Abstract
The discovery of several giant amoeba viruses has opened up a novel area in the field of virology. Despite this, knowledge about ecology of these viruses remains patchy. In this study, we aimed to characterize the diversity of giant viruses in Algeria by inoculating 64 environmental samples on various amoeba strains. After isolation by co-culture with nine amoeba supports, flow cytometry and electron microscopy were used to putatively identify viruses. Definitive identification was performed by PCR and sequencing. Mimiviruses, marseilleviruses, faustoviruses and cedratviruses were the main viruses isolated in this study. Moreover, a new virus, which we named fadolivirus, was also isolated and was found to belong to the recent metagenomic descriptions of Klosneuvirinae. Despite the use of 9 amoeba supports for co-culture, most of the isolates were obtained from two amoebas: Acanthamoeba castellanii Neff and Vermamoeba vermiformis CDC 19. Finally, the viruses most frequently isolated were marseilleviruses (55.5%) and Mimiviruses (22.2%). This work shows that the isolation of viruses previously detected by metagenomic analyses can be tedious, but possible.
Keywords:
Algeria; co-culture; giant viruses; cedratvirus; marseillevirus; mimivirus; faustovirus; fadolivirus; klosneuvirinae 1. Introduction
Megavirales order has recently been proposed to replace the current nucleocytoplasmic large DNA viruses (NCLDV) based on capsid and genome sizes, ancestral genes encoding autonomy transcription and even translation components [1,2,3,4,5,6]. This order contains viruses considered giant viruses and the story began at the beginning of the 21st century with the discovery of Acanthamoeba polyphaga mimivirus (APMV) [1,7]. These viruses modified the definition of what is a virus regarding their genomic contents, size and various phenotypic characteristics [1,7]. Six years later, a second isolation of giant viruses infecting amoeba was reported with the description of marseillevirus [1,7,8]. These last two families are currently recognized by the International Committee of Taxonomy of Viruses (ICTV).
Later, many other giant viruses infecting amoeba have been isolated and described in several geographical areas worldwide, such as Pandoravirus, Pithovirus, Mollivirus, faustovirus, cedratvirus, kaumoebavirus and pacmanvirus which have also recently been included into the proposed order of Megavirales [2,9,10,11,12,13,14,15]. Recently, several high throughput procedures were implemented to speed up the process of isolating giant virus in amoeba co-cultures and flow cytometry sorting was used to separate mixtures of these viruses and putatively identified the viral population [9,16,17,18].
Despite this, their natural hosts remain unknown, they seem to be ubiquitous in the environment and capable of surviving in highly diverse ecosystems [2,4]. They were isolated in Europe, Asia, Africa and America [5,9,14,17]. The viruses isolated to date appear to be the tip of the iceberg, as recent metagenomic studies have led to the discovery of a large number of new families [6,19]. The introduction of novel amoeba is a strategy consisting to potentially increase number of novel isolates. By the introduction of novel amoeba as V. vermiformis we isolated faustovirus follow by others as kaumoebavirus, orpheovirus. In a previous work [17], we compared different Acanthamoeba and we observed variations in the lineages and virus isolated depending the amoeba hosts. Based on those both experiences, we decided to test more novel hosts.
In the present study, we aimed to characterize the diversity of giant viruses in the Algerian environment using co-culture strategy on different amoeba cell supports from different types of environmental samples.
2. Materials and Methods
2.1. Samples Collection
A total of sixty-four environmental samples were collected in the environment of 4 different cities, including Mostaganem (35°55’59.999’’ N, 0°4’59.999’’ E), Chlef (36°8’26.664’’ N, 1°19’50.124’’ E), Sidi bel Abbes (35°12’0’’ N, 0°38’29’’ W) and Tlemcen (34°53’18.262’’ N, 1°19’4.815’’ W) located in north-west Algeria. Samples were collected between May and September 2017. The nature and origin of the samples are summarized in Table 1. All samples were stored in sterile tubes in a dark room at 4 °C until their inoculations. The soil samples were prepared by adding 5 mL of sterile distilled water and by vortex before inoculations.

Table 1.
Distribution of giant virus isolates with their relative amoeba and samples.
2.2. Preparation of Amoeba Support
A collection of nine amoeba was used to perform co-cultures: Acanthamoeba castellanii strain Neff (ATCC® 30010), A. castellanii strain Douglas (ATCC® 50374), Acanthamoeba polyphaga (strain Linc AP1), Acanthamoeba mauritaniensis (ATCC® 50253), Acanthamoeba quina (ATCC® 50241), Acanthamoeba culbertsoni (ATCC® 30171), Acanthamoeba divionensis (ATCC® 50238), Vermamoeba vermiformis (strain CDC 19) and Willaertia magna (ATCC® 50035). Fresh amoeba strains were grown in 75 cm2 cell culture flask (Corning®, Corning, NY, United States) containing 25 mL of peptone-yeast extract-glucose medium (PYG, Eurobio®, France) and incubated at 28 °C during 48 h for Acanthamoeba spp. and at 25 °C during 72 h for V. vermiformis train CDC 19) and W. magna (ATCC® 50035).
2.3. Isolation of Giant Viruses Using Co-Cultures
We used the protocol previously reported by Reteno et al. [12]. Briefly, amoeba cells suspensions were prepared at 5 × 105 cells/mL for Acanthamoeba strains and 1 × 106 cells/mL for V. vermiformis and W. magna. After centrifugation of amoebas (700× g during 10 min), we replaced the medium by starvation medium also named TS (homemade) [12] for all amoebas species except W. magna for which we added 10% of fetal bovine serum (FBS) in the TS medium. Doxycycline (20 µg/mL), vancomycin (10 µg/mL), ciprofloxacin (10 µg/mL), imipenem (10 µg/mL) and voriconazole (20 µg/mL) were added to cell suspension in order to eliminate contaminant overgrowth including intracellular bacteria and fungi. Then, 500 µL of each amoeba suspension was transferred to 24-wells plates and inoculated with 50 µL of each sample then incubated at 30 °C for 4 days for primo-culture. First and second blind subcultures were performed in new plates of 24-well, by inoculating 50 µL of the primo-culture in 500 µL of fresh cell suspension and incubated at 30 °C for 3 days. If fungal contamination was observed, supernatant was passed through 0.80-μm-pore sized filter before subculture. Third, the sub-culture was prepared as described above on fresh amoebae with doxycycline only (20 µg/mL). In each plate, four wells were used as negative controls with sterile water inoculation instead of sample.
2.4. Preliminary Characterization
We used Hemacolor® staining and optical microscopy for screening wells presenting a cytopathic effect. In this case, the contents of the wells where a cytopathic effect was detected were transferred to a 25 cm2 cell culture flask containing amoeba species used for co-culture with 5 mL of appropriate medium (as described above). For optical microscopy observation, 50 μL of amoeba cells before complete lysis were centrifuged and were then stained using Hemacolor® Rapid staining Kit according to the manufacturer’s recommendations. In parallel, 50 μL of the supernatants were passed to flow cytometry, as previously described, and population analyses were superimposed on those of the previously known viruses using FlowJow software [18]. In the event that the viral population did not overlap perfectly, or rare populations were detected, we performed transmission electron microscopy and molecular identification for confirmation.
2.5. Electron Microscopy Observations
We confirmed the nature of amoeba lytic agents (virus/bacteria) by electron microscopy. We performed a negative staining using a fixed supernatant from co-culture with glutaraldehyde following the same procedure previously described [18]. Embedding procedure was carried out as previously reported [15]. Electron micrographs were obtained on a Tecnai G20 F20 TEM (FEI, Bonn, Germany) operated at 200 keV. Image J software was used to determine particle size.
2.6. Molecular Analysis and Sequencing
Regarding flow cytometry profiles obtained (see section above), we identified potential viral sub-populations as described in previous studies [13,15,18]. Then, we confirmed the identification of viral isolates by both real-time using probes and by standard PCR methods and using specific primer to confirm marseilleviruses, mimiviruses, faustoviruses and cedratviruses (Table 2). The procedure of real-time PCRs of DNA extraction and PCR amplification were performed following the work of Ngounga et al. [20]. None sequencing of amplicons was done. Regarding results obtained by flow cytometry and by PCR we decided to do genomes by Next-genome sequencing.

Table 2.
Primer and probe sequences used for qPCR.
2.7. Genomic Sequencing and Phylogenetic Analyses
For viruses not identified by molecular analysis, 3 faustoviruses and cedratvirus N38, we carried out the sequencing of the genome. Cloned viruses were produced in 15 flasks (75 cm2) in starvation medium containing fresh A. castellanii or V. vermiformis monolayer which were cultivated for 48–72 h at 30 °C. After observation of lysis, supernatants were collected and centrifuged at 60,000× g for 45 min. Then, a 25% gradient sucrose was made following the same procedure previously reported [13,18]. Stocks were kept at − 80 °C. Genome sequencing was performed using MiSeq Technology (Illumina, Inc., San Diego, CA, USA) and using the paired-end application in parallel in a 2 × 251 bp run for each bar-coded library. Genome assembly was performed with hybrid spades with default parameter [21]. Phylogenetic tree were built using MEGA 6.0 package (https://www.megasoftware.net), using MUSCLE for alignments and maximum likelihood construction with JTT model in 1000 bootstrap replicates using all sites in the alignments. Sequences of predicted proteins used for phylogenetic analyses were available on NCBI (MT394892-MT394893-MT394894).
3. Results
3.1. Amoeba Lysis and Identification of Viruses
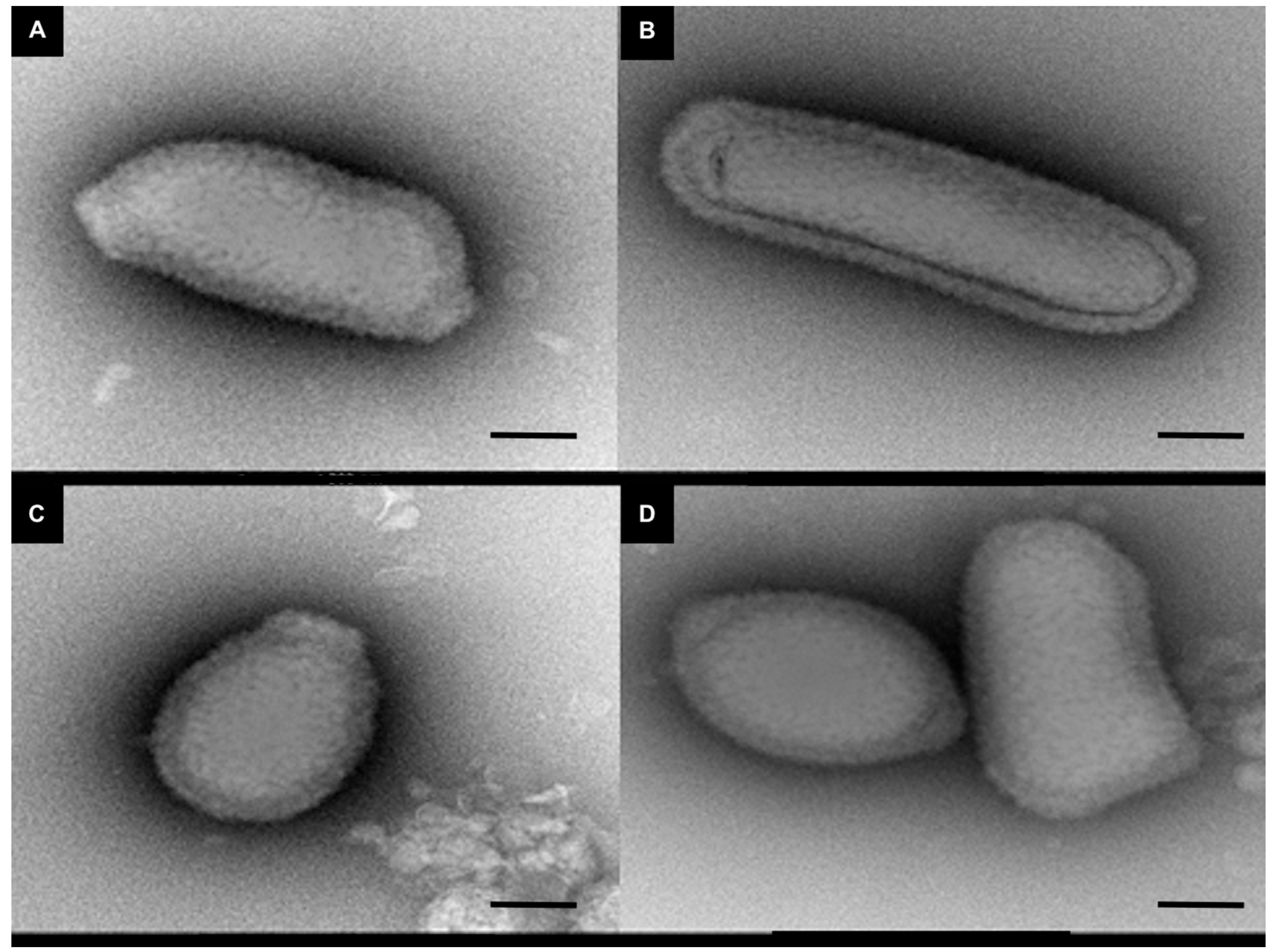
Among the 64 samples inoculated on 9 amoeba hosts, representing 576 wells, a cytopathic effect was observed in 63. Analysis by flow cytometry identified 45 viruses, while other amoeba lyses were due to intracellular bacteria and were not studied. Among these, 25 were identified as marseilleviruses, 10 as mimiviruses, 5 as faustoviruses, 4 as cedratviruses and one was a new virus that we named fadolivirus (Figure 1). The 4 cedratviruses showed a typical morphology of this family of viruses (Figure 2). Rates of isolation as the virus species isolated varied dramatically according to amoeba used as support. Twenty-two viruses (48.9% of the total number of viruses) were isolated in A. castellanii Neff, 10 (22.2%) viruses were isolated in A. polyphaga, 6 (13.3%) in V. vermiformis, 3 (6.7%) in A. castellanii Douglas, 3 (6.7%) in A. mauritaniensis and 1 (2.2%) in A. quina. No viruses were isolated in A. culbertsonii, A. divionensis and W. magna.
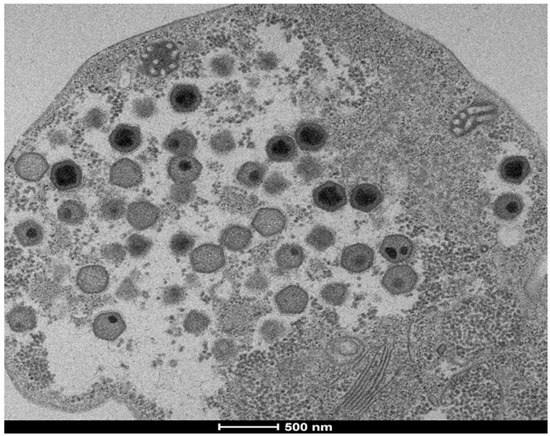
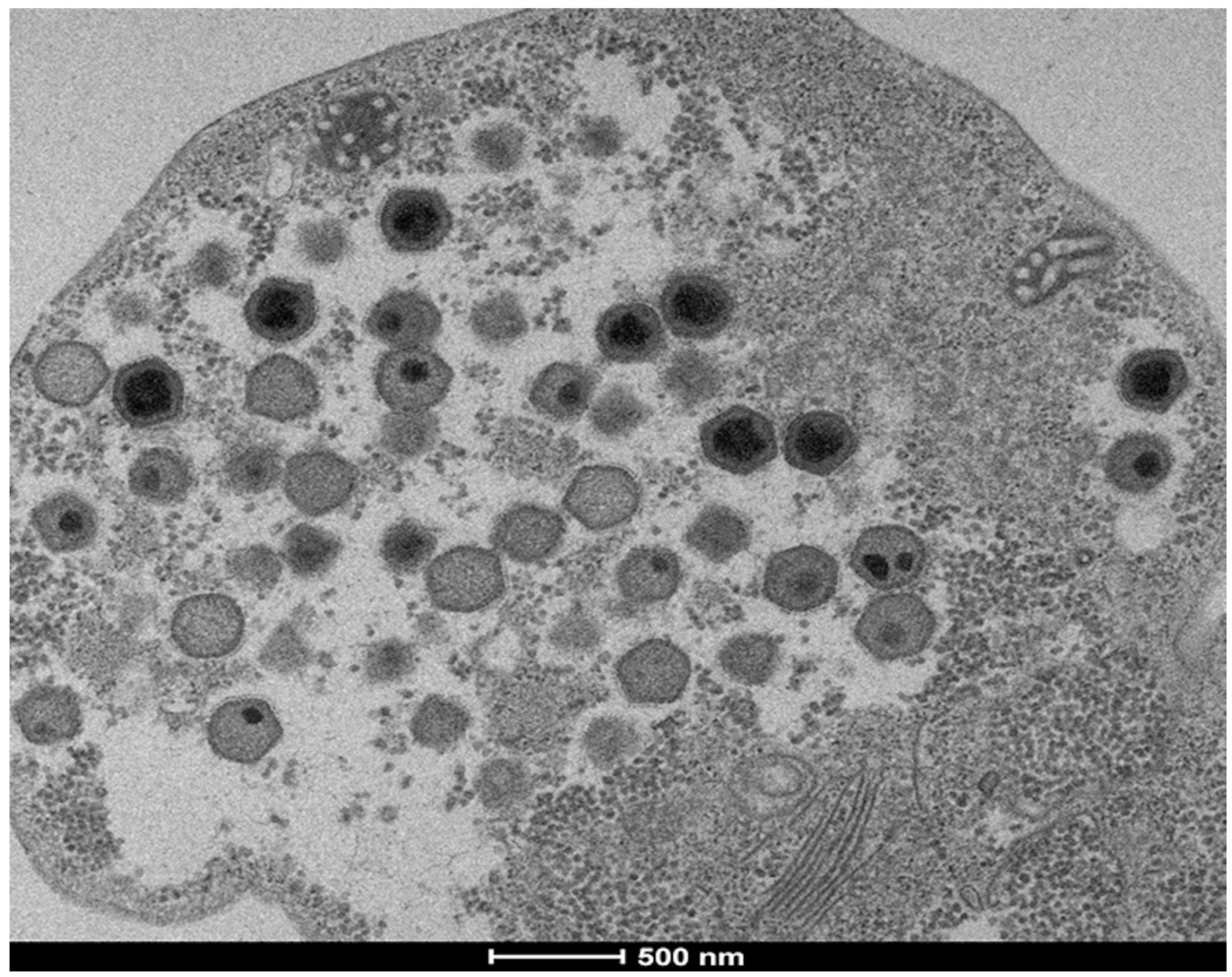
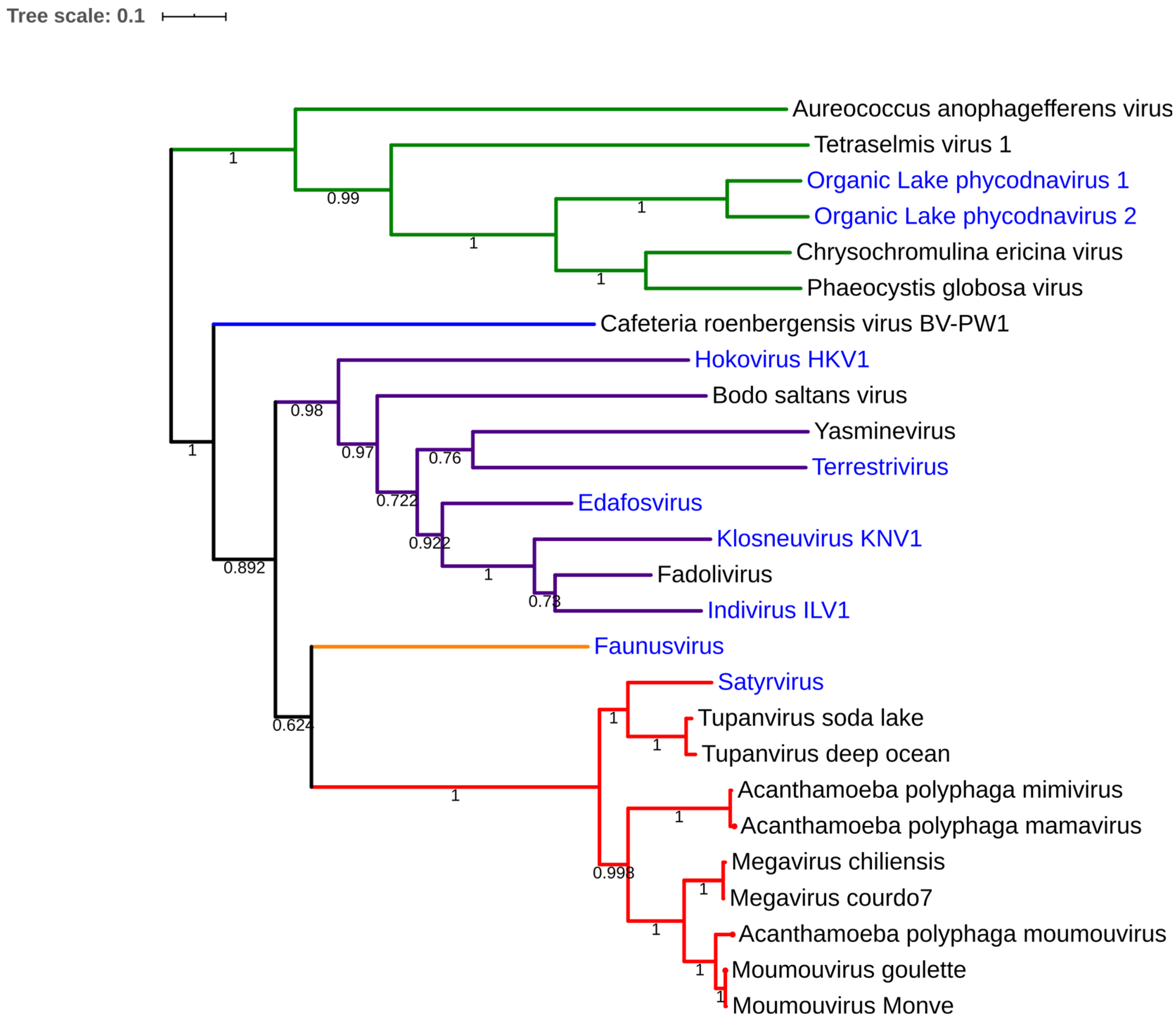
Figure 1.
Ultrathin section of fadolivirus particles inside its host. The observation was made on the host Vermamoeba vermiformis at 36 h post infection. Scale bar is indicated on figure.
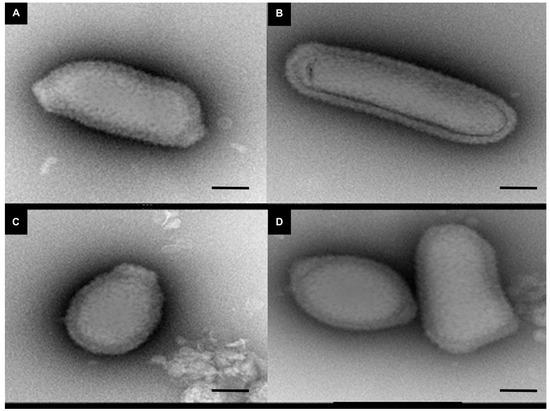
Figure 2.
Negative staining of cedratvirus N38 particles. Black scale bar indicates 200 nm. (A), (B), (C) and (D) show different forms of cedratvirus-like N38 isolated from Chlef City.
Cedratviruses were isolated with A. castellanii Neff only. Faustoviruses and fadolivirus were isolated in V. vermiformis only. Marseilleviruses were also isolated with A. mauritaniensis and A. quina, as well as mimiviruses that were isolated with three different amoeba species: A. polyphaga, A. castellanii Douglas and A. castellanii Neff (Table 2). Among the 31/64 (48.43%) samples positive for virus isolation, 17 were from sewage samples, 10 from water samples and 4 from soil samples. Comparing three types of samples, we found 25 (55.6%) isolates from sewage samples. Of these 25 isolates: 6 mimiviruses, 15 marseilleviruses, 3 faustoviruses and 1 with a new virus, followed by 18 (40%) isolates from water samples: 4 mimiviruses, 9 marseilleviruses, 2 faustoviruses and 3 cedratviruses.
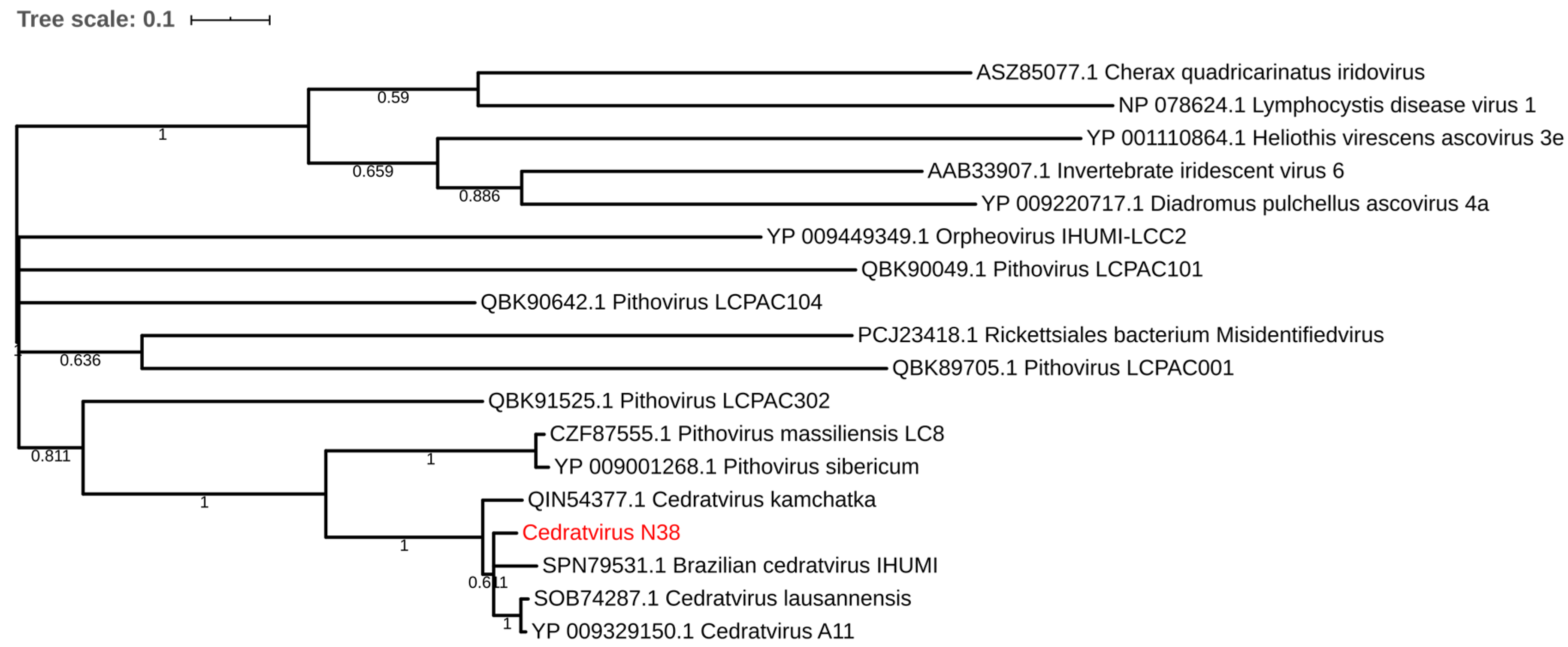
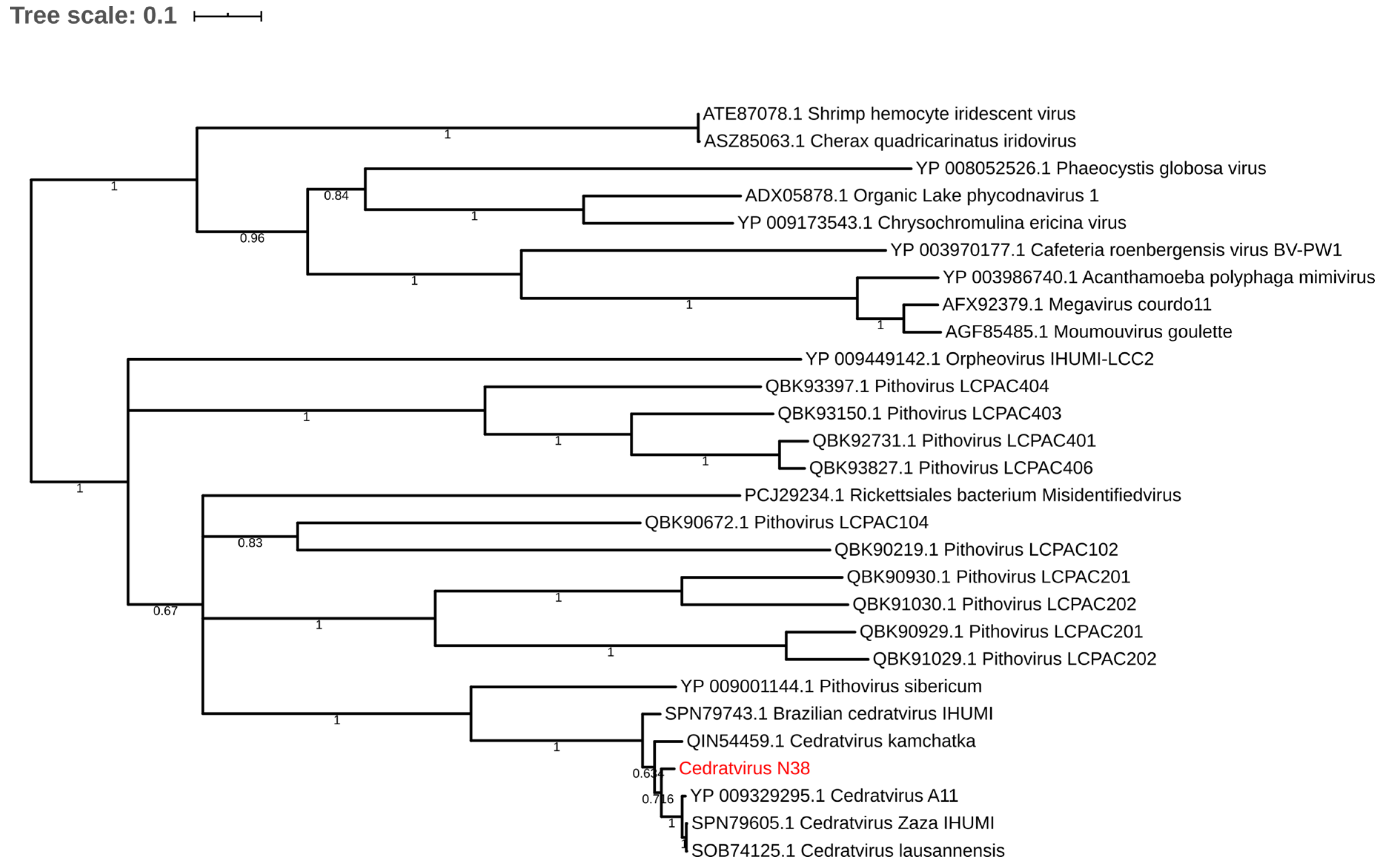
Regarding the distribution of viruses, most isolates came from Mostaganem city 25 (55.6%) followed by Chlef 11 (24.4%), Tlemcen 6 (13.33%) and Sidi bel Abbes 3 (6.66%). For the sample cedratvirus N38, we performed MiSeq sequencing and brief genome analyses. The genome is too partial to lead to a complete analysis. Indeed, we obtained more than 40 scaffolds with an average coverage from 130 to 440 depending scaffold considered and with a range of length from 1600 base pairs to 199,764 base pairs. However, and according to the phylogenetic trees based on the predicted proteins RNA polymerase subunits 1 and 2 (Figure 3 and Figure 4)—we observed the separation between the Brazilian cedratvirus on one side (Lineage B) and the members of lineages A on the other (cedratvirus A11, cedratvirus lausannensis and cedratvirus Zaza) [13,22,23] and another probable lineage with cedratvirus kamchatka (466,767 bp). This suggests that cedratvirus N38 could be a prototype member of a novel lineage and expand the known diversity between cedratvirus strains.
Figure 3.
Maximum-likelihood tree based on the RNA polymerase subunit 1. Branch values lower than a bootstrap value of 0.5 were deleted.
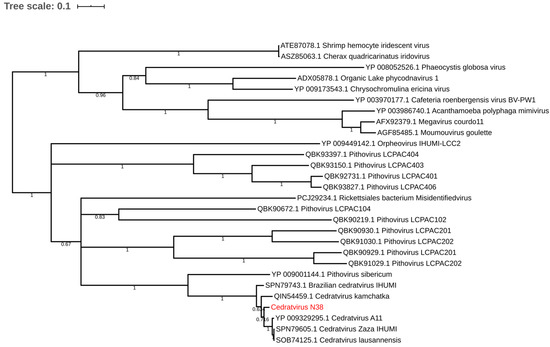
Figure 4.
Maximum-likelihood tree based on the RNA polymerase subunit 2. Branch values lower than a bootstrap value of 0.5 were deleted.
3.2. Isolation of a New Virus Belonging to the Klosneuvirinae Family
Fadolivirus was isolated from a sewage collected in Sidi bel Abbes on V. vermiformis. Ultrathin section permitted to observe icosahedral viral particles of about 300 nm (Figure 1) and seems to possess short fibrils on its capsid. Inside the capsid, the DNA packaging shows a progressive acquisition of density (dark electron dense core) with finally multi-layers, as observed for Bodo saltans virus [24]. The first genomic draft confirms the isolation of Klosneuvirinae-like viruses with a current draft genome higher than 1.5 Megabases and with an average coverage of 121. Indeed, the phylogenetic analysis based on the RNA polymerase subunit 1 shows a relative proximity to viruses recently reconstructed from metagenomics [6,19]. In particular, maximum-likelihood tree of fadolivirus with this protein show a relatively close distance with Indivirus and Klosneuvirus (Figure 5). Currently, this isolation needs further description in order to characterize a clear and complete genomic comparison.
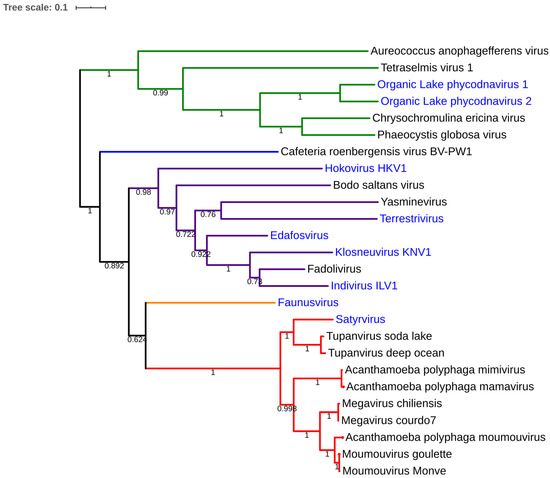
Figure 5.
Maximum-likelihood tree based on the RNA polymerase subunit 1 of Mimiviridae.
Red color indicates nodes containing mimivirus lineages A, B and C and Tupanviruses, orange was used for Faunusvirus, purple for Klosneuvirinae, Blue for Cafeteria roenbergensis virus, green for Mimiviridae infecting algae. We deleted branches supported by bootstrap value of less than 0.6. No homolog of Rpb1 was found in the draft genome of dasosvirus, barrevirus, hyperionvirus. harfovirus, satyrvirus, homavirus, geavirus genomes present a truncated homolog of Rpb1. Blue labels concern viruses detected by metagenomic analyses.
4. Discussion
The results of this study are in agreement with those previously reported regarding giant viruses’ distribution in ecosystems and confirmed their ubiquitous presence [2,18]. We identified 45 giant viruses from 64 Algerian environmental samples. The higher rates of isolation of giant virus were found in sewage samples (54.8%) and are in accordance with those reported in previous studies investigating viruses in the Old world [12,25] and in contrast to studies in the New World [17,26] which showed an increased rate of isolation in water samples. Numerous Mimiviruses and Marseilleviruses were isolated as well as 5 new strains of faustovirus still undergoing genome analysis. In addition, we isolated four strains of cedratvirus with elongated shape, all these cedratvirus-like being identified correctly with real time PCR. Among these, we identified a probable novel lineage of cedratvirus, named cedratvirus N38. These new isolates need a wider complete genome sequencing to determine their exact position. Furthers studies are needed to characterize this novel giant virus named fadolivirus presenting a 300-nm icosahedral capsid and having a close relationship with the recent described putative Klosneuvirinae [19,27]. Recent detection of new families of viruses using a smart approach of mini-metagenomics rather than bulk metagenomics suggests that some of these new families could be in extremely low abundance [6]. As a result, their isolation is probably due to a longer incubation period, very careful well observations and randomness. However, the development of very high throughput procedures for the isolation of giant viruses [28] and the combination with sorting before inoculation, as is done for mini-metagenomics, could be a good strategy in the future to isolate these hidden giant viruses.
Meanwhile, we observed trends similar to those found in a previous study regarding the rate of isolation according to the cell support [17]. Indeed, Dornas et al. reported variation in the mimivirus isolations using 3 different strains of Acanthamoeba sp., A. polyphaga, A. castellanii Neff and A. griffini. Despite this, we observed herein that the rate of isolation could be extremely variable, from about 48% of well lysis with A. castellanii Neff than 0% in W. magna, A. culbertsonii or A. divionensis. This evidence brings complexity in current hypothesis of host prediction. Indeed, bioinformatics and metagenomics studies enable the detection and recovery of many giant viruses [29]. However, the detection of their hosts is currently probabilistic, mainly based on the presence of 18S rRNA genes. We currently do not know their real ability to infect hosts, but our co-culture experience has shown us that this range could be restrictive. To date, only Tupanvirus is capable of infecting two amoebas of different genera, V. vermiformis and Acanthamoeba spp [5]. It is also possible that some amoebae have a high permissiveness to viruses that are not their natural host, as was observed with VERO cells and human viral pathogens.
Author Contributions
Conceptualization, J.A. and B.L.S.; data curation, H.B.; formal analysis, H.B.; supervision, I.B. and B.L.S.; writing—original draft, H.B.; writing—review & editing, J.A., I.B. and B.L.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from the French State managed by the National Research Agency under the "Investissements d’avenir” (Investments for the Future) program with the reference ANR-10-IAHU-03 (Méditerranée Infection) and by the Region Provence-Alpes-Côte d’Azur and European funding FEDER PRIMI.
Acknowledgments
The authors kindly thank Celine Perreal for her kind assistance, Priscilla Jardot for q-PCR and Fabrizio Di Pinto for electronic microscopy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Raoult, D.; Audic, S.; Robert, C.; Abergel, C.; Renesto, P.; Ogata, H.; La Scola, B.; Suzan, M.; Claverie, J.-M. The 1.2-Megabase Genome Sequence of Mimivirus. Science 2004, 306, 1344–1350. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; De Lamballerie, X.; Yutin, N.; Asgari, S.; Bigot, Y.; Bideshi, D.K.; Cheng, X.-W.; Federici, B.A.; Van Etten, J.L.; Koonin, E.V.; et al. “Megavirales” a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. 2013, 158, 2517–2521. [Google Scholar] [CrossRef] [PubMed]
- Legendre, M.; Lartigue, A.; Bertaux, L.; Jeudy, S.; Bartoli, J.; Lescot, M.; Alempic, J.-M.; Ramus, C.; Bruley, C.; Labadie, K.; et al. In-depth study ofMollivirus sibericum, a new 30,000-y-old giant virus Infecting Acanthamoeba. Proc. Natl. Acad. Sci. USA 2015, 112, E5327–E5335. [Google Scholar] [CrossRef] [PubMed]
- Aherfi, S.; Colson, P.; La Scola, B.; Raoult, D. Giant Viruses of Amoebas: An Update. Front. Microbiol. 2016, 7, 12406. [Google Scholar] [CrossRef]
- Abrahao, J.; Silva, L.; Silva, L.S.; Khalil, J.Y.B.; Rodrigues, R.A.L.; Arantes, T.; Assis, F.; Boratto, P.; Andrade, M.; Kroon, E.G.; et al. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nat. Commun. 2018, 9, 749. [Google Scholar] [CrossRef]
- Schulz, F.; Yutin, N.; Ivanova, N.N.; Ortega, D.R.; Lee, T.K.; Vierheilig, J.; Daims, H.; Horn, M.; Wagner, M.; Jensen, G.J.; et al. Giant viruses with an expanded complement of translation system components. Science 2017, 356, 82–85. [Google Scholar] [CrossRef]
- La Scola, B. A Giant Virus in Amoebae. Science 2003, 299, 2033. [Google Scholar] [CrossRef]
- Boyer, M.; Yutin, N.; Pagnier, I.; Barrassi, L.; Fournous, G.; Espinosa, L.; Robert, C.; Azza, S.; Sun, S.; Rossmann, M.G.; et al. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. USA 2009, 106, 21848–21853. [Google Scholar] [CrossRef]
- Boughalmi, M.; Saadi, H.; Pagnier, I.; Colson, P.; Fournous, G.; Raoult, D.; La Scola, B. High-throughput isolation of giant viruses of the Mimiviridae and Marseilleviridae families in the Tunisian environment. Environ. Microbiol. 2013, 15, 2000–2007. [Google Scholar] [CrossRef]
- Campos, R.K.; Boratto, P.V.; De Assis, F.L.; Aguiar, E.; Silva, L.; Albarnaz, J.D.; Dornas, F.; Trindade, G.D.S.; Ferreira, P.C.P.; Marques, J.T.; et al. Samba virus: A novel mimivirus from a giant rain forest, the Brazilian Amazon. Virol. J. 2014, 11, 95. [Google Scholar] [CrossRef]
- Legendre, M.; Bartoli, J.; Shmakova, L.; Jeudy, S.; Labadie, K.; Adrait, A.; Lescot, M.; Poirot, O.; Bertaux, L.; Bruley, C.; et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc. Natl. Acad. Sci. USA 2014, 111, 4274–4279. [Google Scholar] [CrossRef] [PubMed]
- Reteno, D.G.; Benamar, S.; Khalil, J.B.; Andreani, J.; Armstrong, N.; Klose, T.; Rossmann, M.; Colson, P.; Raoult, D.; La Scola, B. Faustovirus, an Asfarvirus-Related New Lineage of Giant Viruses Infecting Amoebae. J. Virol. 2015, 89, 6585–6594. [Google Scholar] [CrossRef] [PubMed]
- Andreani, J.; Aherfi, S.; Khalil, J.Y.; Di Pinto, F.; Bitam, I.; Raoult, D.; Colson, P.; La Scola, B. Cedratvirus, a Double-Cork Structured Giant Virus, is a Distant Relative of Pithoviruses. Viruses 2016, 8, 300. [Google Scholar] [CrossRef] [PubMed]
- Bajrai, L.; Benamar, S.; Azhar, E.I.; Robert, C.; Levasseur, A.; Raoult, D.; La Scola, B. Kaumoebavirus, a New Virus That Clusters with Faustoviruses and Asfarviridae. Viruses 2016, 8, 278. [Google Scholar] [CrossRef] [PubMed]
- Andreani, J.; Khalil, J.Y.B.; Sevvana, M.; Benamar, S.; Di Pinto, F.; Bitam, I.; Colson, P.; Klose, T.; Rossmann, M.G.; Raoult, D.; et al. Pacmanvirus, a New Giant Icosahedral Virus at the Crossroads between Asfarviridae and Faustoviruses. J. Virol. 2017, 91, e00212-17. [Google Scholar] [CrossRef] [PubMed]
- Pagnier, I.; Reteno, D.-G.I.; Saadi, H.; Boughalmi, M.; Gaia, M.; Slimani, M.; Ngounga, T.; Bekliz, M.; Colson, P.; Raoult, D.; et al. A Decade of Improvements in Mimiviridae and Marseilleviridae Isolation from Amoeba. Intervirology 2013, 56, 354–363. [Google Scholar] [CrossRef]
- Dornas, F.P.; Khalil, J.Y.B.; Pagnier, I.; Raoult, D.; Abrahao, J.; La Scola, B. Isolation of new Brazilian giant viruses from environmental samples using a panel of protozoa. Front. Microbiol. 2015, 6, 477. [Google Scholar] [CrossRef]
- Khalil, J.Y.B.; Robert, S.; Reteno, D.G.; Andreani, J.; Raoult, D.; La Scola, B. High-Throughput Isolation of Giant Viruses in Liquid Medium Using Automated Flow Cytometry and Fluorescence Staining. Front. Microbiol. 2016, 7, 1124. [Google Scholar] [CrossRef]
- Schulz, F.; Alteio, L.; Goudeau, D.; Ryan, E.M.; Yu, F.B.; Malmstrom, R.R.; Blanchard, J.; Woyke, T. Hidden diversity of soil giant viruses. Nat. Commun. 2018, 9, 4881. [Google Scholar] [CrossRef]
- Ngounga, T.; Pagnier, I.; Reteno, D.-G.I.; Raoult, D.; La Scola, B.; Colson, P. Real-Time PCR Systems Targeting Giant Viruses of Amoebae and Their Virophages. Intervirology 2013, 56, 413–423. [Google Scholar] [CrossRef]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. hybridSPAdes: An algorithm for hybrid assembly of short and long reads. Bioinformatics 2015, 32, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Mueller, L.; Thomas, V.; Pillonel, T.; Jacquier, N.; Greub, G. Cedratvirus lausannensis—digging into Pithoviridaediversity. Environ. Microbiol. 2017, 19, 4022–4034. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.A.L.; Andrade, A.C.D.S.P.; Boratto, P.V.D.M.; Trindade, G.D.S.; Kroon, E.G.; Abrahão, J. An Anthropocentric View of the Virosphere-Host Relationship. Front. Microbiol. 2017, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Deeg, C.M.; Chow, C.-E.T.; Suttle, C.A. The kinetoplastid-infecting Bodo saltans virus (BsV), a window into the most abundant giant viruses in the sea. eLife 2018, 7, e33014. [Google Scholar] [CrossRef]
- La Scola, B.; Campocasso, A.; N’Dong, R.; Fournous, G.; Barrassi, L.; Flaudrops, C.; Raoult, D. Tentative Characterization of New Environmental Giant Viruses by MALDI-TOF Mass Spectrometry. Intervirology 2010, 53, 344–353. [Google Scholar] [CrossRef]
- Andrade, A.C.D.S.P.; Arantes, T.S.; Rodrigues, R.A.L.; Machado, T.B.; Dornas, F.P.; Landell, M.F.; Furst, C.; Borges, L.G.A.; Dutra, L.A.L.; Almeida, G.M.D.F.; et al. Ubiquitous giants: A plethora of giant viruses found in Brazil and Antarctica. Virol. J. 2018, 15, 22. [Google Scholar] [CrossRef]
- Bajrai, L.H.; Mougari, S.; Andreani, J.; Baptiste, E.; Delerce, J.; Raoult, D.; Azhar, E.I.; La Scola, B.; Levasseur, A. Isolation of Yasminevirus, the First Member of Klosneuvirinae Isolated in Coculture with Vermamoeba vermiformis, Demonstrates an Extended Arsenal of Translational Apparatus Components. J. Virol. 2019, 94. [Google Scholar] [CrossRef]
- Francis, R.; Ominami, Y.; Khalil, J.Y.; La Scola, B. High-throughput isolation of giant viruses using high-content screening. Commun. Biol. 2019, 2, 216. [Google Scholar] [CrossRef]
- Andreani, J.; La Scola, B. Metagenomic binning reconstruction coupled with automatic pipeline annotation and giant viruses: A potential source of mistake in annotations. Virus Res. 2018, 255, 36–38. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).