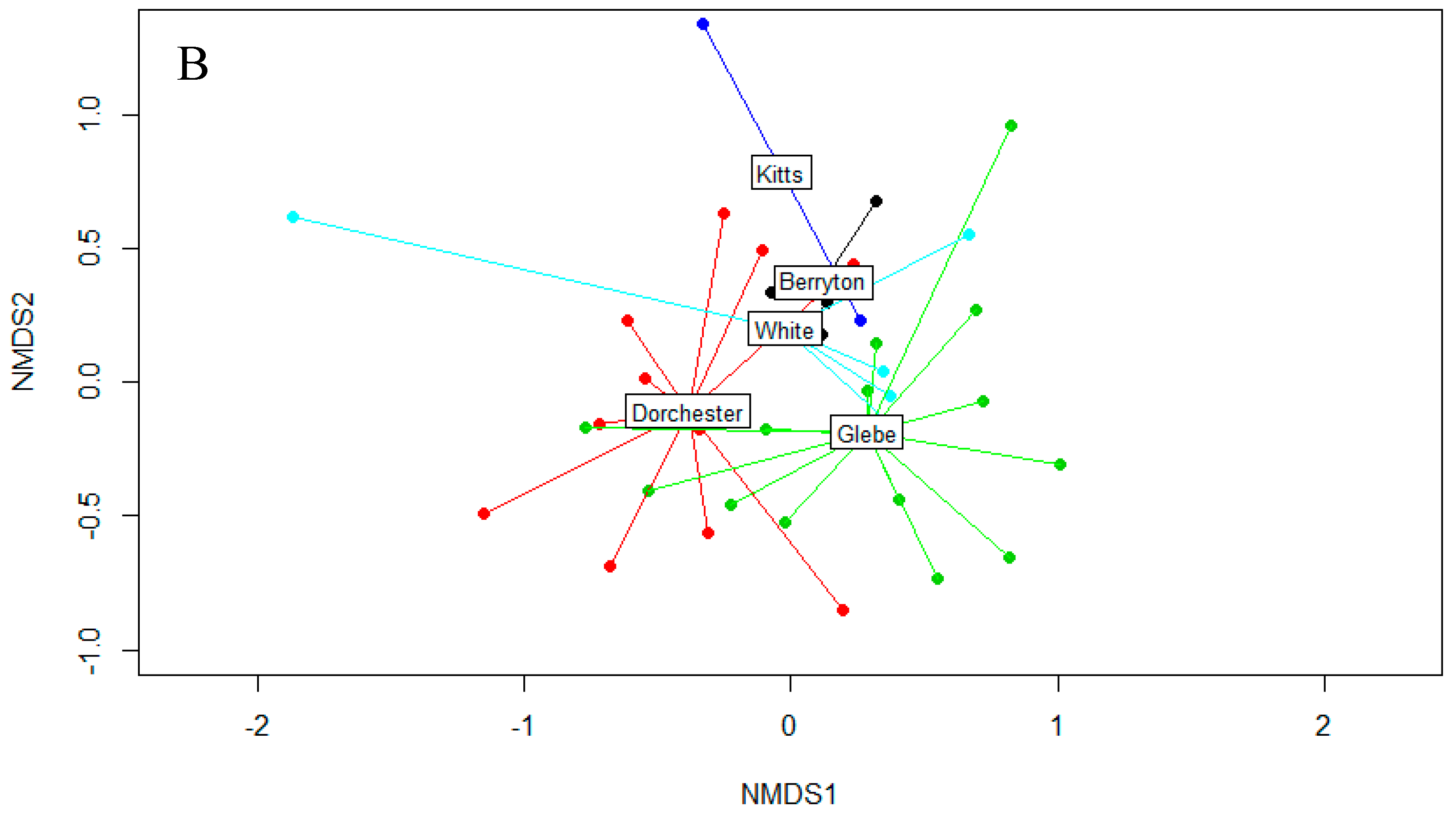
3.1. Fungal Diversity on Cave Walls Compared to Arthropods and Bats
3.2. Fungal Diversity on Cave Walls
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dickson, G.; Kirk, P.W. Distribution of heterotrophic microorganisms in relation to detritivores in Virginia caves (with supplemental bibliography on cave mycology and microbiology). In The Distributional History of the Biota of the Southern Appalachians. IV. Algae and Fungi; Parker, B.C., Roane, M.K., Eds.; University of Virginia Press: Charlottesville, VA, USA, 1976; pp. 205–226. [Google Scholar]
- Nováková, A. Microscopic fungi isolated from the Domica Cave system (Slovak Karst National Park, Slovakia). A review. Int. J. Speleol. 2009, 38, 71–82. [Google Scholar] [CrossRef]
- Vaughan, M.J.; Nelson, W.; Soderlund, C.; Maier, R.M.; Pryor, B.M. Assessing fungal community structure from mineral surfaces in Kartchner Caverns using multiplexed 454 pyrosequencing. Microb. Ecol. 2015, 70, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Grishkan, I.; Nevo, E.; Wasser, S.P. Micromycetes from the Saline Arubotaim Cave: Mount Sedom, The Dead Sea Southwestern Shore, Israel. J. Arid Environ. 2004, 57, 431–443. [Google Scholar] [CrossRef]
- Hasselbring, T.S.; Tansey, M.R.; Jack, M.A. Fungi associated with growing stalagtites. Mycologia 1975, 67, 171–172. [Google Scholar] [CrossRef]
- Man, B.; Wang, H.; Yun, Y.; Xiang, X.; Wang, R.; Duan, Y.; Cheng, X. Diversity of fungal communities in Heshang Cave of Central China revealed by mycobiome-sequencing. Front. Microbiol. 2018, 9, 1400. [Google Scholar] [CrossRef]
- Min, K. Fungus flora of Seongrya Cave in Korea. Trans. Mycol. Soc. Jpn. 1988, 29, 479–487. [Google Scholar]
- Northup, D.E.; Lavoie, K.H. Geomicrobiology of caves: A review. Geomicrobiol. J. 2001, 18, 199–222. [Google Scholar]
- Waring, C.L.; Hankin, S.I.; Griffith, D.W.T.; Kertesz, M.A.; Kobylski, V.; Wilson, N.L.; Coleman, N.V.; Kettlewell, G.; Zlot, R.; Bosse, M.; et al. Seasonal total methane depletion in limestone caves. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Lennon, J.; Nguyễn-thùy, D.; Pham, T.; Drobniak, A.; Ta, P.; Pham, N.; Streil, T.; Webster, K.; Schimmelmann, A. Microbial contributions to subterranean methane sinks. Geobiology 2016, 15, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Burford, E.P.; Kierans, M.; Gadd, G.M. Geomycology: Fungi in mineral substrata. Mycologist 2003, 17, 98–107. [Google Scholar] [CrossRef]
- Gorbushina, A.A. Life on the rocks. Environ. Microbiol. 2007, 9, 1613–1615. [Google Scholar] [CrossRef] [PubMed]
- Viles, H.A.; Gorbushina, A.A. Soiling and microbial colonisation on urban roadside limestone: A three year study in Oxford, England. Build. Environ. 2003, 38, 1217–1224. [Google Scholar] [CrossRef]
- Mcfarlane, D.A.; Keeler, R.C.; Mizutani, H. Ammonia volatilization in a Mexican bat cave ecosystem. Biogeochemistry 1995, 30, 1–8. [Google Scholar] [CrossRef]
- Lorch, J.M.; Meteyer, C.U.; Behr, M.J.; Boyles, J.G.; Cryan, P.M.; Hicks, A.C.; Ballmann, A.E.; Coleman, J.T.H.; Redell, D.N.; Reeder, D.M.; et al. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature 2011, 480, 376–378. [Google Scholar] [CrossRef]
- US Fish and Wildlife Service North American Bat Death Toll Exceeds 5.5 Million from White-Nose Syndrome. Available online: http://static.whitenosesyndrome.org/sites/default/%0Afiles/files/wns_ mortality_2012_nr_final_0.pdf (accessed on 1 January 2013).
- Lorch, J.M.; Lindner, D.L.; Gargas, A.; Muller, L.K.; Minnis, A.M.; Blehert, D.S. A culture-based survey of fungi in soil from bat hibernacula in the eastern United States and its implications for detection of Geomyces destructans, the causal agent of bat white-nose syndrome. Mycologia 2013, 105, 237–252. [Google Scholar] [CrossRef]
- Vanderwolf, K.J.; Malloch, D.; McAlpine, D.F. Detecting viable Pseudogymnoascus destructans (Ascomycota: Pseudeurotiaceae) from walls of bat hibernacula: Effect of culture media. J. Cave Karst Stud. 2016, 78, 158–162. [Google Scholar] [CrossRef]
- Langwig, K.E.; Frick, W.F.; Reynolds, R.; Parise, K.L.; Drees, K.P.; Hoyt, J.R.; Cheng, T.L.; Kunz, T.H.; Foster, J.T.; Marm Kilpatrick, A. Host and pathogen ecology drive the seasonal dynamics of a fungal disease, white-nose syndrome. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142335. [Google Scholar] [CrossRef]
- Vanderwolf, K.J.; Malloch, D.; McAlpine, D.F. Ectomycota associated with arthropods from bat hibernacula in Eastern Canada, with particular reference to Pseudogymnoasucs destructans. Insects 2016, 7, 16. [Google Scholar] [CrossRef]
- Vanderwolf, K.J.; McAlpine, D.F.; Malloch, D.; Forbes, G.J. Ectomycota associated with hibernating bats in Eastern Canadian Caves prior to the emergence of white-nose syndrome. Northeast. Nat. 2013, 20, 115–130. [Google Scholar] [CrossRef]
- Vanderwolf, K.J.; Malloch, D.; McAlpine, D.F. Fungi on white-nose infected bats (Myotis spp.) in Eastern Canada show no decline in diversity associated with Pseudogymnoascus destructans (Ascomycota: Pseudeurotiaceae). Int. J. Speleol. 2016, 45, 43–50. [Google Scholar] [CrossRef]
- Beck, B.F.; Fram, M.; Carvajal Zamora, J.R. The Aguas Buenas Caves, Puerto Rico: Geology, hydrology and ecology with special reference to the histoplasmosis fungus. Natl. Speleol. Soc. Bull. 1976, 38, 1–16. [Google Scholar]
- McAlpine, D.F.; Vanderwolf, K.J.; Forbes, G.J.; Malloch, D. Consumption of bats (Myotis spp.) by Raccoons (Procyon lotor) during an outbreak of white-nose syndrome in New Brunswick, Canada: Implications for estimates of bat mortality. Can. Field Nat. 2011, 125, 257–260. [Google Scholar] [CrossRef]
- Vanderwolf, K.J.; McAlpine, D.F.; Forbes, G.J.; Malloch, D. Bat populations and cave microclimate prior to and at the outbreak of white-nose syndrome in New Brunswick. Can. Field Nat. 2012, 126, 125–134. [Google Scholar] [CrossRef]
- Vanderwolf, K.J.; Malloch, D.; McAlpine, D.F. Fungi associated with over-wintering tricolored bats, Perimyotis subflavus, in a white-nose syndrome region of eastern Canada. J. Cave Karst Stud. 2015, 77, 145–151. [Google Scholar]
- McAlpine, D.F. Status and conservation of solution caves in New Brunswick. New Brunswick Mus. Publ. Nat. Sci. 1983, 1, 1–28. [Google Scholar]
- Janicki, A.F.; Frick, W.F.; Kilpatrick, A.M.; Parise, K.L.; Foster, J.T.; McCracken, G.F. Efficacy of visual surveys for white-nose syndrome at bat hibernacula. PLoS ONE 2015, 10, e0133390. [Google Scholar] [CrossRef] [PubMed]
- US Fish and Wildlife Service Revised Decontamination Protocol: 25 June 2012. Available online: http://www.whitenosesyndrome.org/resource/revised-decontamination-protocol-june-25-2012 (accessed on 10 July 2012).
- Papavizas, G.C.; Davey, C.B. Evaluation of various media and antimicrobial agents for isolation of soil fungi. Soil Sci. 1958, 88, 112–117. [Google Scholar] [CrossRef]
- Tanaka, T.; Kawasaki, K.; Daimon, S.; Kitagawa, W.; Yamamoto, K.; Tamaki, H.; Tanaka, M.; Nakatsu, C.H.; Kamagata, Y. A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl. Environ. Microbiol. 2014, 80, 7659–7666. [Google Scholar] [CrossRef]
- Seifert, K.; Morgan-Jones, G.; Gams, W.; Kendrick, B. The Genera of Hyphomycetes; CBS-KNAW Fungal Biodiversity Centre, CBS Biodiversity Series 8: Utrecht, The Netherlands, 2011. [Google Scholar]
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi, 2nd ed.; IHW-Verlag: Bavaria, Germany, 2007. [Google Scholar]
- Team, R.C. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 1 April 2018).
- Oksanen, J.; Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.; O’Hara, R.; Simpson, G.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-2. Available online: https://cran.r-project.org/package=vegan (accessed on 1 September 2018).
- De Cáceres, M.; Legendre, P. Associations between species and groups of sites: Indices and statistical inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef]
- Ange-Stark, M.; Cheng, T.; Hoyt, J.; Langwig, K.; Parise, K.; Frick, W.F.; Kilpatrick, A.M.; MacManes, M.D.; Foster, J. White-nose syndrome restructures bat skin microbiomes. bioRxiv 2019. [Google Scholar] [CrossRef]
- Zhang, T.; Victor, T.R.; Rajkumar, S.S.; Li, X.; Okoniewski, J.C.; Hicks, A.C.; Davis, A.D.; Broussard, K.; Ladeau, S.L.; Chaturvedi, S. Mycobiome of the bat white-nose syndrome affected caves and mines reveals diversity of fungi and local adaptation by the fungal pathogen Pseudogymnoascus (Geomyces ) destructans. PLoS ONE 2014, 9, e108714. [Google Scholar] [CrossRef] [PubMed]
- Campana, M.G.; Kurata, N.P.; Foster, J.T.; Helgen, L.E.; Reeder, D.M.; Fleischer, R.C.; Helgen, K.M. White-nose syndrome fungus in a 1918 bat specimen from France. Emerg. Infect. Dis. 2017, 23, 1611–1612. [Google Scholar] [CrossRef] [PubMed]
- Vanderwolf, K.J.; McAlpine, D.F.; McGuire, L.P. Hibernacula water chemistry and implications for hibernating bats. J. Mammal. 2017, 98, 1578–1585. [Google Scholar] [CrossRef]
- Jellison, J.; Connolly, J.; Goodell, B.; Doyle, B.; Illman, B.; Fekete, F.; Ostrofsky, A. The role of cations in the biodegradation of wood by the brown rot fungi. Int. Biodeterior. Biodegrad. 1997, 39, 165–179. [Google Scholar] [CrossRef]
- Vanderwolf, K.J.; Malloch, D.; Ivanova, N.V.; McAlpine, D.F. Lack of cave-associated mammals influences the fungal assemblages of insular solution caves in eastern Canada. J. Cave Karst Stud. 2016, 78. [Google Scholar] [CrossRef]
- Sugita, T.; Kikuchi, K.; Makimura, K.; Urata, K.; Someya, T.; Kamei, K.; Niimi, M.; Uehara, Y. Trichosporon species isolated from guano samples obtained from bat-inhabited caves in Japan. Appl. Environ. Microbiol. 2005, 71, 7626–7629. [Google Scholar] [CrossRef] [Green Version]
- Malloch, D.; Sigler, L.; Hambleton, S.; Vanderwolf, K.J.; Gibas, C.F.C.; McAlpine, D.F. Fungi associated with hibernating bats in New Brunswick caves: The genus Leuconeurospora. Botany 2016, 94, 1171–1181. [Google Scholar] [CrossRef] [Green Version]





| Fungal Taxa | Arthropod | Bat | Wall | Inval | p-Value |
|---|---|---|---|---|---|
| Acrodontium spp. | yes | no | no | 0.594 | 0.087 |
| Cladosporium spp. | yes | no | no | 0.798 | 0.054 |
| Verticillium sp. (cf. Gabarnaudia) | yes | no | no | 0.753 | 0.054 |
| Mortierella spp. | no | no | yes | 0.836 | 0.054 |
| Oidiodendron truncatum | no | no | yes | 0.877 | 0.054 |
| Phialophora spp. | no | no | yes | 0.612 | 0.087 |
| Pseudogymnoascus pannorum | no | yes | yes | 0.877 | 0.054 |
| Trichosporiella spp. | no | no | yes | 0.800 | 0.054 |
| Trichosporon spp. | no | no | yes | 0.637 | 0.054 |
| Apiotrichum dulcitum | no | no | yes | 0.802 | 0.054 |
| Total Number of Swabs Done | Glebe | Berryton | Dorchester | White | Kitts |
|---|---|---|---|---|---|
| 30 | 32 | 30 | 20 | 7 | |
| Ascomycota | |||||
| Acaulium caviariformis (Malloch and Hubart) Sandoval-Denis, Guarro and Gene | 6 | 5 | 2 | 6 | 0 |
| Acremonium sp. | 0 | 1 | 5 | 1 | 1 |
| Acremonium rutilum W. Gams | 0 | 0 | 1 | 0 | 0 |
| Acrodontium sp. | 1 | 0 | 0 | 0 | 0 |
| Aphanocladium album (Preuss) W. Gams | 0 | 0 | 1 | 0 | 0 |
| Aphanocladium sp. | 0 | 0 | 1 | 0 | 0 |
| Arachniotus ruber (Tiegh.) J. Schröt. | 0 | 6 | 0 | 1 | 0 |
| Arthroderma silverae Currah, S.P. Abbott and Sigler | 1 | 4 | 0 | 3 | 0 |
| Arthrographis sp. | 0 | 0 | 0 | 1 | 0 |
| Beauveria sp. | 2 | 0 | 0 | 0 | 0 |
| Cadophora sp. | 5 | 0 | 0 | 0 | 1 |
| Cephalotrichum sp. | 0 | 1 | 0 | 0 | 0 |
| Cephalotrichum stemonitis (Pers.) Link | 13 | 10 | 3 | 11 | 2 |
| Chaetomidium sp. | 0 | 0 | 0 | 1 | 0 |
| Chalara sp. | 1 | 0 | 0 | 0 | 0 |
| Chrysosporium sp. | 7 | 1 | 1 | 6 | 0 |
| Cladosporium sp. | 0 | 3 | 1 | 1 | 0 |
| Cordyceps sp. | 0 | 0 | 1 | 1 | 0 |
| Culicinomyces sp. | 1 | 0 | 0 | 0 | 0 |
| Cylindrocarpon sp. | 0 | 0 | 0 | 0 | 3 |
| Fusarium sp. | 3 | 2 | 1 | 1 | 0 |
| Gymnoascus reesii Baran. | 1 | 0 | 0 | 0 | 0 |
| Humicola cf. UAMH 11595 | 8 | 6 | 0 | 11 | 0 |
| Hyphozyma sp. | 1 | 0 | 0 | 0 | 0 |
| Isaria sp. | 1 | 0 | 0 | 1 | 0 |
| Lecythophora sp. | 1 | 0 | 0 | 0 | 0 |
| Leuconeurospora polypaeciloides Malloch, Sigler and Hambleton | 14 | 0 | 0 | 14 | 1 |
| Leuconeurospora capsici (J.F.H. Beyma) Malloch, Sigler and Hambleton | 0 | 20 | 0 | 1 | 0 |
| Mammaria sp. | 0 | 0 | 0 | 1 | 0 |
| Microascus sp. | 1 | 0 | 0 | 0 | 0 |
| Oidiodendron sp. | 1 | 0 | 6 | 1 | 0 |
| Oidiodendron truncatum G.L. Barron | 7 | 17 | 12 | 11 | 2 |
| Paecilomyces sp. | 1 | 2 | 1 | 0 | 0 |
| Penicillium spp. | 12 | 28 | 7 | 12 | 4 |
| Penicillium chrysogenum | 1 | 0 | 0 | 0 | 0 |
| Penicillium glaucoalbidum (Desmazieres) Houbraken and Samson | 1 | 0 | 2 | 0 | 0 |
| Penicillium griseofulvum | 0 | 1 | 0 | 0 | 0 |
| Penicillium thomii Maire | 1 | 0 | 0 | 0 | 0 |
| Phaeoacremonium sp. | 1 | 0 | 0 | 0 | 0 |
| Phaeotrichum hystricinum Cain and M.E. Barr | 12 | 0 | 0 | 3 | 0 |
| Phialophora sp. | 3 | 0 | 0 | 0 | 1 |
| Phoma radicina (McAlpine) Boerema | 1 | 0 | 0 | 0 | 0 |
| Preussia sp. | 3 | 0 | 0 | 1 | 0 |
| Pseudogymnoascus pannorum | 12 | 28 | 15 | 11 | 5 |
| sensu lato (Link) Minnis and D.L. Lindner | |||||
| Pseudogymnoascus destructans | 13 | 8 | 10 | 7 | 0 |
| (Blehert and Gargas) Minnis and D.L. Lindner | |||||
| Pseudogymnoascus roseus Raillo | 1 | 0 | 0 | 8 | 0 |
| Thysanophora sp. | 2 | 0 | 0 | 0 | 0 |
| Tolypocladium sp. | 3 | 0 | 2 | 1 | 1 |
| Tricellula cf. aquatica | 0 | 0 | 0 | 0 | 2 |
| Trichoderma sp. | 1 | 3 | 0 | 0 | 1 |
| Trichophyton sp. | 0 | 0 | 1 | 0 | 0 |
| Trichosporiella sp. | 2 | 1 | 1 | 1 | 1 |
| Verticillium sp. | 2 | 1 | 1 | 0 | 0 |
| Verticillium sp. cf. Gabarnaudia | 0 | 1 | 0 | 0 | 1 |
| Wardomyces spp. | 6 | 2 | 0 | 2 | 0 |
| Wardomyces giganteus (Malloch) Sandoval-Denis, Guarro and Gene | 0 | 10 | 0 | 0 | 0 |
| Wardomyces inflatus (Marchal) Hennebert | 2 | 1 | 0 | 0 | 0 |
| Zalerion sp. | 0 | 0 | 1 | 0 | 0 |
| Basidiomycota | |||||
| Apiotrichum dulcitum (Berkhout) Yurkov and Boekhout | 12 | 6 | 0 | 1 | 3 |
| Asterotremella sp. | 1 | 0 | 0 | 2 | 0 |
| Trichosporon sp. | 6 | 2 | 0 | 2 | 0 |
| Mucoromycota | |||||
| Thamnidium elegans Link | 1 | 0 | 0 | 2 | 0 |
| Mortierella spp. | 20 | 9 | 16 | 5 | 1 |
| Mucor spp. | 15 | 21 | 3 | 12 | 2 |
| Zoopagomycota | |||||
| Kickxella alabastrina Coem. | 1 | 0 | 0 | 0 | 0 |
| Sterile | 9 | 2 | 15 | 7 | 1 |
| Fungal Taxa | 2012 | 2015 |
|---|---|---|
| Pseudogymnoascus pannorum s.l. | 59.4 | 58.8 |
| Penicillium spp. | 50.0 | 53.8 |
| Mortierella spp. | 43.8 | 45.0 |
| Pseudogymnoascus destructans | 40.6 | 31.3 |
| Mucor spp. | 37.5 | 48.8 |
| Oidiodendron truncatum | 31.3 | 46.3 |
| Cephalotrichum stemonitis | 28.1 | 35.0 |
| Leuconeurospora polypaeciloides | 18.8 | 27.5 |
| Site/Medium | # Fungal Genera | # Fungal Taxa/Swab 2012 | # Fungal Taxa/Swab 2015 | Overall Mean |
|---|---|---|---|---|
| Glebe Mine | 38 | 6.3 ± 3.2, n = 10 | 7.8 ± 4.2, n = 20 | 7.3 ± 3.9, n = 30 |
| Berryton Cave | 22 | 4.9 ± 2.8, n = 12 | 6.5 ± 2.9, n = 20 | 6.1 ± 2.9, n = 32 |
| Dorchester Mine | 20 | 3.6 ± 1.8, n = 10 | 3.7 ± 1.7, n = 20 | 3.7 ± 1.7, n = 30 |
| White Cave | 28 | ND | 7.5 ± 4.5, n = 20 | 7.5 ± 4.5, n = 20 |
| SD | 26 | 3.6 ± 1.2, n = 16 | 4.7 ± 2.5, n = 20 | 4.2 ± 2.1, n = 36 |
| MEA | 16 | ND | 3.6 ± 1.2, n = 20 | 3.6 ± 1.2, n = 20 |
| DPYA | 40 | 7.3 ± 3.3, n = 16 | 8.5 ± 4.9, n = 20 | 7.7 ± 4.2, n = 36 |
| modDPYA | 32 | ND | 8.6 ± 2.5, n = 20 | 8.6 ± 2.5, n = 20 |
| Total/Overall Mean | 48 | 5.1 ± 2.8, n = 32 | 6.4 ± 3.8, n = 80 | 6.0 ± 3.6, n = 112 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanderwolf, K.J.; Malloch, D.; McAlpine, D.F. No Change Detected in Culturable Fungal Assemblages on Cave Walls in Eastern Canada with the Introduction of Pseudogymnoascus destructans. Diversity 2019, 11, 222. https://doi.org/10.3390/d11120222
Vanderwolf KJ, Malloch D, McAlpine DF. No Change Detected in Culturable Fungal Assemblages on Cave Walls in Eastern Canada with the Introduction of Pseudogymnoascus destructans. Diversity. 2019; 11(12):222. https://doi.org/10.3390/d11120222
Chicago/Turabian StyleVanderwolf, Karen J., David Malloch, and Donald F. McAlpine. 2019. "No Change Detected in Culturable Fungal Assemblages on Cave Walls in Eastern Canada with the Introduction of Pseudogymnoascus destructans" Diversity 11, no. 12: 222. https://doi.org/10.3390/d11120222
APA StyleVanderwolf, K. J., Malloch, D., & McAlpine, D. F. (2019). No Change Detected in Culturable Fungal Assemblages on Cave Walls in Eastern Canada with the Introduction of Pseudogymnoascus destructans. Diversity, 11(12), 222. https://doi.org/10.3390/d11120222





