Phylogenomic Reconstruction of the Neotropical Poison Frogs (Dendrobatidae) and Their Conservation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Bioinformatics Pipeline
2.3. Phylogenetic Analyses
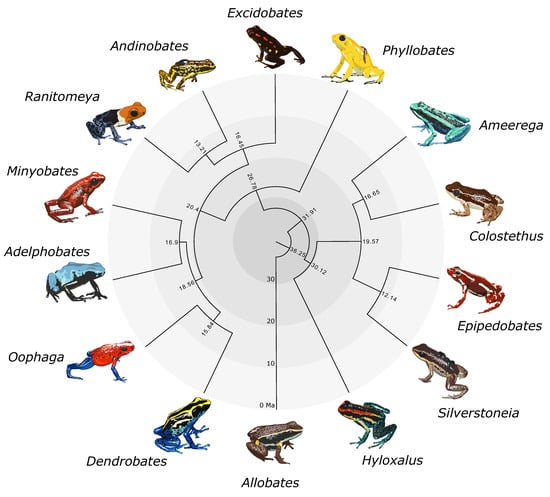
2.4. Divergence Time Estimation
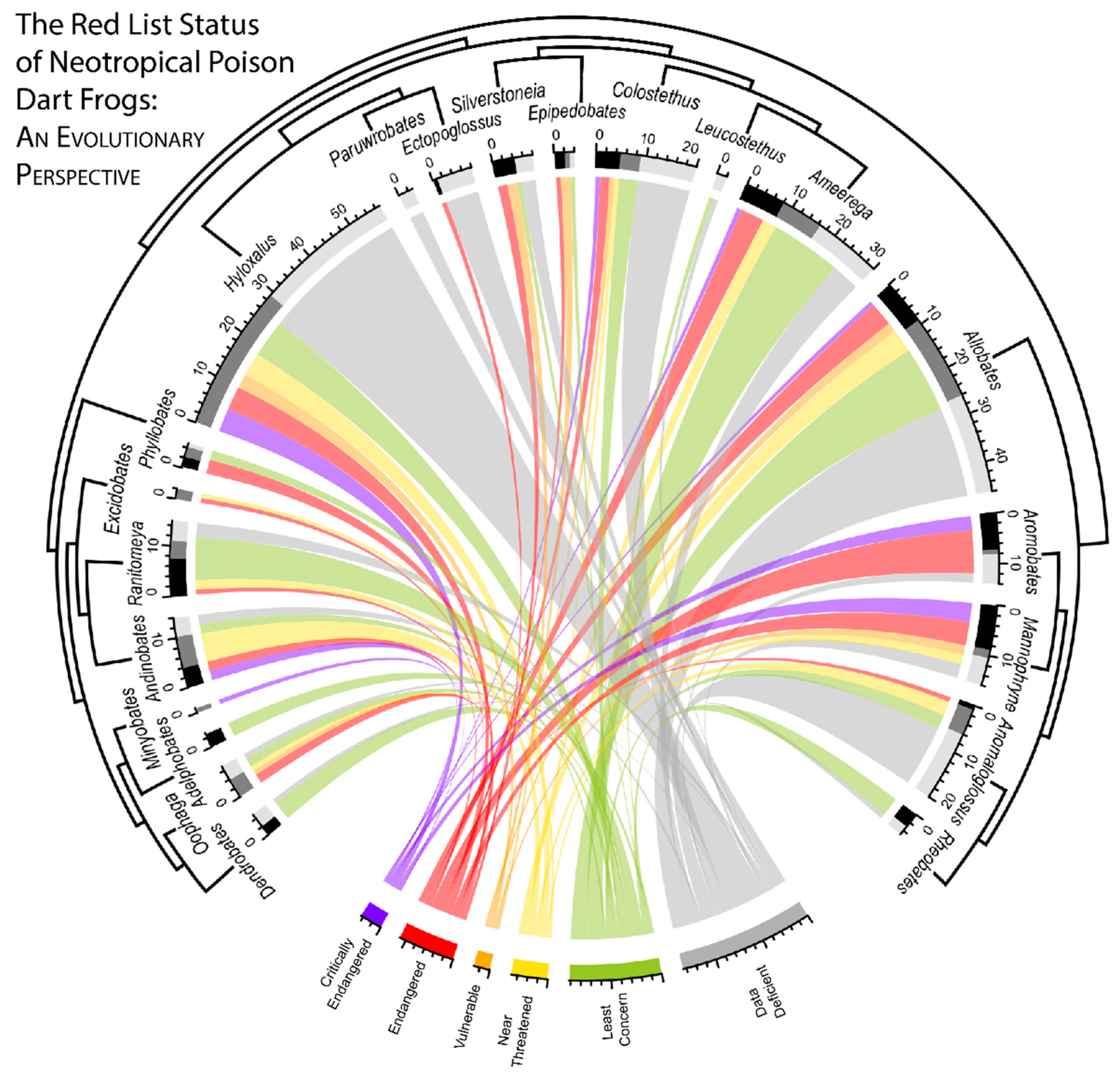
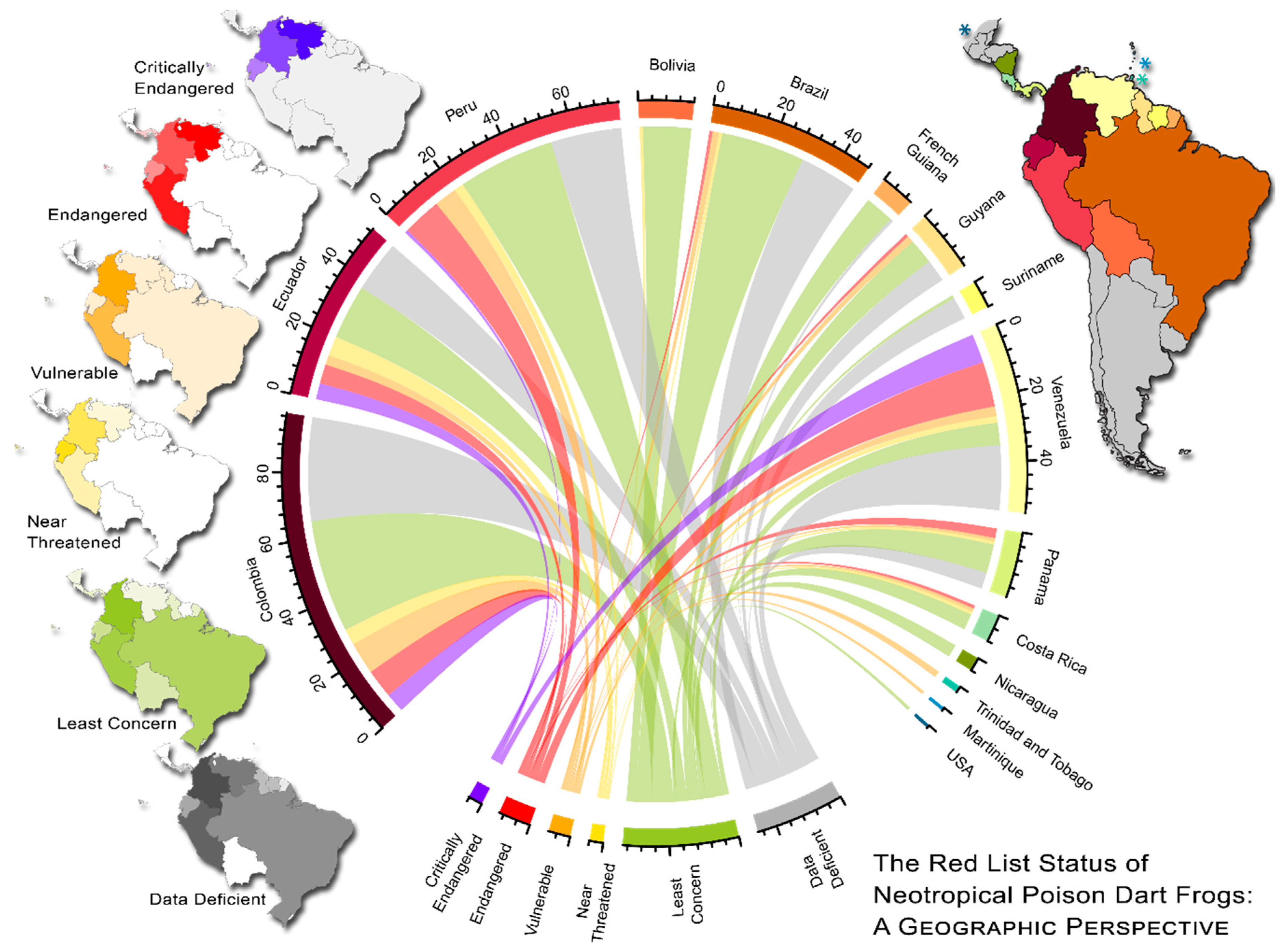
2.5. Visualizing Evolutionary History, Conservation Risk, and Spatial Distributions
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wells, K.D. Behavioral ecology and social organization of a dendrobatid frog (Colostethus inguinalis). Behav. Ecol. Sociobiol. 1980, 6, 199–209. [Google Scholar] [CrossRef]
- Summers, K.; Symula, R.; Clough, M.; Cronin, T. Visual mate choice in poison frogs. Proc. R. Soc. B Biol. Sci. 1999, 266, 2141–2145. [Google Scholar] [CrossRef] [Green Version]
- Daly, J.W.; Myers, C.W. Toxicity of Panamanian poison frogs (Dendrobates): Some biological and chemical aspects. Science 1967, 156, 970–973. [Google Scholar] [CrossRef]
- Daly, J.W.; McNeal, E.T.; Overman, L.E.; Ellison, D.H. A new class of cardiotonic agents: Structure-activity correlations for natural and synthetic analogs of the alkaloid pumiliotoxin B (8-hydroxy-8-methyl-6-alkylidene-1-azabicyclo [4.3.0] nonanes). J. Med. Chem. 1985, 28, 482–486. [Google Scholar] [CrossRef]
- Spande, T.F.; Garraffo, H.M.; Edwards, M.W.; Yeh, H.J.C.; Pannell, L.; Daly, J.W. Epibatidine: A novel (chloropyridyl) azabicycloheptane with potent analgesic activity from an Ecuadoran poison frog. J. Am. Chem. Soc. 1992, 114, 3475–3478. [Google Scholar] [CrossRef]
- Maan, M.E.; Cummings, M.E. Sexual dimorphism and directional sexual selection on aposematic signals in a poison frog. Proc. Natl. Acad. Sci. USA 2009, 106, 19072–19077. [Google Scholar] [CrossRef] [Green Version]
- Wang, I.J.; Shaffer, H.B. Rapid color evolution in an aposematic species: A phylogenetic analysis of color variation in the strikingly polymorphic strawberry poison-dart frog. Evolution 2008, 62, 2742–2759. [Google Scholar] [CrossRef]
- Santos, J.C.; Cannatella, D.C. Phenotypic integration emerges from aposematism and scale in poison frogs. Proc. Natl. Acad. Sci. USA 2011, 108, 6175–6180. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.L.; Twomey, E. Complicated histories: Three new species of poison frogs of the genus Ameerega (Anura: Dendrobatidae) from north-central Peru. Zootaxa 2009, 2049, 38. [Google Scholar] [CrossRef]
- Noonan, B.P.; Gaucher, P. Refugial isolation and secondary contact in the dyeing poison frog Dendrobates tinctorius. Mol. Ecol. 2006, 15, 4425–4435. [Google Scholar] [CrossRef]
- Gehara, M.; Summers, K.; Brown, J.L. Population expansion, isolation and selection: Novel insights on the evolution of color diversity in the strawberry poison frog. Evol. Ecol. 2013, 27, 797–824. [Google Scholar] [CrossRef]
- Young, R.L.; Ferkin, M.H.; Ockendon-Powell, N.F.; Orr, V.N.; Phelps, S.M.; Pogány, Á.; Richards-Zawacki, C.L.; Summers, K.; Székely, T.; Trainor, B.C.; et al. Conserved transcriptomic profiles underpin monogamy across vertebrates. Proc. Natl. Acad. Sci. USA 2019, 116, 1331–1336. [Google Scholar] [CrossRef] [Green Version]
- Tarvin, R.D.; Santos, J.C.; O’Connell, L.A.; Zakon, H.H.; Cannatella, D.C. Convergent substitutions in a sodium channel suggest multiple origins of toxin resistance in poison frogs. Mol. Biol. Evol. 2016, 33, 1068–1081. [Google Scholar] [CrossRef]
- Tarvin, R.D.; Borghese, C.M.; Sachs, W.; Santos, J.C.; Lu, Y.; O’Connell, L.A.; Cannatella, D.C.; Harris, R.A.; Zakon, H.H. Interacting amino acid replacements allow poison frogs to evolve epibatidine resistance. Science 2017, 357, 1261–1266. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, S.M.; Bell, K.E.; Philippi, T.; Sasa, M.; Bolaños, F.; Chaves, G.; Savage, J.M.; Donnelly, M.A. Amphibian and reptile declines over 35 years at La Selva, Costa Rica. Proc. Natl. Acad. Sci. USA 2007, 104, 8352–8356. [Google Scholar] [CrossRef] [Green Version]
- Angulo, A.; von May, R.; Icochea, J. A reassessment of the extinction risk of the critically endangered Oxapampa poison frog Ameerega planipaleae (Dendrobatidae). Oryx 2018, 53, 1–4. [Google Scholar] [CrossRef]
- Auliya, M.; García-Moreno, J.; Schmidt, B.R.; Schmeller, D.S.; Hoogmoed, M.S.; Fisher, M.C.; Pasmans, F.; Henle, K.; Bickford, D.; Martel, A. The global amphibian trade flows through Europe: The need for enforcing and improving legislation. Biodivers. Conserv. 2016, 25, 2581–2595. [Google Scholar] [CrossRef]
- Gorzula, S. The trade in dendrobatid frogs from 1987 to 1993. Herpetol. Rev. 1996, 27, 116–123. [Google Scholar]
- Cuvier, G.L.C.F.D. An VI. In Tableau Élémentaire de l’Histoire Naturelle des Animaux; Baudoin: Paris, France, 1797. [Google Scholar]
- Wagler, J.G. Natürliches System der Amphibien, mit Vorangehender Classification der Säugthiere und Vogel: Ein Neitrag zur Vergleichenden Zoologie; J.G. Cottasche Buchhandlung Nachfolger: Stuttgart, Germany, 1830. [Google Scholar]
- Grant, T.; Frost, D.R.; Caldwell, J.P.; Gagliardo, R.; Haddad, C.F.B.; Kok, P.J.R.; Means, D.B.; Noonan, B.P.; Schargel, W.E.; Wheeler, W.C. Phylogenetic systematics of dart-poison frogs and their relatives (Amphibia: Athesphatanura: Dendrobatidae). Bull. Am. Mus. Nat. Hist. 2006, 299, 1–262. [Google Scholar] [CrossRef]
- Vences, M.; Kosuch, J.; Lötters, S.; Widmer, A.; Jungfer, K.-H.; Köhler, J.; Veith, M. Phylogeny and classification of poison frogs (Amphibia: Dendrobatidae), Based on mitochondrial 16S and 12S ribosomal RNA gene sequences. Mol. Phylogenet. Evol. 2000, 15, 34–40. [Google Scholar] [CrossRef]
- Clough, M.; Summers, K. Phylogenetic systematics and biogeography of the poison frogs: Evidence from mitochondrial DNA sequences. Biol. J. Linn. Soc. 2000, 70, 515–540. [Google Scholar] [CrossRef]
- Widmer, A.; Lötters, S.; Jungfer, K.H. A molecular phylogenetic analysis of the neotropical dart-poison frog genus Phyllobates (Amphibia: Dendrobatidae). Naturwissenschaften 2000, 87, 559–562. [Google Scholar] [CrossRef]
- Summers, K.; Clough, M.E. The evolution of coloration and toxicity in the poison frog family (Dendrobatidae). Proc. Natl. Acad. Sci. USA 2001, 98, 6227–6232. [Google Scholar] [CrossRef] [Green Version]
- La Marca, E.; Vences, M.; Lötters, S. Rediscovery and mitochondrial relationships of the dendrobatid frog Colostethus humilis suggest parallel colonization of the Venezuelan Andes by poison frogs. Stud. Neotrop. Fauna Environ. 2002, 37, 233–240. [Google Scholar] [CrossRef]
- Santos, J.C.; Coloma, L.A.; Cannatella, D.C. Multiple, recurring origins of aposematism and diet specialization in poison frogs. Proc. Natl. Acad. Sci. USA 2003, 100, 12792–12797. [Google Scholar] [CrossRef] [Green Version]
- Vences, M. Convergent evolution of aposematic coloration in Neotropical poison frogs: A molecular phylogenetic perspective. Org. Divers. Evol. 2003, 3, 215–226. [Google Scholar] [CrossRef]
- Roberts, J.L.; Brown, J.L.; von May, R.; Arizabal, W.; Presar, A.; Symula, R.; Schulte, R.; Summers, K. Phylogenetic relationships among poison frogs of the genus Dendrobates (Dendrobatidae): A molecular perspective from increased taxon sampling. 2006, 16, 377–385. 16.
- Frost, D.R.; Grant, T.; Faivovich, J.; Bain, R.H.; Haas, A.; Haddad, C.F.B.; De Sá, R.O.; Channing, A.; Wilkinson, M.; Donnellan, S.C.; et al. The amphibian tree of life. Bull. Am. Mus. Nat. Hist. 2006, 297, 1–291. [Google Scholar] [CrossRef]
- Maddison, W.P. Gene trees in species trees. Syst. Biol. 1997, 46, 523–536. [Google Scholar] [CrossRef]
- Degnan, J.H.; Rosenberg, N.A. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 2009, 24, 332–340. [Google Scholar] [CrossRef]
- Frost, D.R. Amphibian Species of the World 6.0. Available online: http://research.amnh.org/vz/herpetology/amphibia/ (accessed on 19 July 2019).
- Twomey, E.; Brown, J.L.; Amézquita, A.; Mejía-Vargas, D. A taxonomic revision of the Neotropical poison frog genus Ranitomeya (Amphibia: Dendrobatidae). Zootaxa 2011, 3083, 20–39. [Google Scholar]
- Twomey, E.; Brown, J.L. Spotted poison frogs: Rediscovery of a lost species and a new genus (Anura: Dendrobatidae) from northwestern Peru. Herpetologica 2008, 64, 121–137. [Google Scholar] [CrossRef]
- Myers, C.W. New generic names from some neotropical poison frogs (Dendrobatidae). Papéis Avulsos Zool. Mus. Zool. Univ. São Paulo 1987, 36, 301–306. [Google Scholar]
- Bauer, L. New names in the family Dendrobatidae (Anura, Amphibia). RIPA 1994, fall, 1–6. [Google Scholar]
- Bibron, G. pl. 29bis. Type species: Phyllobates bicolor Bibron, 1840, by monotypy. In Atlas de Zoología de Historia Fisica, Politica y Natural de la Isla de Cuba, Segunda Part. Historia Natural; Arthur Bertrand: Paris, France, 1840; p. 8. [Google Scholar]
- Bauer, L. A new genus and a new specific name in the dart poison frog family (Dendrobatidae, Anura, Amphibia). RIPA 1986, 1986, 1–12. [Google Scholar]
- Cope, E.D. Fourth contribution to the herpetology of tropical America. Proc. Acad. Nat. Sci. Phila. 1866, 123–132. [Google Scholar]
- Grant, T.; Rada, M.; Anganoy-Criollo, M.; Batista, A.; Dias, P.H.; Jeckel, A.M.; Machado, D.J.; Rueda-Almonacid, J.V. Phylogenetic systematics of dart-poison frogs and their relatives revisited (Anura: Dendrobatoidea). South. Am. J. Herpetol. 2017, 12, S1–S90. [Google Scholar] [CrossRef]
- Jiménez de la Espada, M. Fauna neotropicalis species quaedam nondum cognitae. J. Sci. Math. Phys. E Nat. 1870, 3, 57–65. [Google Scholar]
- Brown, J.L.; Twomey, E.; Amézquita, A.; de Souza, M.B.; Caldwell, J.P.; Lötters, S.; von May, R.; Melo-Sampaio, P.R.; Mejía-Vargas, D.; Perez-Peña, P.; et al. A taxonomic revision of the Neotropical poison frog genus Ranitomeya (Amphibia: Dendrobatidae). Zootaxa 2011, 3083, 1–120. [Google Scholar] [CrossRef]
- Santos, J.C.; Coloma, L.A.; Summers, K.; Caldwell, J.P.; Ree, R.; Cannatella, D.C. Amazonian amphibian diversity Is primarily derived from late Miocene Andean lineages. PLoS Biol. 2009, 7, e1000056. [Google Scholar] [CrossRef]
- Pyron, A.R.; Wiens, J.J. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 2011, 61, 543–583. [Google Scholar] [CrossRef]
- Faircloth, B.C.; McCormack, J.E.; Crawford, N.G.; Harvey, M.G.; Brumfield, R.T.; Glenn, T.C. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 2012, 61, 717–726. [Google Scholar] [CrossRef]
- Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, S.; Kent, W.J.; Mattick, J.S.; Haussler, D. Ultraconserved elements in the human genome. Science 2004, 304, 1321–1325. [Google Scholar] [CrossRef]
- Siepel, A.; Bejerano, G.; Pedersen, J.S.; Hinrichs, A.S.; Hou, M.; Rosenbloom, K.; Clawson, H.; Spieth, J.; Hillier, L.W.; Richards, S.; et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005, 15, 1034–1050. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, P.S.; Chang, J.; Pan, C.; Sobel, E.; Sinsheimer, J.S.; Faircloth, B.; Alfaro, M.E. Genome-wide ultraconserved elements exhibit higher phylogenetic informativeness than traditional gene markers in percomorph fishes. Mol. Phylogenet. Evol. 2015, 92, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Blaimer, B.B.; Lloyd, M.W.; Guillory, W.X.; Brady, S.G. Sequence capture and phylogenetic utility of genomic ultraconserved elements obtained from pinned insect specimens. PLoS ONE 2016, 11, e0161531. [Google Scholar] [CrossRef]
- McCormack, J.E.; Tsai, W.L.E.; Faircloth, B.C. Sequence capture of ultraconserved elements from bird museum specimens. Mol. Ecol. Resour. 2016, 16, 1189–1203. [Google Scholar] [CrossRef]
- Smith, B.T.; Harvey, M.G.; Faircloth, B.C.; Glenn, T.C.; Brumfield, R.T. Target capture and massively parallel sequencing of ultraconserved elements for comparative studies at shallow evolutionary time scales. Syst. Biol. 2014, 63, 83–95. [Google Scholar] [CrossRef]
- Crawford, N.G.; Faircloth, B.C.; McCormack, J.E.; Brumfield, R.T.; Winker, K.; Glenn, T.C. More than 1000 ultraconserved elements provide evidence that turtles are the sister group of archosaurs. Biol. Lett. 2012, 8, 783–786. [Google Scholar] [CrossRef] [Green Version]
- Crawford, N.G.; Parham, J.F.; Sellas, A.B.; Faircloth, B.C.; Glenn, T.C.; Papenfuss, T.J.; Henderson, J.B.; Hansen, M.H.; Simison, W.B. A phylogenomic analysis of turtles. Mol. Phylogenet. Evol. 2015, 83, 250–257. [Google Scholar] [CrossRef]
- Streicher, J.W.; Wiens, J.J. Phylogenomic analyses of more than 4000 nuclear loci resolve the origin of snakes among lizard families. Biol. Lett. 2017, 13, 20170393. [Google Scholar] [CrossRef]
- Streicher, J.W.; Wiens, J.J. Phylogenomic analyses reveal novel relationships among snake families. Mol. Phylogenet. Evol. 2016, 100, 160–169. [Google Scholar] [CrossRef] [Green Version]
- Streicher, J.W.; Miller, E.C.; Guerrero, P.C.; Correa, C.; Ortiz, J.C.; Crawford, A.J.; Pie, M.R.; Wiens, J.J. Evaluating methods for phylogenomic analyses, and a new phylogeny for a major frog clade (Hyloidea) based on 2214 loci. Mol. Phylogenet. Evol. 2018, 119, 128–143. [Google Scholar] [CrossRef]
- McCormack, J.E.; Faircloth, B.C.; Crawford, N.G.; Gowaty, P.A.; Brumfield, R.T.; Glenn, T.C. Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis. Genome Res. 2012, 22, 746–754. [Google Scholar] [CrossRef]
- Derti, A.; Roth, F.P.; Church, G.M.; Wu, C. Mammalian ultraconserved elements are strongly depleted among segmental duplications and copy number variants. Nat. Genet. 2006, 38, 1216–1220. [Google Scholar] [CrossRef]
- Simons, C.; Pheasant, M.; Makunin, I.V.; Mattick, J.S. Transposon-free regions in mammalian genomes. Genome Res. 2006, 16, 164–172. [Google Scholar] [CrossRef]
- Faircloth, B.C. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 2016, 32, 786–788. [Google Scholar] [CrossRef]
- Faircloth, B.C. Illumiprocessor: A trimmomatic wrapper for parallel adapter and quality trimming. 2013. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Heibl, C. PHYLOCH: R Language Tree Plotting Tools and Interfaces to Diverse Phylogenetic Software Packages. 2008. Available online: http://www.christophheibl.de/Rpackages.html (accessed on 9 June 2019).
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 2018, 19, 153. [Google Scholar] [CrossRef]
- Persons, N.W.; Hosner, P.A.; Meiklejohn, K.A.; Braun, E.L.; Kimball, R.T. Sorting out relationships among the grouse and ptarmigan using intron, mitochondrial, and ultra-conserved element sequences. Mol. Phylogenet. Evol. 2016, 98, 123–132. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Ješovnik, A.; Sosa-Calvo, J.; Lloyd, M.W.; Branstetter, M.G.; Fernández, F.; Schultz, T.R. Phylogenomic species delimitation and host-symbiont coevolution in the fungus-farming ant genus Sericomyrmex Mayr (Hymenoptera: Formicidae): Ultraconserved elements (UCEs) resolve a recent radiation. Syst. Entomol. 2017, 42, 523–542. [Google Scholar] [CrossRef]
- Drummond, A.J.; Bouckaert, R. Bayesian Evolutionary Analysis with BEAST 2; Cambridge University Press: Cambridge, UK, 2015. [Google Scholar]
- Winker, K.; Glenn, T.C.; Faircloth, B.C. Ultraconserved elements (UCEs) illuminate the population genomics of a recent, high-latitude avian speciation event. PeerJ 2018, 6, e5735. [Google Scholar] [CrossRef] [Green Version]
- Hsiang, A.Y.; Field, D.J.; Webster, T.H.; Behlke, A.D.; Davis, M.B.; Racicot, R.A.; Gauthier, J.A. The origin of snakes: Revealing the ecology, behavior, and evolutionary history of early snakes using genomics, phenomics, and the fossil record. BMC Evol. Biol. 2015, 15, 87. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- IUCN 2019. The IUCN Red List of Threatened Species. Version 2019-1. Available online: http://www.iucnredlist.org (accessed on 21 March 2019).
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Swofford, D.L.; Waddell, P.J.; Huelsenbeck, J.P.; Foster, P.G.; Lewis, P.O.; Rogers, J.S. Bias in phylogenetic estimation and its relevance to the choice between parsimony and likelihood methods. Syst. Biol. 2001, 50, 525–539. [Google Scholar] [CrossRef]
- Felsenstein, J. Parsimony in systematics: Biological and statistical issues. Annu. Rev. Ecol. Syst. 1983, 14, 313–333. [Google Scholar] [CrossRef]
- Felsenstein, J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Biol. 1978, 27, 401–410. [Google Scholar] [CrossRef]
- Dos Reis, M.; Donoghue, P.C.J.; Yang, Z. Bayesian molecular clock dating of species divergences in the genomics era. Nat. Rev. Genet. 2016, 17, 71–80. [Google Scholar] [CrossRef]
- Thorne, J.L.; Kishino, H. Divergence time and evolutionary rate estimation with multilocus data. Syst. Biol. 2002, 51, 689–702. [Google Scholar] [CrossRef]
- Schenk, J.J. Consequences of secondary calibrations on divergence time estimates. PLoS ONE 2016, 11, e0148228. [Google Scholar] [CrossRef]
- Rosauer, D.; Laffan, S.W.; Crisp, M.D.; Donnellan, S.C.; Cook, L.G. Phylogenetic endemism: A new approach for identifying geographical concentrations of evolutionary history. Mol. Ecol. 2009, 18, 4061–4072. [Google Scholar] [CrossRef]
- González-del-Pliego, P.; Freckleton, R.P.; Edwards, D.P.; Koo, M.S.; Scheffers, B.R.; Pyron, R.A.; Jetz, W. Phylogenetic and trait-based prediction of extinction risk for data-deficient amphibians. Curr. Biol. 2019, 29, 1557–1563. [Google Scholar] [CrossRef]
| Subfamily | Genus | Authority | Type Species | No. Species | Range |
|---|---|---|---|---|---|
| Dendrobatinae | Adelphobates | Grant et al. 2006 [21] | castaneoticus | 3 | Amazonia |
| Andinobates | Twomey et al. 2011 [34] | bombetes | 15 | N Amazonia | |
| Dendrobates | Wagler 1830 [20] | tinctorius | 5 | C America, N Amazonia, Hawaii (introduced) | |
| Excidobates | Twomey and Brown 2008 [35] | mysteriosus | 3 | NW Peru | |
| Minyobates | Myers 1987 [36] | steyermarki | 1 | Venezuela | |
| Oophaga | Bauer 1994 [37] | pumilio | 12 | C America, W Andean versant | |
| Phyllobates | Bibron 1840 [38] | bicolor | 5 | C America, Colombia | |
| Ranitomeya | Bauer 1986 [39] | reticulata | 16 | Amazonia | |
| Colostethinae | Ameerega | Bauer 1986 [39] | trivittata | 30 | Amazonia, Bolivia |
| Colostethus | Cope 1866 [40] | latinasus | 15 | Panama, NW S America | |
| Epipedobates | Myers 1987 [36] | tricolor | 8 | W Andean versant | |
| Leucostethus | Grant et al. 2017 [41] | argyrogaster | 6 | W Amazonia | |
| Silverstoneia | Grant et al. 2006 [21] | nubicola | 8 | C America, Colombia | |
| Hyloxalinae | Ectopoglossus | Grant et al. 2017 [41] | saxatilis | 7 | C America, W Andean versant |
| Hyloxalus | Jiménez de la Espada 1870 [42] | fuliginosus | 60 | Panama, W Andean versant, NW Amazonia | |
| Paruwrobates | Bauer 1994 [37] | andinus | 3 | W Andean versant |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guillory, W.X.; Muell, M.R.; Summers, K.; Brown, J.L. Phylogenomic Reconstruction of the Neotropical Poison Frogs (Dendrobatidae) and Their Conservation. Diversity 2019, 11, 126. https://doi.org/10.3390/d11080126
Guillory WX, Muell MR, Summers K, Brown JL. Phylogenomic Reconstruction of the Neotropical Poison Frogs (Dendrobatidae) and Their Conservation. Diversity. 2019; 11(8):126. https://doi.org/10.3390/d11080126
Chicago/Turabian StyleGuillory, Wilson X., Morgan R. Muell, Kyle Summers, and Jason L. Brown. 2019. "Phylogenomic Reconstruction of the Neotropical Poison Frogs (Dendrobatidae) and Their Conservation" Diversity 11, no. 8: 126. https://doi.org/10.3390/d11080126
APA StyleGuillory, W. X., Muell, M. R., Summers, K., & Brown, J. L. (2019). Phylogenomic Reconstruction of the Neotropical Poison Frogs (Dendrobatidae) and Their Conservation. Diversity, 11(8), 126. https://doi.org/10.3390/d11080126