Community Structures of Bacteria, Archaea, and Eukaryotic Microbes in the Freshwater Glacier Lake Yukidori-Ike in Langhovde, East Antarctica
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling
2.2. Nutrient Analysis
2.3. Bacterial 16S rRNA Clone Library Analysis
2.4. Archaeal 16S rRNA Clone Library Analysis
2.5. Eukaryotic 18S rRNA Clone Library Analysis
2.6. Phylogenetic Analysis
2.7. Coverage and Diversity Indices
2.8. Nucleotide Sequences
3. Results
3.1. Nutrients
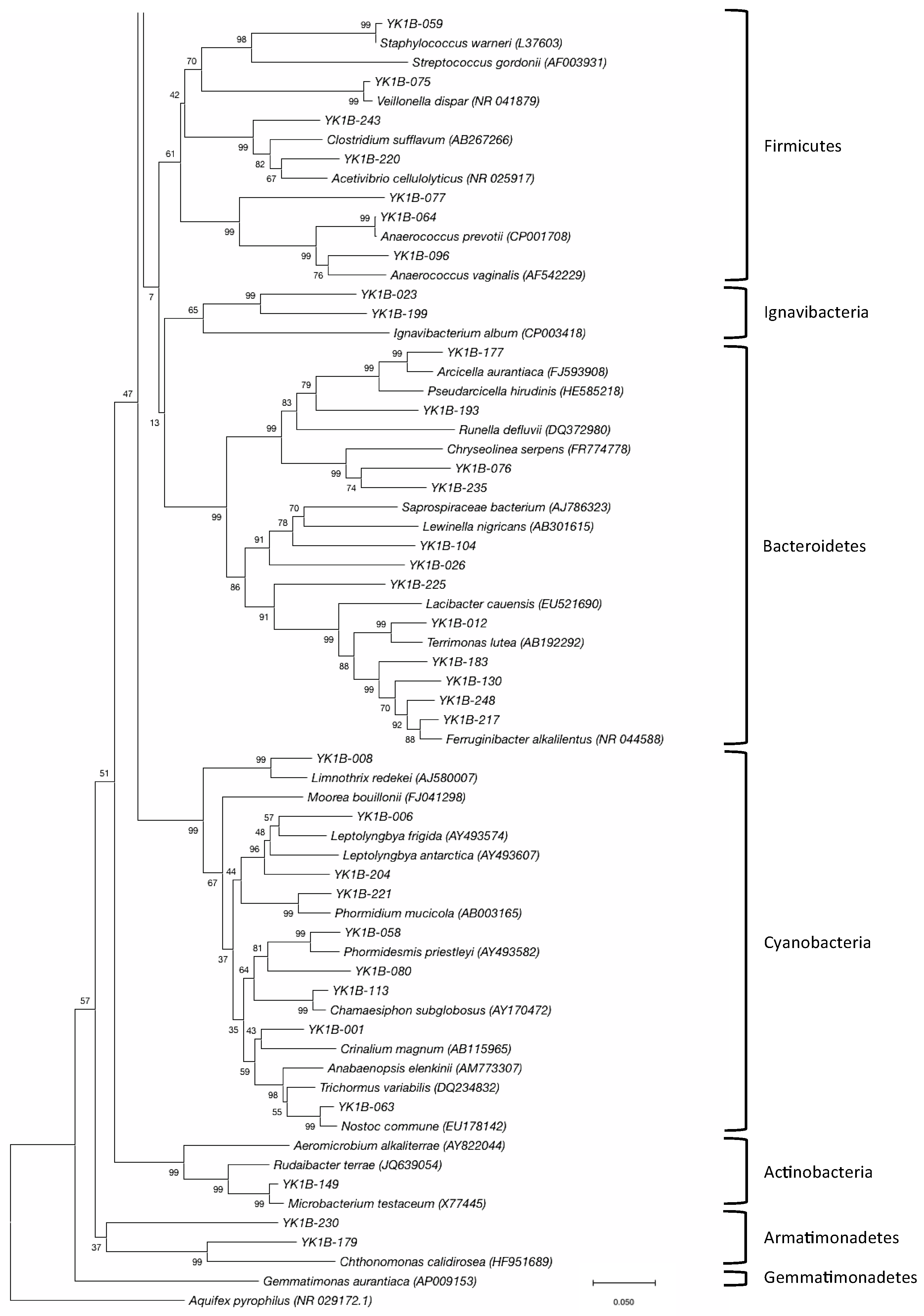
3.2. Bacterial Community
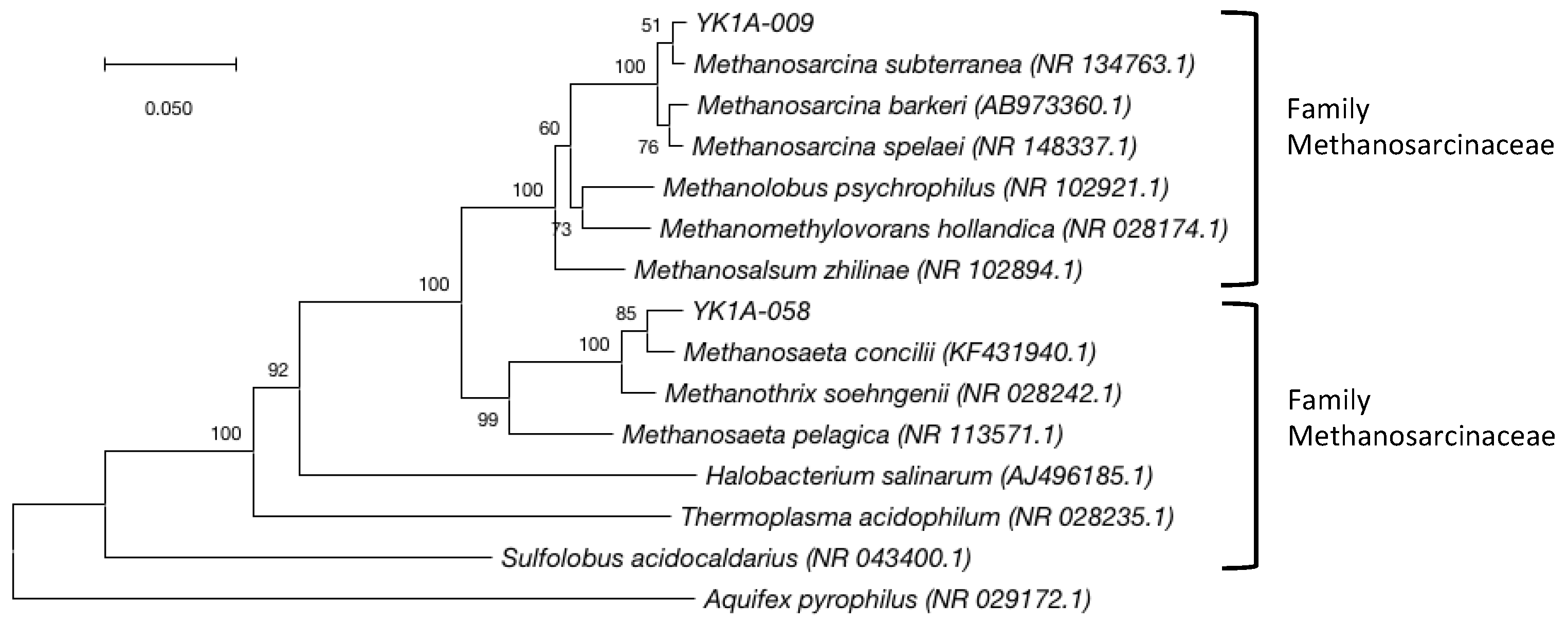
3.3. Archaeal Community
3.4. Eukaryotic Community
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kimura, S.; Ban, S.; Imura, S.; Kudoh, S.; Matsuzaki, M. Limnological characteristics of vertical structure in the lakes of Syowa oasis, East Antarctica. Polar Sci. 2010, 3, 262–271. [Google Scholar] [CrossRef]
- Franzmann, P.D. Examination of Antarctic prokaryotic diversity through molecular comparisons. Biodivers. Conserv. 1996, 5, 1295–1305. [Google Scholar] [CrossRef]
- Bowman, J.P.; McCammon, S.A.; Rea, S.M.; McMeekin, T.A. The microbial composition of three limnologically disparate hypersaline Antarctic lakes. FEMS Microbiol. Lett. 2000, 183, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Sjöling, S.; Cowan, D.A. High 16S rDNA bacterial diversity in glacial meltwater lake sediment, Bratina Island, Antarctica. Extremophiles 2003, 7, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, N.; Sato, S.; Kawarabayasi, Y.; Imura, S.; Naganuma, T. Archaeal and bacterial community structures in the anoxic sediment of Antarctic meromictic lake Nurume-Ike. Polar Sci. 2010, 4, 421–429. [Google Scholar] [CrossRef]
- Michaud, L.; Caruso, C.; Mangano, S.; Interdonato, F.; Bruni, V.; Giudice, A.L. Predominance of Flavobacterium, Pseudomonas, and Polaromonas within the prokaryotic community of freshwater shallow lakes in the northern Victoria Land, East Antarctica. FEMS Microbiol. Ecol. 2012, 82, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.S. Identification of Microbial Dark Matter in Antarctic Environments. Front. Microbiol. 2018, 9, 3165. [Google Scholar] [CrossRef]
- Kanda, H.; Inoue, M.; Mochida, Y.; Sugawara, H.; Ino, Y.; Ohtani, S.; Ohyama, Y. Biological studies on ecosystems in the Yukidori Valley, Langhovde, East Antarctica. Antarct. Rec. 1990, 34, 76–93. [Google Scholar]
- Kanda, H.; Inoue, M. Ecological monitoring of moss and lichen vegetation in the Syowa Station area, Antarctica. Mem. NIPR Symp. Polar Biol. 1994, 7, 221–231. [Google Scholar]
- Tanabe, Y.; Yasui, S.; Osono, T.; Uchida, M.; Kudoh, S.; Yamamuro, M. Abundant deposits of nutrients inside lakebeds of Antarctic oligotrophic lakes. Polar Biol. 2017, 40, 603–613. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, K.L.; Barns, S.M.; Reysenbach, A.L.; Dawson, S.C.; Pace, N.R. Wide diversity of Crenarchaeota. Nature 1996, 384, 420. [Google Scholar] [CrossRef] [PubMed]
- Jurgens, G.; Glöckner, F.O.; Amann, R.; Saano, A.; Montonen, L.; Likolammi, M.; Münster, U. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 2000, 34, 45–56. [Google Scholar] [CrossRef]
- López-García, P.; Rodriguez-Valera, F.; Pedrós-Alió, C.; Moreira, D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 2001, 409, 603–607. [Google Scholar] [CrossRef] [PubMed]
- López-García, P.; Philippe, H.; Gail, F.; Moreira, D. Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc. Natl. Acad. Sci. USA 2003, 100, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Good, I.J. The population frequencies of species and the estimation of population parameters. Biometrika 1953, 40, 237–264. [Google Scholar] [CrossRef]
- Colwell, R.K. EstimateS: Statistical Estimation of Species Richness and Shared Species from Samples; Version 9; User’s Guide and application [WWW document] 2013. Available online: http://purl.oclc.org/estimates (accessed on 2 June 2019).
- Zwart, G.; Crump, B.C.; Kamst-van Agterveld, M.P.; Hagen, F.; Han, S.K. Typical freshwater bacteria: An analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 2002, 28, 141–155. [Google Scholar] [CrossRef]
- Garneau, M.È.; Vicent, W.F.; Alonso-Sáez, L.; Gratton, Y.; Lovejoy, C. Prokaryotic community structure and heterotrophic production in a river-influenced coastal arctic ecosystem. Aquat. Microb. Ecol. 2006, 42, 27–40. [Google Scholar] [CrossRef]
- Cheng, S.M.; Foght, J.M. Cultivation-independent and-dependent characterization of bacteria resident beneath John Evans Glacier. FEMS Microbiol. Ecol. 2007, 59, 318–330. [Google Scholar] [CrossRef]
- Galand, P.E.; Lovejoy, C.; Pouliot, J.; Garneau, M.È.; Vincent, W.F. Microbial community diversity and heterotrophic production in a coastal Arctic ecosystem: A stamukhi lake and its source waters. Limnol. Oceanogr. 2008, 53, 813–823. [Google Scholar] [CrossRef]
- Pienitz, R.; Doran, P.T.; Lamoureux, S.F. Origin and geomorphology of lakes in the polar regions. In Polar Lakes and Rivers: Limnology of Arctic and Antarctic Aquatic Ecosystems; Vincent, W.F., Laybourn-Parry, J., Eds.; Oxford University Press: Oxford, UK, 2008; pp. 25–41. [Google Scholar]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Cameron, K.A.; Hodson, A.J.; Osborn, A.M. Structure and diversity of bacterial, eukaryotic and archaeal communities in glacial cryoconite holes from the Arctic and the Antarctic. FEMS Microbiol. Ecol. 2012, 82, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Unrein, F.; Izaguirre, I.; Massana, R.; Balagué, V.; Gasol, J.M. Nanoplankton assemblages in maritime Antarctic lakes: Characterisation and molecular fingerprinting comparison. Aquat. Microb. Ecol. 2005, 40, 269–282. [Google Scholar] [CrossRef]
- Nakai, R.; Abe, T.; Baba, T.; Imura, S.; Kagoshima, H.; Kanda, H.; Naganuma, T. Eukaryotic phylotypes in aquatic moss pillars inhabiting a freshwater lake in East Antarctica, based on 18S rRNA gene analysis. Polar Biol. 2012, 35, 1495–1504. [Google Scholar] [CrossRef]
- Velasco-Castrillón, A.; Gibson, J.A.E.; Stevens, M.I. A review of current Antarctic limno-terrestrial microfauna. Polar Biol. 2014, 37, 1517–1531. [Google Scholar] [CrossRef]
- McInnes, S.J. Tardigrades from Signy Island, South Orkney Islands, with particular reference to freshwater species. J. Nat. Hist. 1995, 29, 1419–1445. [Google Scholar] [CrossRef]
- Gibson, J.A.E.; Comer, L.; Agius, J.T.; McInnes, S.J.; Marley, N.J. Tardigrade eggs and exuviae in Antarctic lake sediments: Insights into Holocene dynamics and origins of the fauna. J. Limnol. 2007, 66, 65–71. [Google Scholar] [CrossRef]
- Tsujimoto, M.; McInnes, S.J.; Convey, P.; Imura, S. Preliminary description of tardigrade species diversity and distribution pattern around coastal Syowa Station and inland Sør Rondane Mountains, Dronning Maud Land, East Antarctica. Polar Biol. 2014, 37, 1361–1367. [Google Scholar] [CrossRef]
- Yang, B.; Wang, Y.; Qian, P.Y. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinform. 2016, 17, 135. [Google Scholar] [CrossRef]
- Hadziavdic, K.; Lekang, K.; Lanzen, A.; Jonassen, I.; Thompson, E.M.; Troedsson, C. Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS ONE 2014, 9, e87624. [Google Scholar] [CrossRef] [PubMed]

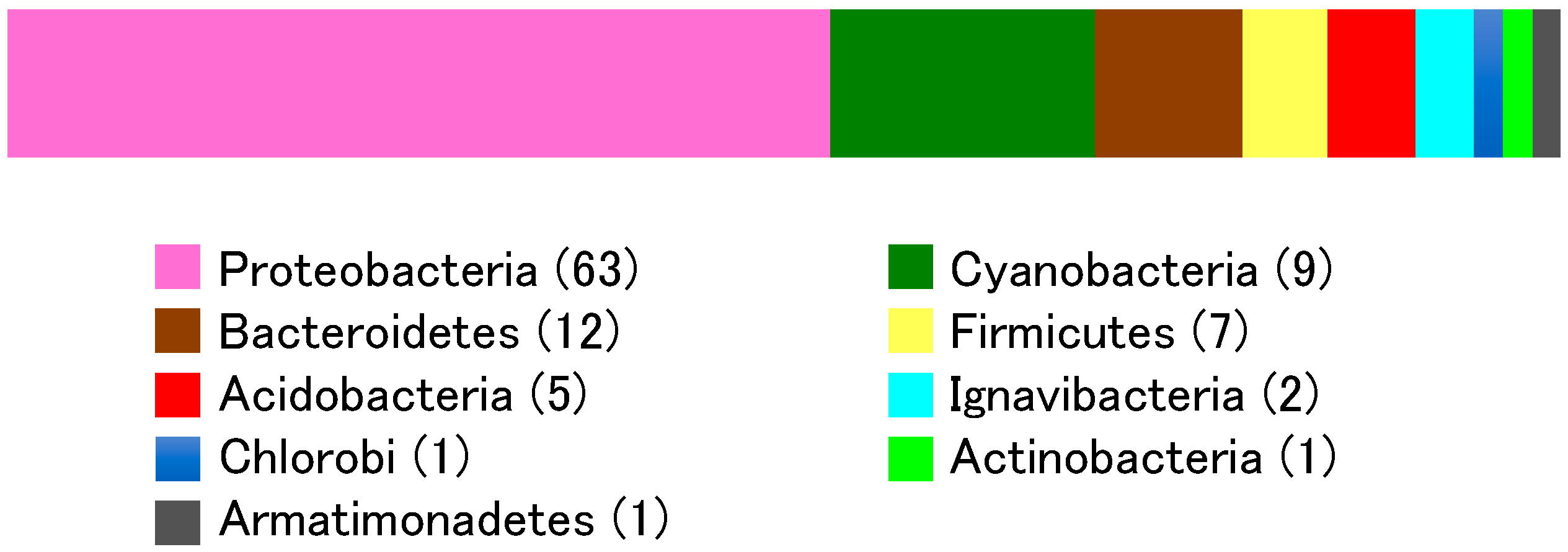

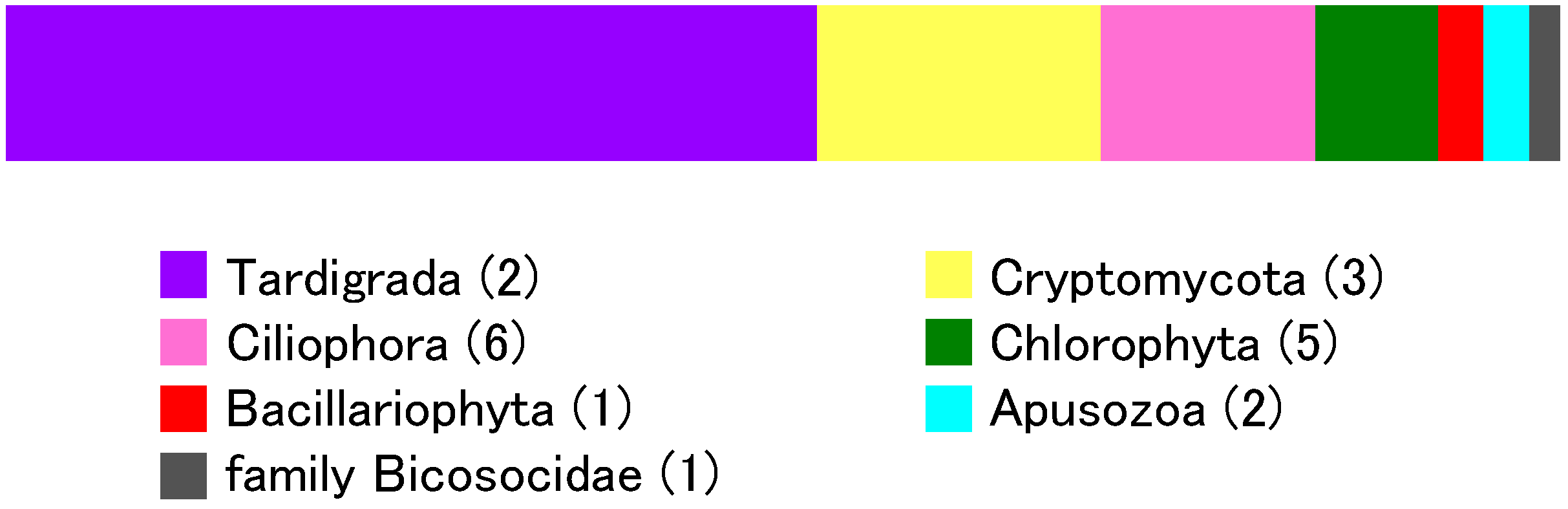
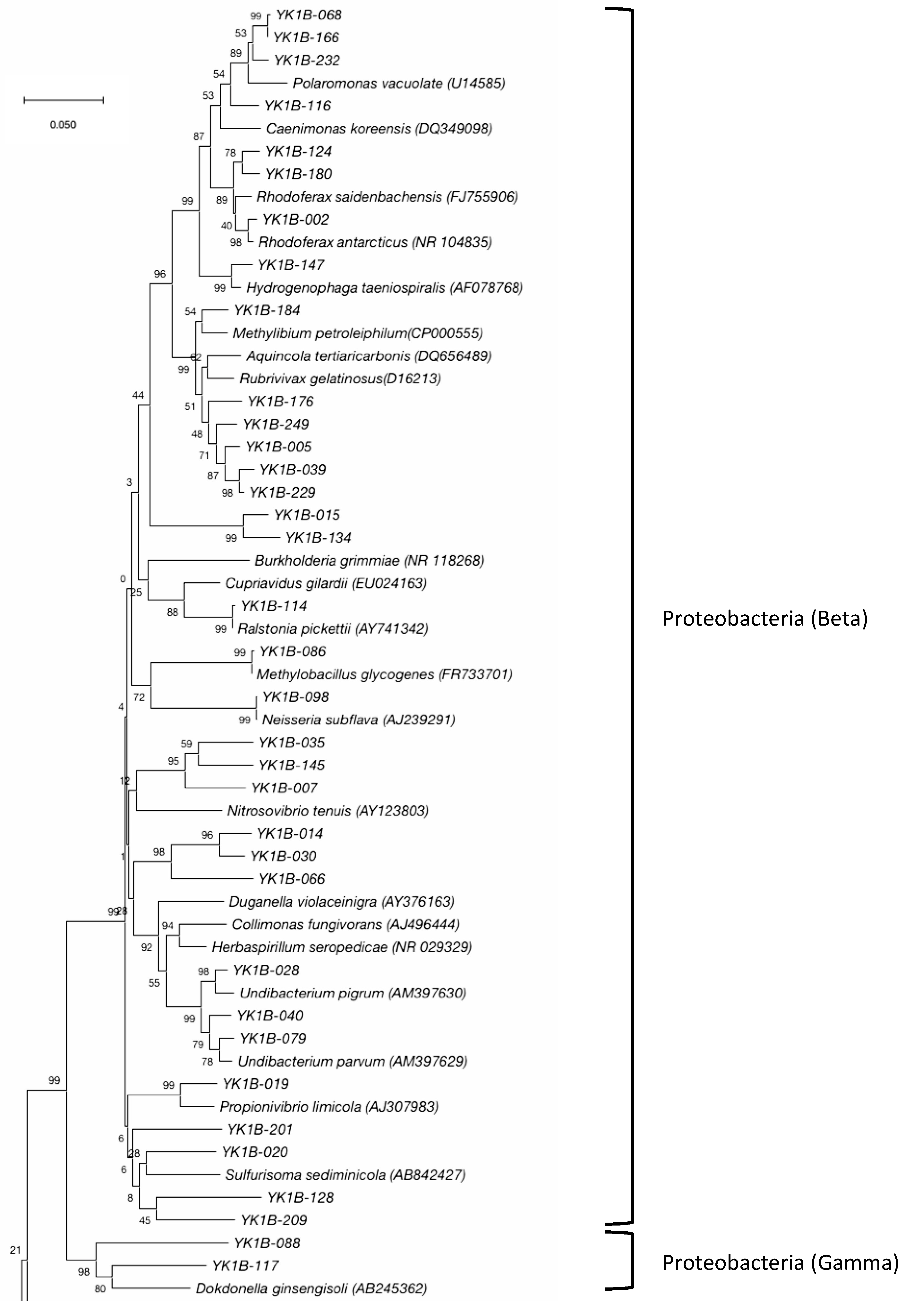
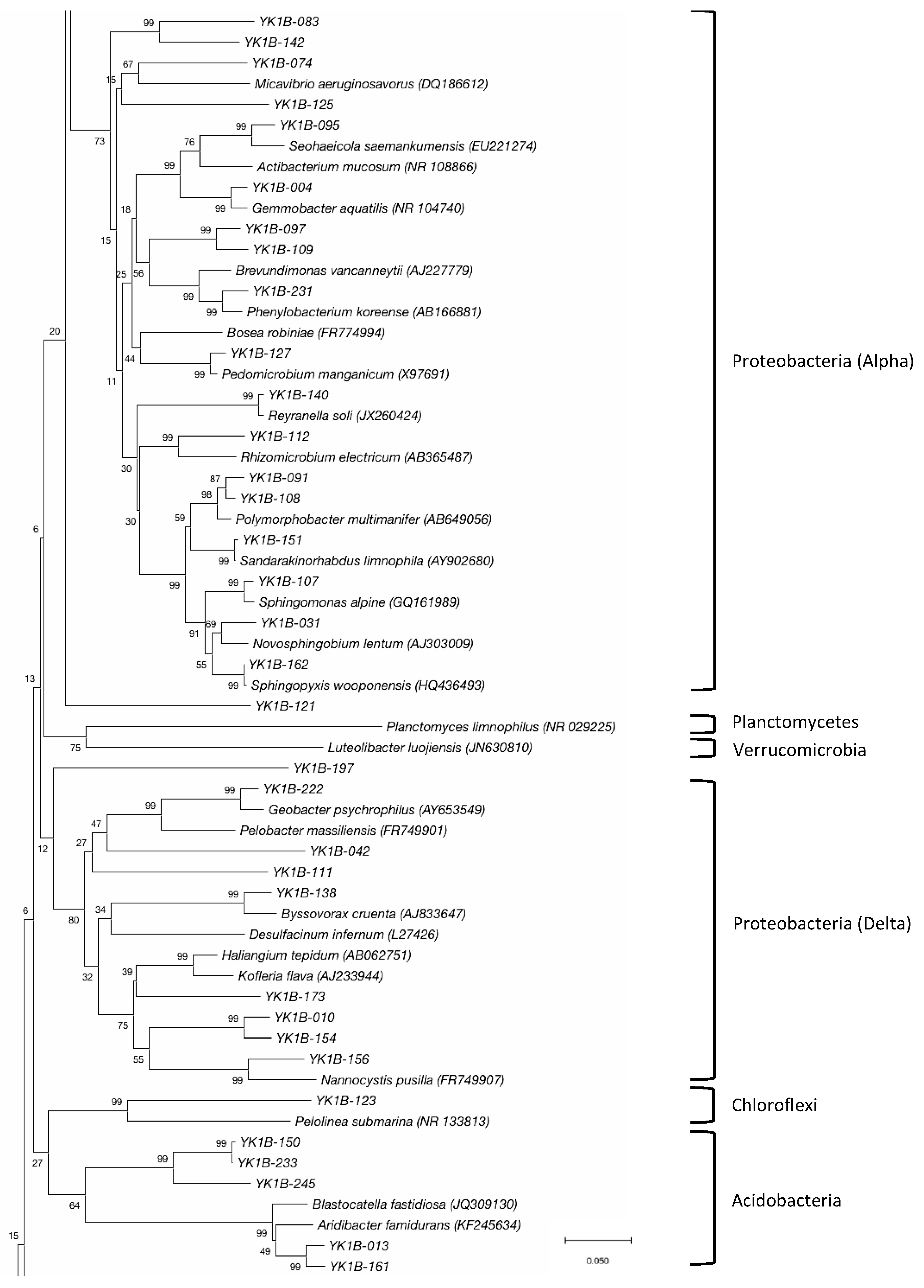

| Phylum | Class | Oder | Family | Genus | Number of Clones | Number of OTUs |
|---|---|---|---|---|---|---|
| Proteobacteria | α-Proteobacteria | Sphingomonadales | Sphingomonadaceae | Polymorphobacter | 3 | 2 |
| Novosphingobium | 2 | 1 | ||||
| Sphingomonas | 1 | 1 | ||||
| Sandarakinorhabdus | 1 | 1 | ||||
| Sphingopyxis | 1 | 1 | ||||
| Rhodobacterales | Rhodobacteraceae | Gemmobacter | 2 | 1 | ||
| Seohaeicola | 1 | 1 | ||||
| Rhodospirillales | No rank | Reyranella | 1 | 1 | ||
| Micropepsales | Micropepsaceae | Unclassified | 1 | 1 | ||
| Rhizobiales | Hyphomicrobiaceae | Pedomicrobium | 1 | 1 | ||
| Caulobacterales | Caulobacteraceae | Phenylobacterium | 1 | 1 | ||
| Unclassified | Unclassified | 2 | 2 | |||
| Unclassified | Unclassified | Unclassified | 6 | 4 | ||
| β-Proteobacteria | Burkholderiales | Comamonadaceae | Polaromonas | 8 | 3 | |
| Rhodoferax | 7 | 3 | ||||
| Hydrogenophaga | 1 | 1 | ||||
| Unclassified | 1 | 1 | ||||
| No rank | Methylibium | 1 | 1 | |||
| Burkholderiaceae | Ralstonia | 2 | 1 | |||
| Oxalobacteraceae | Undibacterium | 6 | 3 | |||
| Unclassified | Unclassified | 6 | 5 | |||
| Rhodocyclales | Rhodocyclaceae | Propionivibrio | 3 | 1 | ||
| Nitrosomonadales | Methylophilaceae | Methylobacillus | 1 | 1 | ||
| Unclassified | Unclassified | 8 | 7 | |||
| Neisseriales | Neisseriaceae | Neisseria | 1 | 1 | ||
| Unclassified | Unclassified | Unclassified | 7 | 5 | ||
| δ-Proteobacteria | Myxococcales | Polyangiaceae | Byssovorax | 1 | 1 | |
| Nannocystaceae | Unclassified | 2 | 1 | |||
| Unclassified | Unclassified | 3 | 3 | |||
| Desulfuromonadales | Geobacteraceae | Geobacter | 1 | 1 | ||
| Unclassified | Unclassified | 2 | 2 | |||
| γ-Proteobacteria | Xanthomonadales | Rhodanobacteraceae | Unclassified | 3 | 1 | |
| Unclassified | Unclassified | 1 | 1 | |||
| Unclassified | Unclassified | Unclassified | Unclassified | 2 | 2 | |
| Cyanobacteria | Cyanobacteria | Oscillatoriales | Gomontiellaceae | Crinalium | 12 | 1 |
| Oscillatoriaceae | Phormidium | 1 | 1 | |||
| Synechococcales | Leptolyngbyaceae | Leptolyngbya | 3 | 2 | ||
| Phormidesmis | 2 | 1 | ||||
| Unclassified | 6 | 1 | ||||
| Pseudanabaenaceae | Limnothrix | 1 | 1 | |||
| Chamaesiphonaceae | Chamaesiphon | 1 | 1 | |||
| Nostocales | Nostocaceae | Nostoc | 3 | 1 | ||
| Bacteroidetes | Cytophagia | Cytophagales | Cytophagaceae | Arcicella | 1 | 1 |
| Unclassified | Unclassified | 3 | 3 | |||
| Chitinophagia | Chitinophagales | Chitinophagaceae | Ferruginibacter | 1 | 1 | |
| Terrimonas | 3 | 1 | ||||
| Unclassified | 3 | 3 | ||||
| Unclassified | Unclassified | Unclassified | 6 | 3 | ||
| Firmicutes | Tissierellia | Tissierellales | Peptoniphilaceae | Anaerococcus | 3 | 2 |
| Unclassified | Unclassified | 1 | 1 | |||
| Clostridia | Clostridiales | Ruminococcaceae | Acetivibrio | 1 | 1 | |
| Clostridiaceae | Unclassified | 1 | 1 | |||
| Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | 3 | 1 | |
| Negativicutes | Veillonellales | Veillonellaceae | Veillonella | 1 | 1 | |
| Acidobacteria | Blastocatellia | Blastocatellales | Blastocatellaceae | Aridibacter | 5 | 2 |
| Unclassified | Unclassified | Unclassified | Unclassified | 4 | 3 | |
| Ignavibacteria | Unclassified | Unclassified | Unclassified | Unclassified | 2 | 2 |
| Chlorobi | Unclassified | Unclassified | Unclassified | Unclassified | 1 | 1 |
| Actinobacteria | Actinobacteria | Micrococcales | Microbacteriaceae | Microbacterium | 1 | 1 |
| Armatimonadetes | Chthonomonadetes | Unclassified | Unclassified | Unclassified | 1 | 1 |
| Total | 160 | 101 |
| Phylum | Class | Oder | Family | Genus | Number of Clones | Number of OTUs |
|---|---|---|---|---|---|---|
| Euryarchaeota | Methanomicrobia | Methanosarcinales | Methanosarcinaceae | Methanosarcina | 70 | 1 |
| Methanosaetaceae | Methanosaeta | 2 | 1 | |||
| Total | 72 | 2 |
| Phylum | Class | Oder | Family | Genus | Number of Clones | Number of OTUs |
|---|---|---|---|---|---|---|
| Tardigrada | Eutardigrada | Parachela | Hypsibiidae | Diphascon | 40 | 1 |
| Acutuncus | 8 | 1 | ||||
| Ciliophora | Stichotrichia | Sporadotrichida | Oxytrichidae | Onychodromopsis | 6 | 1 |
| Rigidocortex | 2 | 1 | ||||
| Oligohymenophorea | Peritrichia | Vorticellidae | Vorticellides | 2 | 1 | |
| Philasterida | Philasteridae | Unclassified | 1 | 1 | ||
| Litostomatea | Haptorida | Spathidiidae | Spathidium | 1 | 1 | |
| Phyllopharyngia | Chlamydodontida | Chilodonellidae | Phascolodon | 1 | 1 | |
| Bacillariophyta | Bacillariophyceae | Bacillariophycidae | Naviculales | Humidophila | 2 | 1 |
| no rank | no rank | Bicosoecida | Bicosoecidae | Unclassified | 1 | 1 |
| Chlorophyta | Chlorophyceae | Chlamydomonadales | Haematococcaceae | Chlorogonium | 3 | 1 |
| Chlamydomonadaceae | Oogamochlamys | 1 | 1 | |||
| Actinochloridaceae | Macrochloris | 1 | 1 | |||
| Unclassified | Unclassified | 2 | 2 | |||
| Cryptomycota (Fungi) | Unclassified | Unclassified | Unclassified | Unclassified | 11 | 1 |
| Unclassified | Unclassified | Unclassified | Unclassified | 5 | 1 | |
| Unclassified | Unclassified | Unclassified | Unclassified | 1 | 1 | |
| Apusozoa | no rank | no rank | Apusomonadidae | Apusomonas | 2 | 2 |
| Total | 90 | 20 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaya, A.; Kurosawa, N.; Kawamata, A.; Kosugi, M.; Imura, S. Community Structures of Bacteria, Archaea, and Eukaryotic Microbes in the Freshwater Glacier Lake Yukidori-Ike in Langhovde, East Antarctica. Diversity 2019, 11, 105. https://doi.org/10.3390/d11070105
Chaya A, Kurosawa N, Kawamata A, Kosugi M, Imura S. Community Structures of Bacteria, Archaea, and Eukaryotic Microbes in the Freshwater Glacier Lake Yukidori-Ike in Langhovde, East Antarctica. Diversity. 2019; 11(7):105. https://doi.org/10.3390/d11070105
Chicago/Turabian StyleChaya, Aoi, Norio Kurosawa, Akinori Kawamata, Makiko Kosugi, and Satoshi Imura. 2019. "Community Structures of Bacteria, Archaea, and Eukaryotic Microbes in the Freshwater Glacier Lake Yukidori-Ike in Langhovde, East Antarctica" Diversity 11, no. 7: 105. https://doi.org/10.3390/d11070105
APA StyleChaya, A., Kurosawa, N., Kawamata, A., Kosugi, M., & Imura, S. (2019). Community Structures of Bacteria, Archaea, and Eukaryotic Microbes in the Freshwater Glacier Lake Yukidori-Ike in Langhovde, East Antarctica. Diversity, 11(7), 105. https://doi.org/10.3390/d11070105






