Highlighting the Crude Oil Bioremediation Potential of Marine Fungi Isolated from the Port of Oran (Algeria)
Abstract
:1. Introduction
2. Material and Methods
2.1. Sampling and Isolation
2.2. Identification of Fungal Isolates
2.3. Screening of Fungal Growth on Crude Oil
2.4. Crude Oil Degradation Assay
2.5. Biosurfactant Production
3. Results
3.1. Diversity of Culturable Fungal Communities in the Port of Oran
3.2. Ability to Grow on Crude Oil
3.3. Crude Oil Degradation
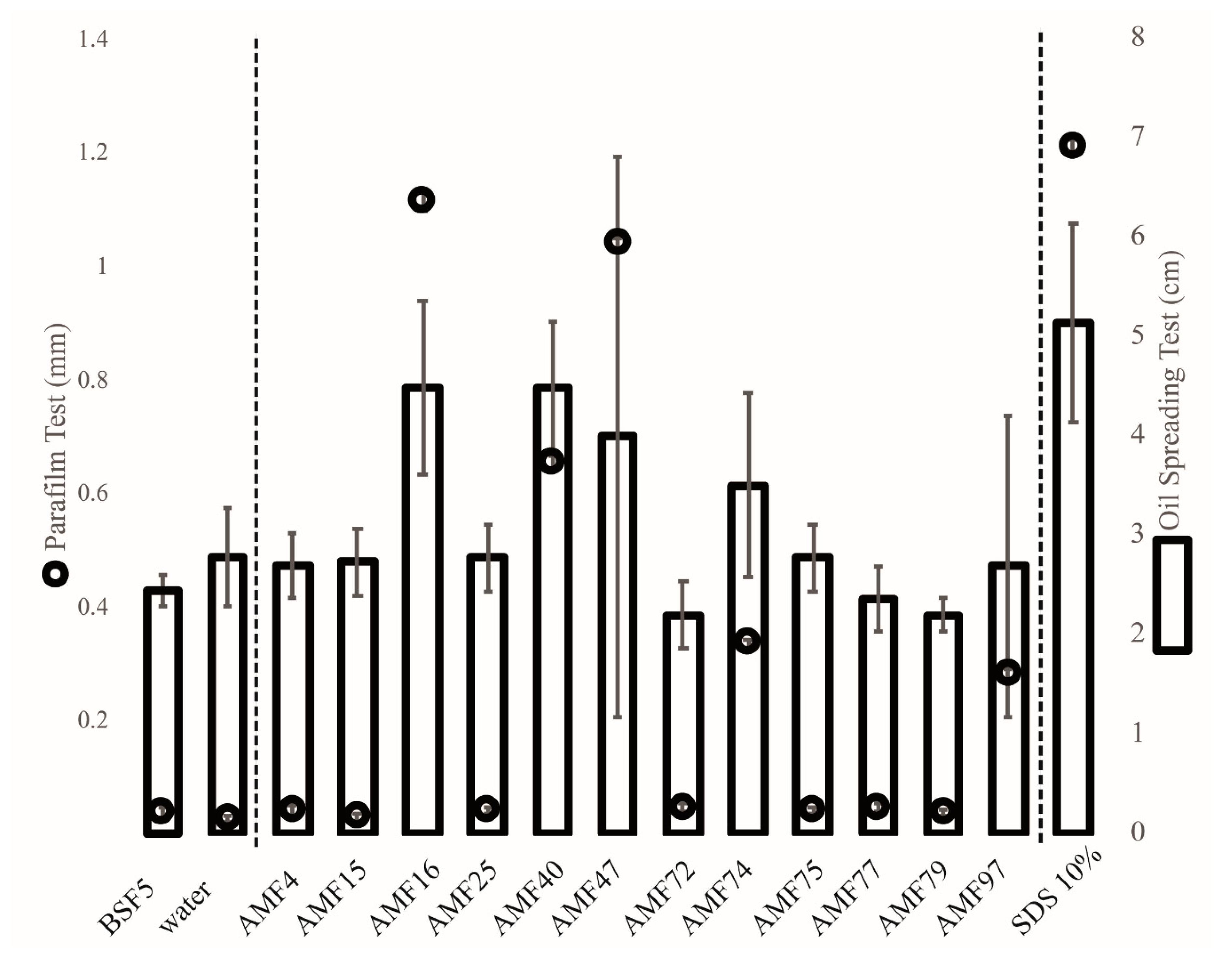
3.4. Biosurfactant Production
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cerniglia, C.E.; Sutherland, J.B. Degradation of Polycyclic Aromatic Hydrocarbons by Fungi. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin, Germany, 2010; pp. 2079–2110. [Google Scholar]
- Passarini, M.R.Z.; Rodrigues, M.V.N.; da Silva, M.; Sette, L.D. Marine-derived filamentous fungi and their potential application for polycyclic aromatic hydrocarbon bioremediation. Mar. Pollut. Bull. 2011, 62, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Hassanshahian, M.; Cappello, S. Crude Oil Biodegradation in the Marine Environments. Eng. Technol. 2013, 5, 101–135. [Google Scholar]
- Catania, V.; Santisi, S.; Signa, G.; Vizzini, S.; Mazzola, A.; Cappello, S.; Yakimov, M.M.; Quatrini, P. Intrinsic bioremediation potential of a chronically polluted marine coastal area. Mar. Pollut. Bull. 2015, 99, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Kumari, B.; Singh, G.; Sinam, G.; Singh, D.P. Microbial Remediation of Crude Oil-Contaminated Sites, Environmental Concerns and Sustainable Development; Shukla, V., Kumar, N., Eds.; Springer: Singapore, 2020; pp. 333–351. [Google Scholar]
- Head, I.M.; Jones, D.M.; Röling, W.F. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 2006, 4, 173–182. [Google Scholar] [CrossRef]
- Yun, T.; Yuan-rong, L.; Tian-ling, Z.; Li-zhe, C.; Xiao-xing, C.; Chong-ling, Y. Contamination and potential biodegradation of polycyclic aromatic hydrocarbons in mangrove sediments of Xiamen, China. Mar. Pollut. Bull. 2008, 56, 1184–1191. [Google Scholar]
- Ezeji, U.E.; Anyadoh, S.O.; Ibekwe, V.I. Clean up of Crude oil-Contaminated Soil. Terr. Aquat. Environ. Toxicolgy 2007, 1, 54–59. [Google Scholar]
- Das, N.; Chandran, P. Microbial Degradation of Petroleum Hydrocarbon Contaminants: An Overview. Biotechnol. Res. Int. 2011, 2011, 13. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.; Yu, Y.; Bai, Y.; Wang, L.; Wu, Y. Marine Oil-Degrading Microorganisms and Biodegradation Process of Petroleum Hydrocarbon in Marine Environments: A Review. Curr. Microbiol. 2015, 71, 220–228. [Google Scholar] [CrossRef]
- Mahjoubi, M.; Cappello, S.; Souissi, Y.; Jaouani, A.; Cherif, A. Microbial Bioremediation of Petroleum Hydrocarbon–Contaminated Marine Environments. M. Zoveidavianpoor (Ed.), In Recent Insights in Petroleum Science and Engineering; IntechOpen: London, UK, 2018. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Johnson, Z.I.; Wang, G. Molecular characterization of the spatial diversity and novel lineages of mycoplankton in Hawaiian coastal waters. ISME J. 2010, 4, 111–120. [Google Scholar] [CrossRef]
- Gutiérrez, M.H.; Pantoja, S.; Tejos, E.; Quiñones, R.A. The role of fungi in processing marine organic matter in the upwelling ecosystem off Chile. Mar. Biol. 2011, 158, 205–219. [Google Scholar] [CrossRef]
- Taylor, J.D.; Cunliffe, M. Multi-year assessment of coastal planktonic fungi reveals environmental drivers of diversity and abundance. ISME J. 2016, 10, 2118–2128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgcomb, V.P.; Beaudoin, D.; Gast, R.; Biddle, J.F.; Teske, A. Marine subsurface eukaryotes: The fungal majority. Environ. Microbiol. 2011, 13, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Rédou, V.; Navarri, M.; Meslet-Cladière, L.; Barbier, G.; Burgaud, G. Species richness and adaptation of marine fungi from deep-subseafloor sediments. Appl. Environ. Microbiol. 2015, 81, 3571–3583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pachiadaki, M.G.; Rédou, V.; Beaudoin, D.J.; Burgaud, G.; Edgcomb, V.P. Fungal and prokaryotic activities in the marine subsurface biosphere at Peru Margin and Canterbury Basin inferred from RNA-based analyses and microscopy. Front. Microbiol. 2016, 7, 846. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, M.; Bian, X.; Guo, J.; Cai, L. A High-Level Fungal Diversity in the Intertidal Sediment of Chinese Seas Presents the Spatial Variation of Community Composition. Front. Microbiol 2016, 7, 2098. [Google Scholar] [CrossRef] [Green Version]
- Park, M.S.; Oh, S.-Y.; Fong, J.J.; Houbraken, J.; Lim, Y.W. The diversity and ecological roles of Penicillium in intertidal zones. Sci. Rep. 2019, 9, 13540. [Google Scholar] [CrossRef] [Green Version]
- Rämä, T.; Nordén, J.; Davey, M.L.; Mathiassen, G.H.; Spatafora, J.W.; Kauserud, H. Fungi ahoy! Diversity on marine wooden substrata in the high North. Fungal Ecol. 2014, 8, 46–58. [Google Scholar] [CrossRef] [Green Version]
- Gnavi, G.; Garzoli, L.; Poli, A.; Prigione, V.; Burgaud, G.; Varese, G.C. The culturable mycobiota of Flabellia petiolata: First survey of marine fungi associated to a Mediterranean green alga. PLoS ONE 2017, 12, 1–20. [Google Scholar] [CrossRef]
- Hatai, K. Diseases of fish and shellfish caused by marine fungi. In Biology of Marine Fungi; Springer: Berlin/Heidelberg, Germany, 2012; pp. 15–52. [Google Scholar]
- Van Dover, C.L.; Ward, M.E.; Scott, J.L.; Underdown, J.; Anderson, B.; Gustafson, C.; Carnegie, R.B. A fungal epizootic in mussels at a deep-sea hydrothermal vent. Mar. Ecol. 2007, 28, 54–62. [Google Scholar] [CrossRef]
- Cunliffe, M.; Hollingsworth, A.; Bain, C.; Sharma, V.; Taylor, J.D. Algal polysaccharide utilisation by saprotrophic planktonic marine fungi. Fungal Ecol. 2017, 30, 135–138. [Google Scholar] [CrossRef]
- Arifeen, M.Z.U.; Liu, C.H. Novel enzymes isolated from marine-derived fungi and its potential applications. United J. Biochem. Biotechnol 2018, 1, 1–11. [Google Scholar]
- Paço, A.; Duarte, K.; da Costa, J.P.; Santos, P.S.; Pereira, R.; Pereira, M.E.; Rocha-Santos, T.A. Biodegradation of polyethylene microplastics by the marine fungus Zalerion maritimum. Sci. Total Environ. 2017, 586, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, A.L.; Proietti, M.C.; Secchi, E.R.; Taylor, J.D. Diverse groups of fungi are associated with plastics in the surface waters of the Western South Atlantic and the Antarctic Peninsula. Mol. Ecol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Prince, R.C.; Walters, C.C. Biodegradation of Oil Hydrocarbons and its Implications for Source Identification. In Oil Spill Environmental Forensics, USA; Wang, Z., Stout, S.A., Eds.; Academic Press Publication: Cambridge, CA, USA, 2007; pp. 349–403. [Google Scholar]
- Khelil, F.; Matallah-Boutiba, A.; Chemlal-Kherraz, D.; Boutiba, Z. Characterization of hydrocarbonoclastes bacteria isolated from marine waters west algeria: Evolution analysis in presence of crude oil. J. Curr. Res. Sci. 2014, 2, 557–563. [Google Scholar]
- Prince, R. The Microbiology of Marine Oil Spill Bioremediation. In Petroleum Microbiology; Ollivier, B., Magot, M., Eds.; ASM Press: Washington, DC, USA, 2005; pp. 317–335. [Google Scholar]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the Marine Environment: Open questions and unsolved problems. mBio 2019, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Brooijmans, R.J.W.; Pastink, M.I.; Siezen, R.J. Hydrocarbon-degrading bacteria: The oil-spill clean-up crew. Microb. Biotechnol. 2009, 2, 587–594. [Google Scholar] [CrossRef]
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 2011, 9, 177–192. [Google Scholar] [CrossRef]
- Marco-Urrea, E.; García-Romera, I.; Aranda, E. Potential of non-ligninolytic fungi in bioremediation of chlorinated and polycyclic aromatic hydrocarbons. New Biotechnol. 2015, 32, 620–628. [Google Scholar] [CrossRef]
- Steliga, T. Role of Fungi in Biodegradation of Petroleum Hydrocarbons in Drill Waste. Pol. J. Environ. Stud 2012, 21, 471–479. [Google Scholar]
- In der Wiesche, C.; Martens, R.; Zadrazil, F. The effect of interaction between white-rot fungi and indigenous microorganisms on degradation of polycyclic aromatic hydrocarbons in soil. WaterAirSoil Pollut. 2003, 3, 73–79. [Google Scholar]
- Zheng, Z.; Obbard, J.P. Effect of non-ionic surfactants on elimination of polycyclic aromatic hydrocarbons (PAHs) in soil-slurry by Phanerochaete chrysosporium. J. Chem. Technol. Biotechnol. 2001, 76, 423–429. [Google Scholar] [CrossRef]
- Yadav, J.S.; Reddy, C.A. Degradation of benzene, toluene, ethylbenzene, and xylenes (BTEX) by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 1993, 59, 756–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capotorti, G.; Digianvincenzo, P.; Cesti, P.; Bernardi, A.; Guglielmetti, G. Pyrene and benzo(a)pyrene metabolism by an Aspergillus terreus strain isolated from a polycylic aromatic hydrocarbons polluted soil. Biodegradation 2004, 15, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Reyes-César, A.; Absalon, A.E.; Fernandez, F.J.; Gonzalez, J.M.; Cortès-Espinosa, D.V. Biodegradation of a mixture of PAHs by non-ligninolytic fungal strains isolated from crude oil-contaminated soil. World J. Microbiol. Biotechnol. 2014, 30, 999–1009. [Google Scholar] [CrossRef] [PubMed]
- Godoy, P.; Reina, R.; Calderón, A.; Wittich, R.M.; García-Romera, I.; Aranda, E. Exploring the potential of fungi isolated from PAH-polluted soil as a source of xenobiotics-degrading fungi. Environ. Sci. Pollut. Res. 2016, 23, 20985–20996. [Google Scholar] [CrossRef]
- Hassanshahian, M.; Tebyanian, H.; Cappello, S. Isolation and characterization of two crude oil-degrading yeast strains, Yarrowia lipolytica PG-20 and PG-32, from the Persian Gulf. Mar. Pollut. Bull. 2012, 64, 1386–1391. [Google Scholar] [CrossRef]
- McGenity, T.J.; Folwell, B.D.; McKew, B.A.; Sanni, G.O. Marine crude-oil biodegradation: A central role for interspecies interactions. Aquat. Biosyst. 2012, 8, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Simister, R.L.; Poutasse, C.M.; Thurston, A.M.; Reeve, J.L.; Baker, M.C.; White, H.K. Degradation of oil by fungi isolated from Gulf of Mexico beaches. Mar. Pollut. Bull. 2015, 100, 327–333. [Google Scholar] [CrossRef]
- Bovio, E.; Gnavi, G.; Prigione, V.; Spina, F.; Denaro, R.; Yakimov, M.; Calogero, R.; Crisafi, F.; Varese, G.C. The culturable mycobiota of a Mediterranean marine site after an oil spill: Isolation, identification and potential application in bioremediation. Sci. Total Environ. 2017, 576, 310–318. [Google Scholar] [CrossRef]
- Das, T.T.; Gohel, H.R.; Panchal, M.R.; Ghosh, S.K.; Braganza, V.J. Determination of crude oil degradation efficiency of glass biofilm of isolated bacterium and fungus. Int. Res. J. Biol. Sci. 2014, 3, 67–69. [Google Scholar]
- Sen, S.; Borah, S.N.; Bora, A.; Deka, S. Production, characterization, and antifungal activity of a biosurfactant produced by Rhodotorula babjevae YS3. Microb. Cell Factories 2017, 16, 1–14. [Google Scholar]
- Banat, I.M.; Satpute, S.K.; Cameotra, S.S.; Patil, R.; Nyayanit, N.V. Cost effective technologies and renewable substrates for biosurfactants production. Front. Microbiol. 2014, 5, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Damare, S.; Singh, P.; Raghukumar, S. Biotechnology of Marine Fungi, Biology of Marine Fungi. In Progress in Molecular and Subcellular Biology; Raghukumar, C., Ed.; Springer: Berlin, Germany, 2012; pp. 277–297. [Google Scholar]
- Shekhar, S.; Sundaramanickam, A.; Balasubramanian, T. Biosurfactant Producing Microbes and their Potential Applications: A Review. J. Crit. Rev. Environ. Sci. Technol. 2015, 45, 1522–1554. [Google Scholar] [CrossRef]
- Soberón-Chávez, G. (Ed.) Biosurfactants: From Genes to Applications; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Rufino, R.D.; de Luna, J.M.; de Campos Sarubbo, T.G.M.L.A. Characterization and properties of the biosurfactant produced by Candida lipolytica UCP 0988. Electron. J. Biotechnol. 2014, 17, 34–38. [Google Scholar] [CrossRef] [Green Version]
- Sajna, K.V.; Sukumaran, R.K.; Gottumukkala, L.D.; Pandey, A. Crude oil biodegradation aided by biosurfactants from Pseudozyma sp. NII 08165 or its culture broth. Bioresour. Technol. 2015, 191, 133–139. [Google Scholar] [CrossRef]
- Andrade Silva, N.R.; Luna, M.A.C.; Santiago, A.L.C.M.A.; Franco, L.O.; Silva, G.K.B.; de Souza, P.M.; Okada, K.; Albuquerque, C.D.C.; Alves da Silva, C.A.; Campos-Takaki, G.M. Biosurfactant-and-Bioemulsifier Produced by a Promising Cunninghamella echinulata Isolated from Caatinga Soil in the Northeast of Brazil. Int. J. Mol. Sci. 2014, 15, 15377–15395. [Google Scholar] [CrossRef] [Green Version]
- Lima, J.M.S.; Pereira, J.O.; Batista, I.H.; Costa Neto, P.Q.; dos Santos, J.C.; de Araújo, S.P.; Pantoja, M.C.; da Mota, A.J.; de Azevedo, J.L. Potential biosurfactant producing endophytic and epiphytic fungi, isolated from macrophytes in the Negro River in Manaus, Amazonas, Brazil. Afr. J. Biotechnol. 2016, 15, 1217–1223. [Google Scholar]
- Patowary, K.; Patowary, R.; Kalita, M.C.; Deka, S. Characterization of Biosurfactant Produced during Degradation of Hydrocarbons Using Crude Oil As Sole Source of Carbon. Front. Microbiol. 2017, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: Orlando, FL, USA, 1990; pp. 315–322. [Google Scholar]
- Buchanan, R.L.; Whiting, R.C.; Damert, W.C. When is simple good enough: A comparison of the Gompertz, Baranyi, and three-phase linear models for fitting bacterial growth curves. Food Microbiol. 1997, 14, 313–326. [Google Scholar] [CrossRef]
- Dagnas, S.; Membré, J.-M. Predicting and Preventing Mold Spoilage of Food Products. J. Food Prot. 2013, 76, 538–551. [Google Scholar] [CrossRef]
- Varjani, S.J.; Upasani, V.N. Comparative studies on bacterial consortia for hydrocarbon degradation. Int. J. Innov. Res. Sci. Eng. Technol. 2013, 2, 5377–5383. [Google Scholar]
- Bhardwaj, G.; Cameotra, S.S.; Chopra, H.K. Biosurfactants from fungi: A review. J. Pet. Envrion. Biotechnol. 2013, 4, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Bodour, A.A.; Miller-Maier, R.M. Application of a modified drop-collapse technique for surfactant quantitation and screening of biosurfactant-producing microorganisms. J. Microbiol. Methods 1998, 32, 273–280. [Google Scholar] [CrossRef]
- Rodrigues, L.R.; Teixeira, J.A.; Van der Mei, H.C.; Oliveira, R. Physicochemical and functional characterization of a biosurfactant produced by Lactococcus lactis 53. Colloids Surf. B Biointerfaces 2006, 49, 79–86. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.F.; Zhou, L.M.; Chen, S.T.; Yang, B.; Liao, S.R.; Kong, F.D.; Lin, X.P.; Wang, F.Z.; Zhou, X.F.; Liu, Y.H. New chlorinated diphenyl ethers and xanthones from a deep-sea-derived fungus Penicillium chrysogenum SCSIO 41001. Fitoterapia 2018, 125, 49–54. [Google Scholar] [CrossRef]
- Luo, J.J.; Ding, J.F.; Li, G.W.; Zheng, T.L.; Luo, Z.H. Characterization of a formaldehyde degrading fungus Penicillium chrysogenum DY-F2 isolated from deep sea sediment. Int. Biodeterior. Biodegrad. 2014, 89, 45–49. [Google Scholar] [CrossRef]
- Elshafie, A.; AlKindi, A.Y.; Al-Busaidi, S.; Bakheit, C.; Albahry, S.N. Biodegradation of crude oil and n-alkanes by fungi isolated from Oman. Mar. Pollut. Bull. 2007, 54, 1692–1696. [Google Scholar] [CrossRef]
- Alwakeel, S.S. Molecular identification of fungi isolated from coastal regions of Red Sea, Jeddah, Saudi Arabia. J. Assoc. Arab Univ. Basic Appl. Sci. 2017, 24, 115–119. [Google Scholar] [CrossRef] [Green Version]
- Marchese, P.; Gnavi, G.; Garzoli, L.; Bouraoui, A.; Murphy, M.; Barry, F.; Mehiri, M.; Prigione, V. Marine fungi from Holothuria poli (Della Chiaje, 1823): Diversity and extracts bioactivity. Biol. Mar. Mediterr. 2016, 23, 290–291. [Google Scholar]
- Barnes, N.M.; Khodse, V.B.; Lotlikar, N.P.; Meena, R.M.; Damare, S.R. Bioremediation potential of hydrocarbon-utilizing fungi from select marine niches of India. 3 Biotech 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Zuluaga-Montero, A.; Ramirez-Camejo, L.; Rauscher, J.; Bayman, P. Marine isolates of Aspergillus flavus: Denizens of the deep or lost at sea? Fungal Ecol. 2010, 3, 386–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visamsetti, A.; Ramachandrana, S.S.; Kandasamy, D. Penicillium chrysogenum DSOA associated with marine sponge (Tedania anhelans) exhibit antimycobacterial activity. Microbiol. Res. 2016, 185, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.S.; Li, X.M.; Li, C.S.; Proksch, P.; Wang, B.G. Penicisteroids A and B, antifungal and cytotoxic polyoxygenated steroids from the marine alga-derived endophytic fungus Penicillium chrysogenum QEN-24S. Bioorg. Med. Chem. Lett. 2011, 21, 2894–2897. [Google Scholar] [CrossRef]
- Paz, Z.; Komon-Zelazowska, M.; Druzhinina, I.S.; Aveskamp, M.M.; Shnaiderman, A.; Aluma, Y.; Carmeli, S.; Ilan, M.; Yarden, O. Diversity and potential antifungal properties of fungi associated with a Mediterranean sponge. Fungal Divers. 2010, 42, 17–26. [Google Scholar] [CrossRef]
- Passarini, M.R.Z.; Santos, C.; Lima, N.; Berlinck, R.G.S.; Sette, L.D. Filamentous fungi from the Atlantic marine sponge Dragmacidon reticulatum. Arch. Microbiol. 2013, 195, 99–111. [Google Scholar] [CrossRef]
- Ding, B.; Yin, Y.; Zhang, F.; Li, Z. Recovery and phylogenetic diversity of culturable fungi associated with marine sponges Clathrina luteoculcitella and Holoxea sp. in the South China Sea. Mar. Biotechnol. 2011, 13, 713–721. [Google Scholar] [CrossRef]
- Pivkin, M.V.; Aleshko, S.A.; Krasokhin, V.B.; Khudyakova, Y.V. Fungal Assemblages Associated with Sponges of the Southern Coast of Sakhalin Island. Russ. J. Mar. Biol. 2006, 32, 207–213. [Google Scholar] [CrossRef]
- Wiese, J.; Ohlendorf, B.; Blümel, M.; Schmaljohann, R.; Imhoff, J.F. Phylogenetic identificationof fungi isolated from the marine sponge Tethya aurantium and identification of their secondary metabolites. Mar. Drugs 2011, 9, 561–585. [Google Scholar] [CrossRef]
- Panno, L.; Bruno, M.; Voyron, S.; Anastasi, A.; Gnavi, G.; Miserere, L.; Varese, G.C. Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanica. New Biotechnol. 2013, 30, 685–694. [Google Scholar] [CrossRef]
- Bovio, E.; Garzoli, L.; Poli, A.; Luganini, A.; Villa, P.; Musumeci, R.; McCormack, G.P.; Cocuzza, C.E.; Gribaudo, G.; Mehiri, M.; et al. Marine Fungi from the Sponge Grantia compressa: Biodiversity, Chemodiversity, and Biotechnological Potential. Mar. Drugs 2019, 17, 220. [Google Scholar] [CrossRef] [Green Version]
- Neethu, S.; Midhun, S.J.; Radhakrishnan, E.K.; Jyothis, M. Green synthesized silver nanoparticles by marine endophytic fungus Penicillium polonicum and its antibacterial efficacy against biofilm forming, multidrug-resistant Acinetobacter baumanii. Microb. Pathog. 2018, 116, 263–272. [Google Scholar] [CrossRef]
- Garzoli, L.; Poli, A.; Prigione, V.; Gnavi, G.; Varese, G.C. Peacock’s tail with a fungal cocktail: First assessment of the mycobiota associated with the brown alga Padina pavonica. Fungal Ecol. 2018, 35, 87–97. [Google Scholar] [CrossRef]
- López-Legentil, S.; Erwin, P.M.; Turon, M.; Yarden, O. Diversity of fungi isolated from three temperate ascidians. Symbiosis 2015, 66, 99–106. [Google Scholar] [CrossRef]
- Mabrouk, A.M.; Kheiralla, Z.H.; Hamed, E.R.; Youssry, A.A.; Abd El Aty, A.A. Screening of some marine-derived fungal isolates for lignin degrading enzymes (LDEs) production. Agric. Biol. J. N. Am. 2010, 1, 591–599. [Google Scholar]
- Godinho, V.M.; de Paula, M.T.R.; Silva, D.A.S.; Paresque, K.; Martins, A.P.; Colepicolo, P.; Rosa, C.A.; Rosa, L.H. Diversity and distribution of hidden cultivable fungi associated with marine animals of Antarctica. Fungal Biol. 2019, 123, 507–516. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Suetrong, S.; Sakayaroj, J.; Bahkali, A.H.; Abdel-Wahab, M.A.; Boekhout, T.; Pang, K.L. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Divers. 2015, 73, 1–72. [Google Scholar] [CrossRef]
- Raghukumar, C.; Ravindran, J. Fungi and Their Role in Corals and Coral Reef Ecosystems. In Biology of Marine Fungi; Raghukumar, C., Ed.; Progress in Molecular and Subcellular Biology; Springer: Berlin/Heidelberg, Germany, 2012; pp. 89–113. [Google Scholar]
- Lin, A.; Lu, X.; Fang, Y.; Zhu, T.; Gu, Q.; Zhu, W. Two New 5-Hydroxy-2-pyrone Derivatives Isolated from a Marine-derived Fungus Aspergillus flavus. J. Antibiot. 2008, 61, 245–249. [Google Scholar] [CrossRef]
- Sun, H.-F.; Li, X.-M.; Meng, L.; Cui, C.-M.; Gao, S.-S.; Li, C.-S.; Huang, C.-G.; Wang, B.-G. Asperolides A–C, tetranorlabdane diterpenoids from the marine alga-derived endophytic fungus Aspergillus wentii EN-48. J. Nat. Prod. 2012, 75, 148–152. [Google Scholar] [CrossRef]
- Shigemori, H.; Kasai, Y.; Komatsu, K.; Tsuda, M.; Mikami, Y.; Kobayashi, J. Sporiolides A and B, New Cytotoxic Twelve-Membered Macrolides from a Marine-Derived Fungus Cladosporium Species. Mar. Drugs 2004, 2, 164–169. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.-S.; Li, X.-M.; Wang, B.-G. Perylene Derivatives Produced by Alternaria alternata, an Endophytic Fungus Isolated from Laurencia Species. Nat. Prod. Commun. 2009, 4, 1477–1480. [Google Scholar] [CrossRef] [Green Version]
- Shaaban, M.; Shaaban, K.A.; Abdel-Aziz, M.S. Seven naphtho-g-pyrones from the marine derived fungus Alternaria alternata: Structure elucidation and biological properties. Org. Med. Chem. Lett. 2012, 2, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Terpenes with Rarely Occurring Frameworks from the Marine-Alga-Epiphytic Fungus Alternaria alternata k21-1. J. Nat. Prod. 2017, 80, 2524–2529. [CrossRef]
- Wang, S.; Li, X.-M.; Teuscher, F.; Li, D.-L.; Diesel, A.; Ebel, R.; Proksch, P.; Wang, B.-G. Chaetopyranin, a Benzaldehyde Derivative, and Other Related Metabolites from Chaetomium globosum, an Endophytic Fungus Derived from the Marine Red Alga Polysiphonia urceolata. J. Nat. Prod. 2006, 69, 1622–1625. [Google Scholar] [CrossRef]
- Cui, C.-M.; Li, X.-M.; Li, C.-S.; Proksch, P.; Wang, B.-G.; Cytoglobosins, A.-G. Cytochalasans from a Marine-Derived Endophytic Fungus, Chaetomium globosum QEN-14. J. Nat. Prod. 2010, 73, 729–733. [Google Scholar] [CrossRef]
- Yamada, T.; Muroga, Y.; Tanaka, R. New Azaphilones, Seco-Chaetomugilins A and D, Produced by a Marine-Fish-Derived Chaetomium Globosum. Mar. Drugs 2009, 7, 249–257. [Google Scholar] [CrossRef] [Green Version]
- Muroga, Y.; Yamada, T.; Numata, A.; Tanaka, R. 11-and 4′-Epimers of Chaetomugilin A, Novel Cytostatic Metabolites from Marine Fish-Derived Fungus Chaetomium Globosum. Helv. Chim. Acta 2010, 93, 542–549. [Google Scholar] [CrossRef]
- Henríquez, M.; Vergara, K.; Norambuena, J.; Beiza, A.; Maza, F.; Ubilla, P.; Araya, I.; Chavez, R.; San-Martin, A.; Darias, J.; et al. Diversity of cultivable fungi associated with Antarctic marine sponges and screening for their antimicrobial, antitumoral and antioxidant potential. World J. Microbiol. Biotechnol. 2014, 30, 65–76. [Google Scholar] [CrossRef]
- Rozas, E.E.; Albano, R.M.; Lôbo-Hajdu, G.; Müller, W.E.G.; Schröder, H.-C.; Custódio, M.R. Isolation and cultivation of fungal strains from in vitro cell cultures of two marine sponges (Porifera: Halichondrida and Haplosclerida). Braz. J. Microbiol. 2011, 42, 1560–1568. [Google Scholar] [CrossRef] [Green Version]
- Damare, S.; Raghukumar, C.; Raghukumar, S. Fungi in deep-sea sediments of the Central Indian Basin. Deep-Sea Res. I 2006, 53, 14–27. [Google Scholar]
- Pindi, P.K. Diversity of fungi at various depths of marine water. Res. Biotechnol. 2012, 3, 62–66. [Google Scholar]
- Shao, Z.; Sun, F. Intracellular sequestration of manganese and phosphorus in a metal-resistant fungus Cladosporium cladosporioides from deep-sea sediment. Extremophiles 2007, 11, 435–443. [Google Scholar] [CrossRef]
- Manríquez, V.; Galdámez, A.; Veliz, B.; Rovirosa, J.; Díaz-Marrero, A.R.; Cuetob, M.; Darias, J.; Martínez, C.; San-Martín, A. N-Methyl-1h-Indole-2-Carboxamide From The Marine Fungus Cladosporium Cladosporioides. J. Chil. Chem. Soc 2009, 54, 314–316. [Google Scholar] [CrossRef] [Green Version]
- San-Martin, A.; Painemal, K.; Díaz, Y.; Martinez, C.; Rovirosa, J. Metabolites from the marine fungus Cladosporium cladosporioides. J. Argent. Chem. Soc. 2005, 93, 247–251. [Google Scholar]
- Sayed, M.A.E.; Abd El-Rahman, T.M.A.; El-Diwany, A.I.; Sayed, S.M. Biodiversity and bioactivity of red sea sponge associated endophytic fungi. Int. J. Adv. Res. Eng. Appl. Sci. 2016, 5, 1–15. [Google Scholar]
- Bonugli-Santos, R.C.; Durrant, L.R.; da Silva, M.; Sette, L.D. Production of laccase, manganese peroxidase and lignin peroxidase by Brazilian marine-derived fungi. Enzym. Microb. Technol. 2010, 46, 32–37. [Google Scholar] [CrossRef]
- Hulikere, M.M.; Joshi, C.G. Characterization, antioxidant and antimicrobial activity of silver nanoparticles synthesized using marine endophytic fungus-Cladosporium cladosporioides. Process Biochem. 2019, 82, 199–204. [Google Scholar] [CrossRef]
- Namikoshi, M.; Negishi, R.; Nagai, H.; Dmitrenok, A.; Kobayashi, H. Three New Chlorine Containing Antibiotics from a Marine-derived Fungus Aspergillus ostianus Collected in Pohnpei. J. Antibiot. 2003, 56, 755–761. [Google Scholar] [CrossRef] [Green Version]
- Xin, Z.H.; Fang, Y.; Du, L.; Zhu, T.; Duan, L.; Chen, J.; Gu, Q.-Q.; Zhu, W.-M. Aurantiomides A–C, Quinazoline Alkaloids from the Sponge-Derived Fungus Penicillium aurantiogriseum SP0-19. J. Nat. Prod. 2007, 70, 853–855. [Google Scholar] [CrossRef]
- Song, F.; Ren, B.; Yu, K.; Chen, C.; Guo, H.; Yang, N.; Gao, H.; Liu, X.; Liu, M.; Tong, Y.; et al. Quinazolin-4-one Coupled with Pyrrolidin-2-iminium Alkaloids from Marine-Derived Fungus Penicillium Aurantiogriseum. Mar. Drugs 2012, 10, 1297–1306. [Google Scholar] [CrossRef] [Green Version]
- Yu, K.; Ren, B.; Wei, J.; Chen, C.; Sun, J.; Song, F.; Dai, H.; Zhang, L. Verrucisidinol and Verrucosidinol Acetate, Two Pyrone-Type Polyketides Isolated from a Marine Derived Fungus, Penicillium Aurantiogriseum. Mar. Drugs 2010, 8, 2744–2754. [Google Scholar] [CrossRef] [Green Version]
- Abo-Kadoum, M.A.; Abo-Dahab, N.F.; Awad, M.F.; Abdel-Hadi, A.M. Marine-derived fungus, Penicillium aurantiogriseum AUMC 9757: A producer of bioactive secondary metabolites. J. Basic Appl. Mycol. 2013, 4, 77–83. [Google Scholar]
- Chung, D.; Baek, K.; Bae, S.S.; Jung, J. Identification and characterization of a marine-derived chitinolytic fungus, Acremonium sp. YS2-2. J. Microbiol. 2019, 57, 372–380. [Google Scholar] [CrossRef]
- Zuccaro, A.; Summerbell, R.C.; Gams, W.; Schroers, H.-J.; Mitchell, J.I. A new Acremonium species associated with Fucus spp. and its affinity with a phylogenetically distinct marine Emericellopsis clade. Stud. Mycol. 2004, 50, 283–297. [Google Scholar]
- Smetanina, O.F.; Kalinovskii, A.I.; Khudyakova, Y.V.; Slinkina, N.N.; Pivkin, M.V.; Kuznetsova, T.A. Metabolites from The Marine Fungus Eurotium repens. Chem. Nat. Compd. 2007, 43, 395–398. [Google Scholar] [CrossRef]
- Lee, S.; Park, M.S.; Lim, Y.W. Diversity of Marine-Derived Aspergillus from Tidal Mudflats and Sea Sand in Korea. Mycobiology 2016, 44, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Velez, P.; Gasca-Pineda, J.; Riquelme, M. Cultivable fungi from deep-sea oil reserves in the Gulf of Mexico: Genetic signatures in response to hydrocarbons. Mar. Environ. Res. 2020, 153, 1–11. [Google Scholar] [CrossRef]
- Nagano, Y.; Miura, T.; Nishi, S.; Lima, A.O.; Nakayama, C.; Pellizari, V.H.; Fujikura, K. Fungal diversity in deep-sea sediments associated with asphalt seeps at the Sao Paulo Plateau. Deep-Sea Res. Part I 2017, 146, 59–67. [Google Scholar] [CrossRef]
- Li, C.J.; Chen, P.N.; Li, H.J.; Mahmud, T.; Wu, D.L.; Xu, J.; Lan, W.J. Potential Antidiabetic Fumiquinazoline Alkaloids from the Marine-Derived Fungus Scedosporium apiospermum F41-1. J. Nat. Prod. 2020, 83, 1082–1091. [Google Scholar] [CrossRef]
- Raghukumar, S. Fungi in Coastal and Oceanic Marine Ecosystems; Springer: New York, NY, USA, 2017; pp. 1–364. [Google Scholar]
- Marchand, C.; St-Arnaud, M.; Hogland, W.; Bell, T.H.; Hijri, M. Petroleum biodegradation capacity of bacteria and fungi isolated from petroleum-contaminated soil. Int. Biodeterior. Biodegrad. 2017, 116, 48–57. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Timmis, K.N.; Golyshin, P.N. Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 2007, 18, 257–266. [Google Scholar] [CrossRef]
- Wang, B.; Lai, Q.; Cui, Z.; Tan, T.; Shao, Z. A pyrene-degrading consortium from deep-sea sediment of the West Pacific and its key member Cycloclasticus sp.P1. Environ. Microbiol. 2008, 10, 1948–1963. [Google Scholar] [CrossRef] [PubMed]
- Hassanshahian, M.; Emtiazi, G.; Cappello, S. Isolation and characterization of crude-oil-degrading bacteria from the Persian Gulf and the Caspian Sea. Mar. Pollut. Bull. 2012, 64, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Shahaliyan, F.; Safahieh, A.; Abyar, H. Evaluation of Emulsification Index in Marine Bacteria Pseudomonas sp. and Bacillus sp. Arab. J. Sci. Eng. 2015, 40, 1849–1854. [Google Scholar] [CrossRef]
- Abbasian, F.; Lockington, R.; Mallavarapu, M.; Naidu, R. A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699. [Google Scholar] [CrossRef]
- Bik, H.M.; Halanych, K.M.; Sharma, J.; Thomas, W.K. Dramatic shifts in benthic microbial eukaryote communities following the Deepwater Horizon oil spill. PLoS ONE 2012, 7, 1–6. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Sakayaroj, J.; Suetrong, S.; Somrithipol, S.; Pang, K.L. Classification of marine Ascomycota, anamorphic taxa and Basidiomycota. Fungal Divers. 2009, 35, 1–187. [Google Scholar]
- Garzoli, L.; Gnavi, G.; Poli, A.P.V.; Varese, G.C. Marine Fungi in the Mediterranean Sea–hidden Biodiversity and Taxonomical Challenges, Second International Workshop on Ascomycete Systematics. 2015. Available online: http://www.wi.knaw.nl/images/Meetings/Abstracts_book_CBS_symp_2015.pdf (accessed on 14 May 2020).
- Matallah-Boutiba, A.; Ruiz, N.; Sallenave-Namont, C.; Grovel, O.; Amiard, J.-C.; Pouchus, Y.F.; Boutiba, Z. Screening for toxigenic marine-derived fungi in Algerian mussels and their immediate environment. Aquaculture 2012, 342–343, 75–79. [Google Scholar] [CrossRef]
- Sadaba, R.B.; Sarinas, B.G.S. Fungal communities in bunker C oil-impacted sites off southern Guimaras, Philippines: A post-spill assessment of Solar 1 oil spill. Bot. Mar. 2010, 53, 565–575. [Google Scholar] [CrossRef]
- Vansteelandt, M.; Roullier, C.; Blanchet, E.; Guitton, Y.; Pouchus, Y.F.; Ruiz, N.; Grovel, O. Impact of Marine-Derived Penicillium Species in the Discovery of New Potential Antitumor Drugs. In Outstanding Marine Molecules: Chemistry, Biology, Analysis; La Barre, S., Kornprobst, J.-M., Eds.; Wiley-VCH: Weinheim, Germany, 2014; pp. 45–84. [Google Scholar]
- Da Silva, M.; Passarini, M.R.Z.; Bonugli, R.C.; Sette, L.D. Cnidarian-Derived Filamentous Fungi From Brazil: Isolation, Characterisation And Rbbr Decolourisation Screening. Environ. Technol. 2008, 29, 1331–1339. [Google Scholar] [CrossRef]
- Birolli, W.G.; Santos, D.D.A.; Alvarenga, N.; Garcia, A.C.F.S.; Romão, L.P.C.; Porto, A.L.M. Biodegradation of anthracene and several PAHs by the marine-derived fungus Cladosporium sp. CBMAI 1237. Mariune Pollut. Bull. 2018, 129, 525–533. [Google Scholar] [CrossRef]
- Bovio, E.; Garzoli, L.; Poli, A.; Prigione, V.; Firsova, D.; McCormack, G.P.; Varese, G.C. The culturable mycobiota associated with three Atlantic sponges, including two new species: Thelebolus balaustiformis and T. spongiae. Fungal Syst. Evol. 2018, 1, 141–167. [Google Scholar] [CrossRef] [Green Version]
- Obire, O.; Anyanwu, E.C.; Okigbo, R.N. Saprophytic and crude oil degrading fungi from cow dung and poultry droppings as bioremediation agents. J. Agric. Technol. 2008, 4, 81–89. [Google Scholar]
- Bartha, R.; Atlas, R.M. The microbiology of aquatic oil spills. Adv. Appl. Microbiol. 1977, 22, 225–266. [Google Scholar]
- Godinho, V.M.; Gonçalves, V.N.; Santiago, I.F.; Figueredo, H.M.; Vitoreli, G.A.; Schaefer, C.E.G.R.; Barbosa, E.C.; Oliveira, J.G.; Alves, T.M.A.; Zani, C.L.; et al. Diversity and bioprospection of fungal community present in oligotrophic soil of continental Antarctica. Extremophiles 2015, 19, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Qadri, M.; Johri, S.; Shah, B.A.; Khajuria, A.; Sidiq, T.; Lattoo, S.K.; Abdin, M.Z.; Riyaz-Ul-Hassan, S. Identification and bioactive potential of endophytic fungi isolated from selected plants of the Western Himalayas. SpringerPlus 2013, 2, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nandhini, M.; Rajinia, S.B.; Udayashankar, A.C.; Niranjana, S.R.; Lund, O.S.; Shetty, H.S.; Prakash, H.S. Diversity, plant growth promoting and downy mildew disease suppression potential of cultivable endophytic fungal communities associated with pearl millet. Biol. Control 2018, 127, 127–138. [Google Scholar] [CrossRef]
- Pang, K.L.; Overy, D.P.; Jones, E.G.; da Luz Calado, M.; Burgaud, G.; Walker, A.K.; Bills, G.F. ‘Marine fungi’ and ‘marine-derived fungi’ in natural product chemistry research: Toward a new consensual definition. Fungal Biol. Rev. 2016, 30, 163–175. [Google Scholar] [CrossRef]
- Al-Nasrawi, H. Biodegradation of crude oil by fungi isolated from Gulf of Mexico. J. Bioremed. Biodegrad. 2012, 3, 1–6. [Google Scholar] [CrossRef]
- Kiran, G.S.; Hema, T.A.; Gandhimathi, R.; Selvin, J.; Thomas, T.A.; Ravji, T.R.; Natarajaseenivasan, K. Optimization and production of a biosurfactant from the sponge-associated marine fungus Aspergillus ustus MSF3. Colloids Surf. B Biointerfaces 2009, 73, 250–256. [Google Scholar] [CrossRef]
- Sena, H.H.; Sanches, M.A.; Silva Rocha, D.F.; Segundo, W.O.P.F.; de Souza, E.S.; de Souza, J.V.B. Production of biosurfactants by soil fungi isolated from the Amazon forest. Int. J. Microbiol. 2018, 2018, 8. [Google Scholar] [CrossRef]
- Cai, Q.; Zhu, Z.; Chen, B.; Zhang, B. Oil-in-water emulsion breaking marine bacteria for demulsifying oily wastewater. Water Res. 2019, 149, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Lee, H.; Kwon, B.-O.; Khim, J.S.; Yim, U.H.; Kim, B.S.; Kim, J.-J. Biosurfactant-assisted bioremediation of crude oil by indigenous bacteria isolated from Taean beach sediment. Environ. Pollut. 2018, 241, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Thavasi, R.; Sharma, S.; Jayalakshmi, S. Evaluation of Screening Methods for the Isolation of Biosurfactant Producing Marine Bacteria. J. Pet. Envrion. Biotechnol. 2011, S1. [Google Scholar] [CrossRef] [Green Version]
- Dusane, D.H.; Matkar, P.; Venugopalan, V.P.; Kumar, A.R.; Zinjarde, S.S. Cross-Species Induction of Antimicrobial Compounds, Biosurfactants and Quorum-Sensing Inhibitors in Tropical Marine Epibiotic Bacteria by Pathogens and Biofouling Microorganisms. Curr. Microbiol. 2011, 62, 974–980. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, M.; Hirata, Y.; Imanaka, T. A study on the structure-function relationship of lipopeptide biosurfactants. Biochim. Biophys. Acta 2000, 1488, 211–218. [Google Scholar] [CrossRef]
- Saravanan, R.; Sivakumar, T. Biodiversity and biodegradation potentials of fungi isolated from marine systems of East Coast of Tamil Nadu, India. Int. J. Curr. Microbiol. Appl. Sci. 2013, 2, 192–201. [Google Scholar]
- Leitão, A.L. Potential of Penicillium Species in the Bioremediation Field. Int. J. Environ. Res. Public Health 2009, 6, 1393–1417. [Google Scholar] [CrossRef] [Green Version]
- Covino, S.; D’Annibale, A.; Stazi, S.R.; Cajthaml, T.; Čvančarová, M.; Stella, T.; Petruccioli, M. Assessment of degradation potential of aliphatic hydrocarbons by autochthonous filamentous fungi from a historically polluted clay soil. Sci. Total Environ. 2015, 505, 545–554. [Google Scholar] [CrossRef]
- Al-Hawash, A.B.; Alkooranee, J.T.; Abbood, H.A.; Zhang, J.; Suna, J.; Zhang, X.; Ma, F. Isolation and characterization of two crude oil-degrading fungi strains from Rumaila oil field, Iraq. Biotechnol. Rep. 2018, 17, 104–109. [Google Scholar] [CrossRef]



| Sample Type | Number of Isolates | Seawater/Sediments | Fish/Marine Invertebrates/Algae/Marine Plant (Posidonia oceanica) | |
|---|---|---|---|---|
| Fungal Species | ||||
| Acremonium sp. | 1 (AMF100) | (53), (52) | (54) | |
| Alternaria alternata | 2 (AMF7, AMF24) | (3), (51) | (16), (30), (31), (32) | |
| Aspergillus cibarius | 5 (AMF19, AMF93, AMF26, AMF45, AMF105) | - | - | |
| Aspergillus flavus | 7 (AMF29, AMF22, AMF20, AMF46, AMF57 AMF80, AMF90) | (5), (7), (8), (51) | (8), (14), (27) | |
| Aspergillus ochraceopetaliformis | 1(AMF68) | - | - | |
| Aspergillus ostianus | 3 (AMF12, AMF8, AMF3) | - | (47) | |
| Aspergillus pseudoglaucus | 6 (AMF23, AMF21, AMF82, AMF83, AMF94, AMF43) | (56) | (55) | |
| Aspergillus ruber | 5 (AMF17, AMF18, AMF81, AMF48, AMF54,) | - | - | |
| Aspergillus sydowii | 2 (AMF15 AMF77) | (5), (7), (39), (56) | (11), (24) | |
| Aspergillus tritici | 3 (AMF44, AMF78, AMF106) | - | - | |
| Aspergillus versicolor | 4 (AMF75, AMF97, AMF92, AMF104) | (3), (7), (51) | (11), (14), (16) | |
| Aspergillus wentii * | 1 (AMF61) | - | (28) | |
| Aspergillus westerdijkiae | 2 (AMF42, AMF60) | (3), (56) | - | |
| Chaetomium globosum* | 1 (AMF31) | - | (33), (34), (35), (36) | |
| Cladosporium aggregatocicatricatum | 1 (AMF65) | - | - | |
| Cladosporium cladosporioides | 6 (AMF91, AMF36, AMF87, AMF95, AMF63, AMF89) | (3), (40), (41) | (37), (38), (42), (43), (44), (45), (46) | |
| Cladosporium dominicanum | 3 (AMF88, AMF96, AMF51 | - | - | |
| Cladosporium herbarum* | 1 (AMF85) | - | (29) (16) | |
| Cladosporium limoniforme | 1 (AMF35) | - | - | |
| Penicillium brevicompactum | 2 (AMF34, AMF86) | (3), (6), (57) | (20), (11), (14), (16), (21), (22), (12) (23), (19), (24) | |
| Penicillium chrysogenum | 4 (AMF4, AMF33, AMF40, AMF76) | (1), (2), (3), (4), (5), (40), (58) | (9), (10), (11), (12), (13), (14), (15), (16), (17) | |
| Penicillium citreonigrum | 3 (AMF39, AMF102, AMF103) | (3) | (15), (25), (26) | |
| Penicillium cyclopium | 3 (AMF47, AMF73 AMF74) | - | - | |
| Penicillium glabrum* | 1 (AMF56) | - | (11) | |
| Penicillium mononematosum | 2 (AMF27, AMF79) | - | - | |
| Penicillium olsonii | 1 (AMF72) | (3) | (14) | |
| Penicillium polonicum* | 1 (AMF16) | (49), (50), (51) | (14), (18), (48) | |
| Penicillium steckii | 2 (AMF25, AMF66) | (6) | (11), (19) | |
| Phialemonium inflatum | 1 (AMF9) | - | - | |
| Pleosporales sp. | 6 (AMF10, AMF6, AMF14, AMF11, AMF37, AMF38) | n.d | n.d | |
| Scedosporium sp. | 3 (AMF67, AMF71, AMF28) | (59) | (60) | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maamar, A.; Lucchesi, M.-E.; Debaets, S.; Nguyen van Long, N.; Quemener, M.; Coton, E.; Bouderbala, M.; Burgaud, G.; Matallah-Boutiba, A. Highlighting the Crude Oil Bioremediation Potential of Marine Fungi Isolated from the Port of Oran (Algeria). Diversity 2020, 12, 196. https://doi.org/10.3390/d12050196
Maamar A, Lucchesi M-E, Debaets S, Nguyen van Long N, Quemener M, Coton E, Bouderbala M, Burgaud G, Matallah-Boutiba A. Highlighting the Crude Oil Bioremediation Potential of Marine Fungi Isolated from the Port of Oran (Algeria). Diversity. 2020; 12(5):196. https://doi.org/10.3390/d12050196
Chicago/Turabian StyleMaamar, Ahlem, Marie-Elisabeth Lucchesi, Stella Debaets, Nicolas Nguyen van Long, Maxence Quemener, Emmanuel Coton, Mohammed Bouderbala, Gaëtan Burgaud, and Amaria Matallah-Boutiba. 2020. "Highlighting the Crude Oil Bioremediation Potential of Marine Fungi Isolated from the Port of Oran (Algeria)" Diversity 12, no. 5: 196. https://doi.org/10.3390/d12050196
APA StyleMaamar, A., Lucchesi, M.-E., Debaets, S., Nguyen van Long, N., Quemener, M., Coton, E., Bouderbala, M., Burgaud, G., & Matallah-Boutiba, A. (2020). Highlighting the Crude Oil Bioremediation Potential of Marine Fungi Isolated from the Port of Oran (Algeria). Diversity, 12(5), 196. https://doi.org/10.3390/d12050196







