Genetic Diversity and Population Structure of the Spring Orchid Cymbidium goeringii in Korean Distant Islands
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Investigation
2.2. DNA Purification and Microsatellite Genotyping
2.3. Determination of Genetic Diversity and Genetic Distance
2.4. Determination of Population Structure
3. Results
3.1. Comparison of Genetic Diversity among Spring Orchids from Three Regions
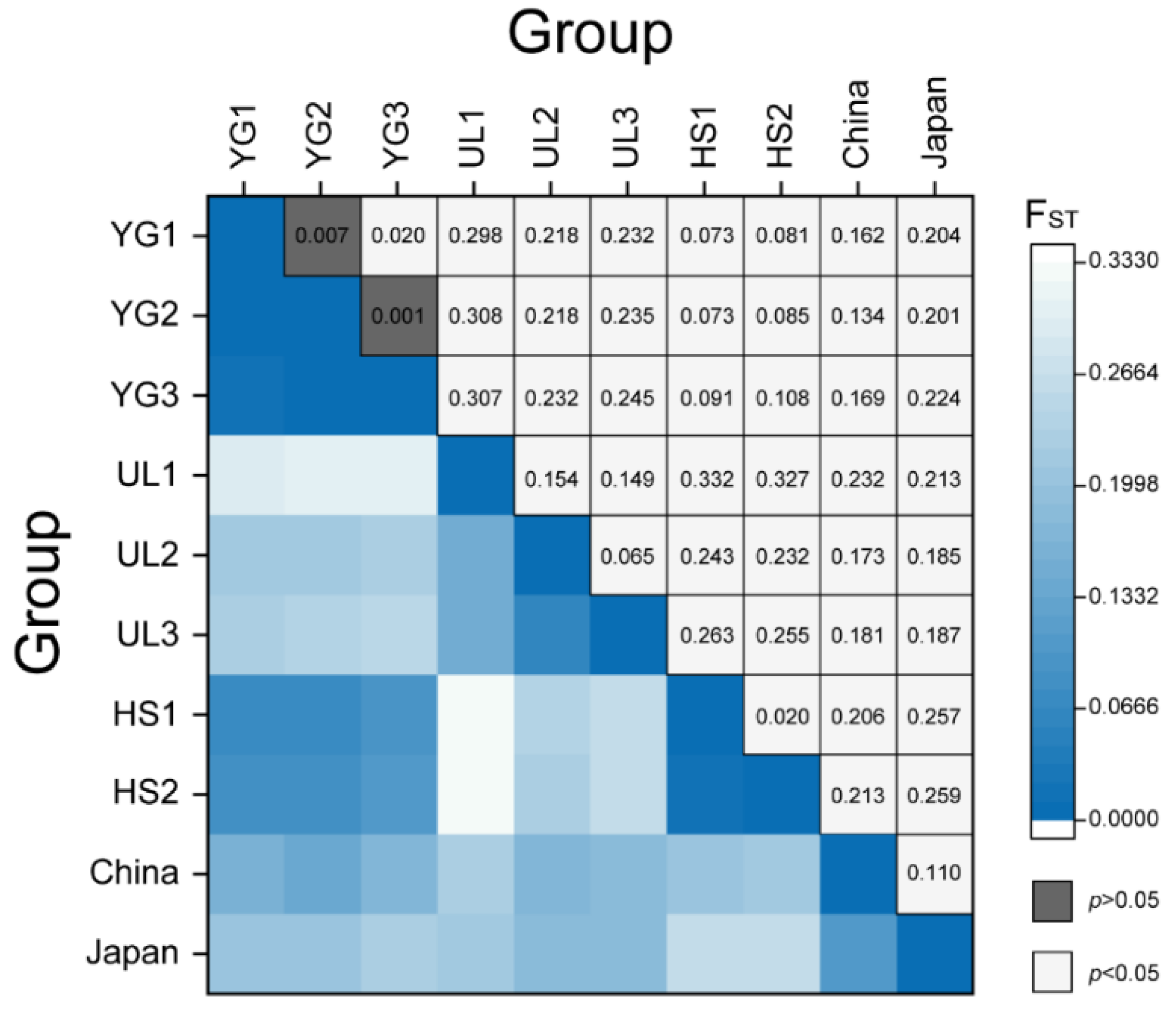
3.2. Pairwise Comparison of the Population Differentiation in Five Sites
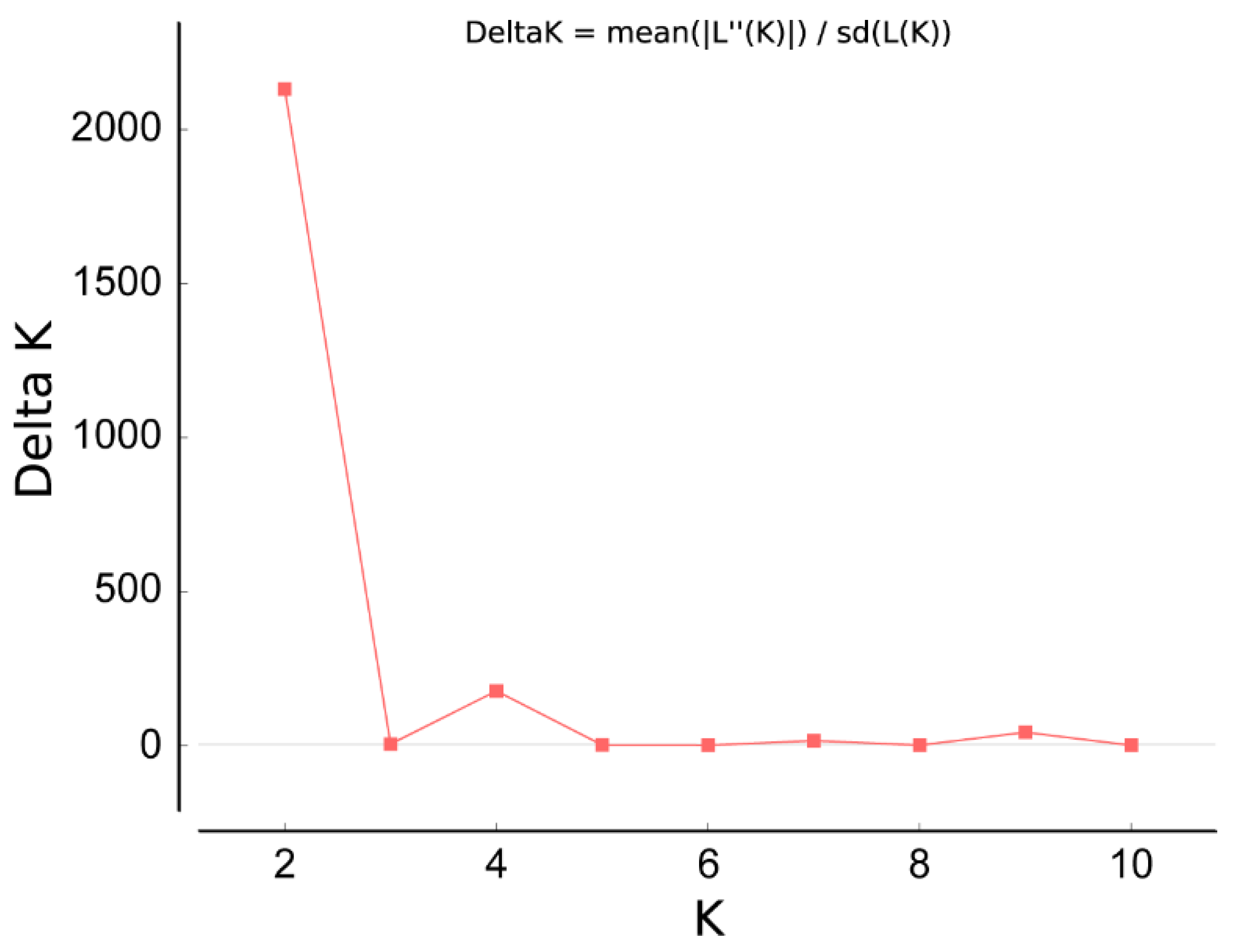
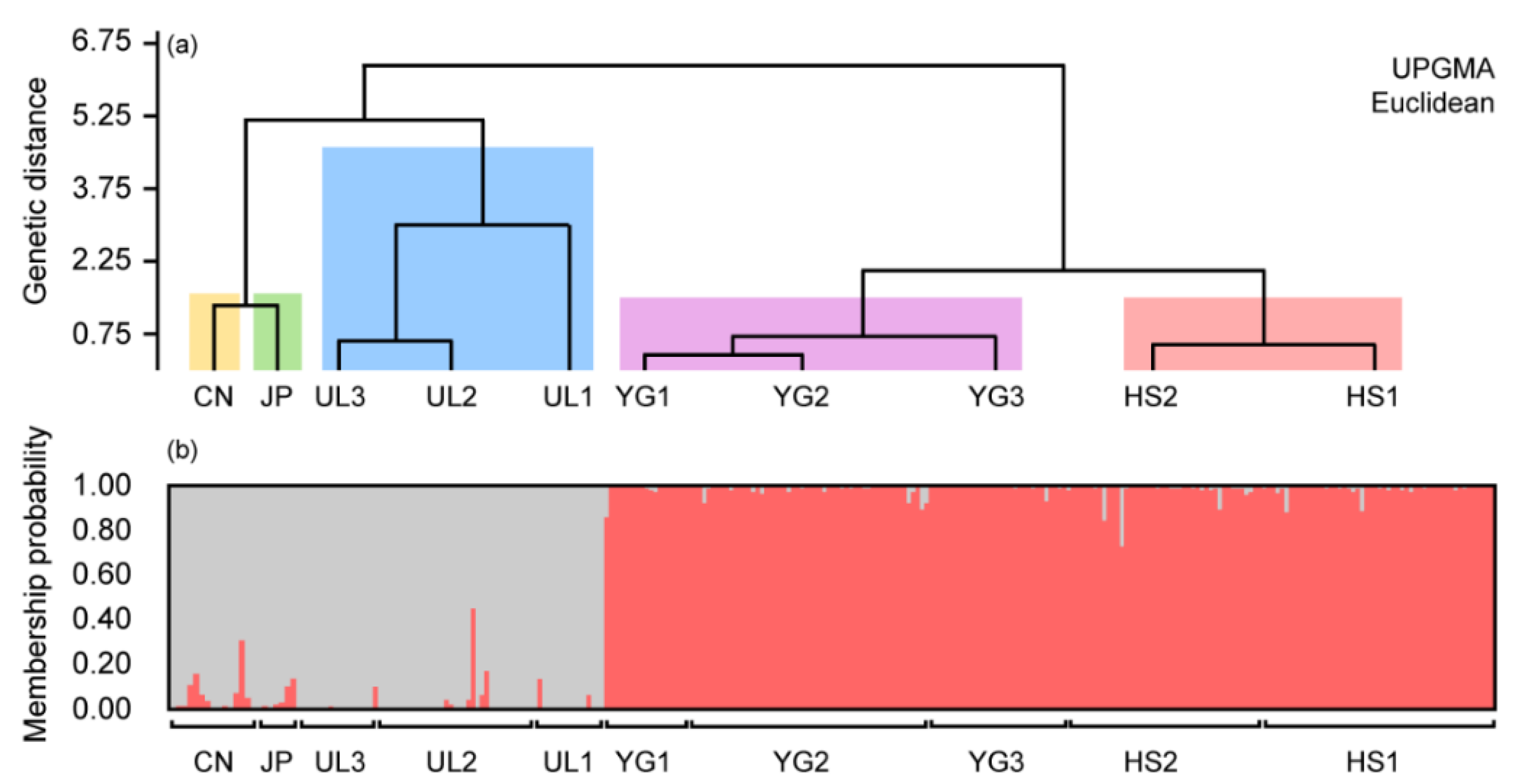
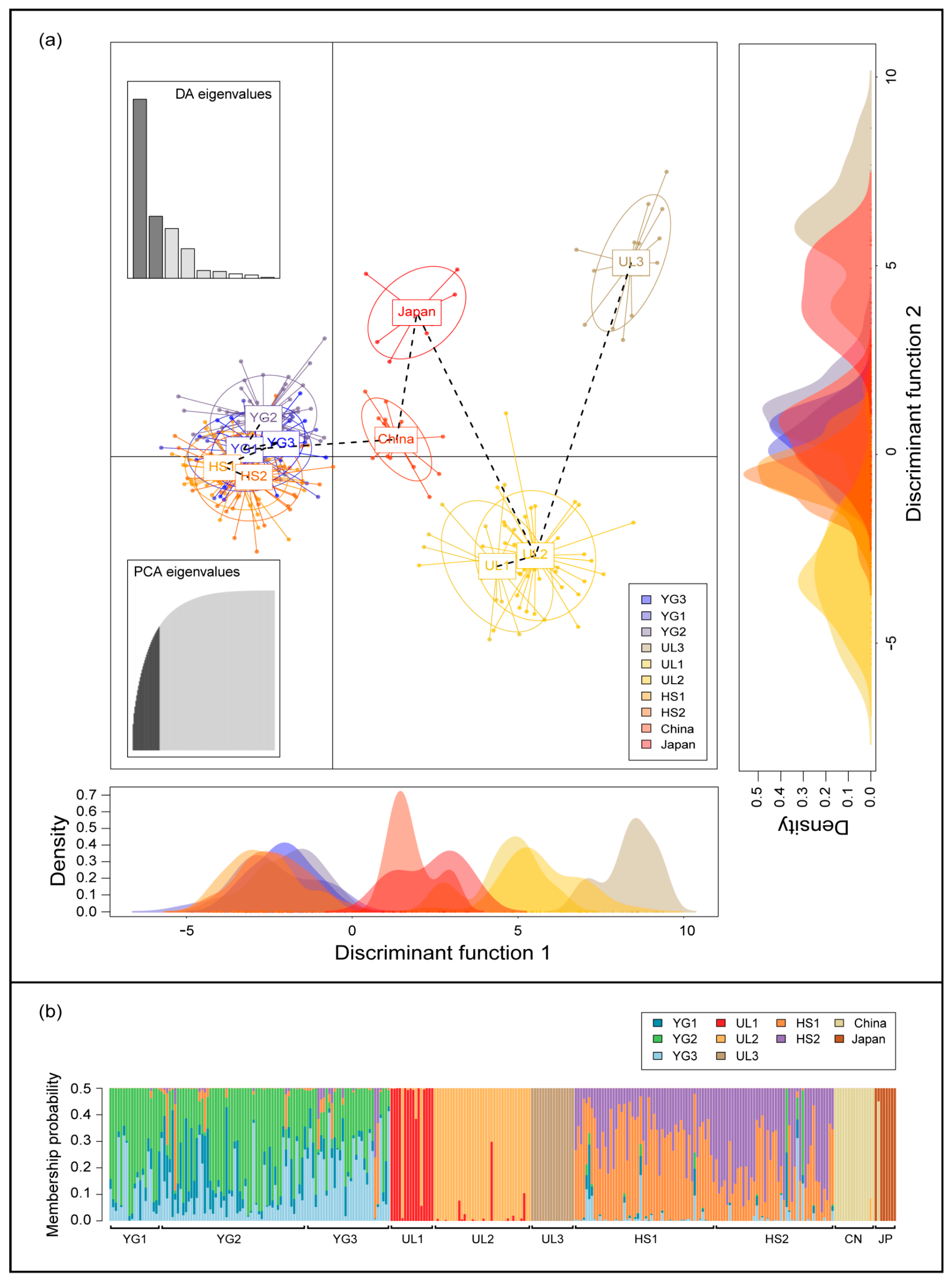
3.3. Analysis of Population Structure from Five Sites
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DuPuy, D.; Cribb, P.J. The Genus Cymbidium; Christopher Helm: London, UK, 1988. [Google Scholar]
- Dressler, R.L. Phylogeny and Classification of the Orchid Family; Cambridge University Press: Cambridge, UK, 1993. [Google Scholar]
- Obara-Okeyo, P.; Kako, S. Genetic diversity and identification of Cymbidium cultivars as measured by random amplified polymorphic DNA (RAPD) markers. Euphytica 1998, 99, 95–101. [Google Scholar] [CrossRef]
- Suetsugu, K. Autonomous self-pollination and insect visitors in partially and fully mycoheterotrophic species of Cymbidium (Orchidaceae). J. Plant Res. 2015, 128, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Moe, K.T.; Zhao, W.; Song, H.S.; Kim, Y.H.; Chung, J.W.; Cho, Y.I.; Park, P.H.; Park, H.S.; Chae, S.C.; Park, Y.J. Development of SSR markers to study diversity in the genus Cymbidium. Biochem. Syst. Ecol. 2010, 38, 585–594. [Google Scholar] [CrossRef]
- Huang, J.L.; Zeng, C.X.; Li, H.T.; Yang, J.B. Isolation and characterization of 15 microsatellite markers from the spring orchid (Cymbidium goeringii) (Orchidaceae). Am. J. Bot. 2011, 98, e76–e77. [Google Scholar] [CrossRef]
- Hyun, Y.S.; Kim, J.; Chung, K.W. Development of polymorphic microsatellite markers for Cymbidium goeringii (Orchidaceae). Am. J. Bot. 2012, 99, e193–e198. [Google Scholar] [CrossRef]
- Tang, M.; Zeng, C.X.; Bi, Y.F.; Yang, J.B. Microsatellite markers for the Chinese endangered and endemic orchid Cymbidium tortisepalum (Orchidaceae). Am. J. Bot. 2012, 99, e11–e13. [Google Scholar] [CrossRef]
- Li, X.; Jin, F.; Jin, L.; Jackson, A.; Huang, C.; Li, K.; Shu, X. Development of Cymbidium ensifolium genic-SSR markers and their utility in genetic diversity and population structure analysis in cymbidiums. BMC Genet. 2014, 15, 124. [Google Scholar] [CrossRef]
- Liu, X.; Huang, Y.; Li, F.; Xu, C.; Chen, K. Genetic diversity of 129 spring orchid (Cymbidium goeringii) cultivars and its relationship to horticultural types as assessed by EST-SSR markers Author links open overlay panel. Sci. Hort. 2014, 174, 178–184. [Google Scholar] [CrossRef]
- Lee, H.J.; Park, H.R.; Lee, A.J.; Nam, D.E.; Lee, D.G.; Do, Y.; Chung, K.W. Genetic authentication of cultivars with flower-variant types using SSR markers in spring orchid Cymbidium goeringii. Hort. Environ. Biotechnol. 2020, 61, 577–590. [Google Scholar] [CrossRef]
- Kang, D.; Kim, J.E. Fine, ultrafine, and yellow dust: Emerging health problems in Korea. J. Korean Med. Sci. 2014, 29, 621–622. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F. Genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Peakall, P.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A. Structure harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Waraniak, J.M.; Fisher, J.D.L.; Purcell, K.; Mushet, D.M.; Stockwell, C.A. Landscape genetics reveal broad and fine-scale population structure due to landscape features and climate history in the northern leopard frog (Rana pipiens) in North Dakota. Ecol. Evol. 2019, 9, 1041–1060. [Google Scholar] [CrossRef] [PubMed]
- Pruett, C.L.; Winker, K. The effects of sample size on population genetic diversity estimates in song sparrows Melospiza melodia. J. Avian Biol. 2008, 39, 252–256. [Google Scholar] [CrossRef]
- Honnay, O.; Jacquemyn, H.; Bossuyt, B.; Hermy, M. Forest fragmentation effects on patch occupancy and population viability of herbaceous plant species. New Phytol. 2005, 166, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Lowe, A.J.; Boshier, D.; Ward, M.; Bacles, C.F.E.; Navarro, C. Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees. Heredity 2005, 95, 255–273. [Google Scholar] [CrossRef] [PubMed]
- Honnay, O.; Jacquemyn, H. Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv. Biol. 2007, 21, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, R.; Quesada, M.; Ashworth, L.; Herrerias-Diego, Y.; Lobo, J. Genetic consequences of habitat fragmentation in plant populations: Susceptible signals in plant traits and methodological approaches. Mol. Ecol. 2008, 17, 5177–5188. [Google Scholar] [CrossRef]
- Chung, M.Y.; Nason, J.D.; Chung, M.G. Significant demographic and fine-scale genetic structure in expanding and senescing populations of the terrestrial orchid Cymbidium goeringii (Orchidaceae). Am. J. Bot. 2011, 98, 2027–2039. [Google Scholar] [CrossRef]
- Liu, Z.J.; Chen, S.C.; Ru, Z.Z.; Chen, L.J. The Genus Cymbidium in China; Science Press: Beijing, China, 2006; pp. 195–198. [Google Scholar]
- Wang, H.Z.; Wu, Z.X.; Lu, J.J.; Shi, N.N.; Zhao, Y.; Zhang, Z.T.; Liu, J.J. Molecular diversity and relationships among Cymbidium goeringii cultivars based on inter-simple sequence repeat (ISSR) markers. Genetica 2009, 136, 391–399. [Google Scholar] [CrossRef]


| Group | N | Na | Ne | Ho | He | I | FIS | h |
|---|---|---|---|---|---|---|---|---|
| YG1 | 18 | 6.636 | 3.387 | 0.641 | 0.657 | 1.377 | 0.037 | 0.711 |
| YG2 | 54 | 9.818 | 3.594 | 0.636 | 0.689 | 1.550 | 0.093 | 0.674 |
| YG3 | 32 | 8.182 | 3.504 | 0.571 | 0.656 | 1.451 | 0.127 | 0.676 |
| UL1 | 16 | 3.636 | 2.400 | 0.580 | 0.542 | 0.957 | −0.060 | 0.636 |
| UL2 | 36 | 5.818 | 3.172 | 0.389 | 0.637 | 1.271 | 0.362 | 0.660 |
| UL3 | 16 | 5.091 | 3.286 | 0.449 | 0.641 | 1.261 | 0.268 | 0.560 |
| HS1 | 52 | 5.545 | 2.899 | 0.556 | 0.589 | 1.194 | 0.051 | 0.595 |
| HS2 | 44 | 5.818 | 2.903 | 0.477 | 0.591 | 1.195 | 0.244 | 0.597 |
| Overall | 6.318 | 3.143 | 0.537 | 0.625 | 1.282 | 0.14 | 0.639 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noh, S.W.; Park, J.-K.; Yu, J.S.; Nam, D.E.; Do, Y.; Chung, K.W. Genetic Diversity and Population Structure of the Spring Orchid Cymbidium goeringii in Korean Distant Islands. Diversity 2020, 12, 486. https://doi.org/10.3390/d12120486
Noh SW, Park J-K, Yu JS, Nam DE, Do Y, Chung KW. Genetic Diversity and Population Structure of the Spring Orchid Cymbidium goeringii in Korean Distant Islands. Diversity. 2020; 12(12):486. https://doi.org/10.3390/d12120486
Chicago/Turabian StyleNoh, Seung Woo, Jun-Kyu Park, Jin Seok Yu, Da Eun Nam, Yuno Do, and Ki Wha Chung. 2020. "Genetic Diversity and Population Structure of the Spring Orchid Cymbidium goeringii in Korean Distant Islands" Diversity 12, no. 12: 486. https://doi.org/10.3390/d12120486
APA StyleNoh, S. W., Park, J.-K., Yu, J. S., Nam, D. E., Do, Y., & Chung, K. W. (2020). Genetic Diversity and Population Structure of the Spring Orchid Cymbidium goeringii in Korean Distant Islands. Diversity, 12(12), 486. https://doi.org/10.3390/d12120486





