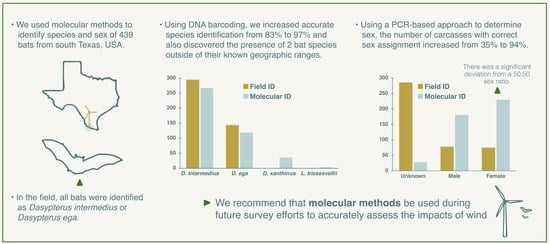
Genetic Approaches Are Necessary to Accurately Understand Bat-Wind Turbine Impacts
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. DNA Barcoding
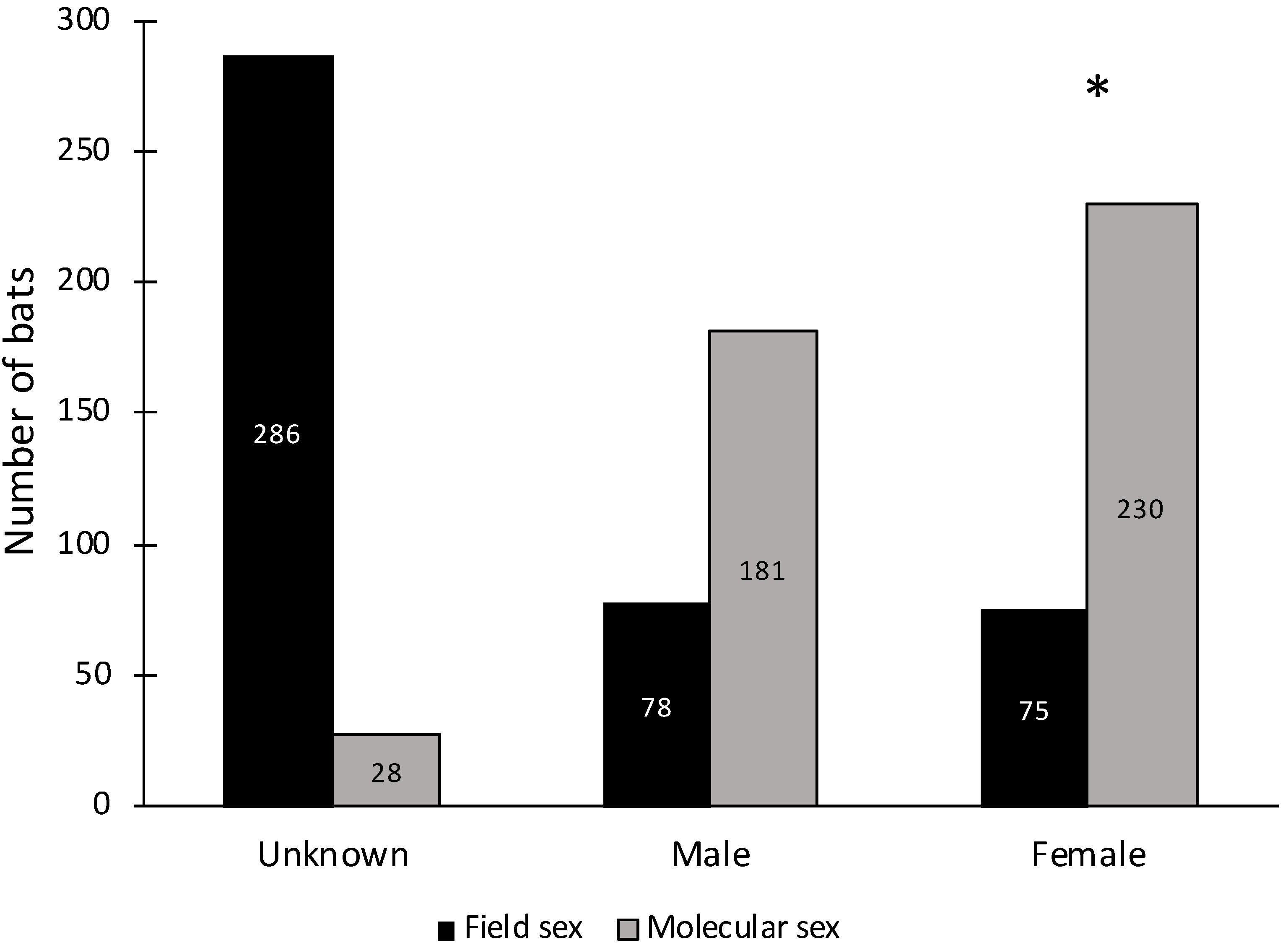
3.2. Molecular Sex Determination
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boyles, J.C.; Cryan, P.M.; McCracken, G.F.; Kunz, T.H. Economic importance of bats in agriculture. Science 2011, 332, 41–42. [Google Scholar] [CrossRef] [PubMed]
- Kunz, T.H.; de Torrez, E.B.; Bauer, D.; Lobova, T.; Fleming, T.H. Ecosystem services provided by bats. Ann. N. Y. Acad. Sci. 2011, 1233, 1–38. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, T.J.; Cryan, P.M.; Hayman, D.T.S.; Plowright, R.K.; Steicker, D.G. Multiple mortality events in bats: A global review. Mamm. Rev. 2016, 46, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Frick, W.F.; Kingston, T.; Flanders, J. A review of the major threats and challenges to global bat conservation. Ann. N. Y. Acad. Sci. 2019, 1–21. [Google Scholar] [CrossRef]
- Tuttle, M.D. Fear of bats and its consequences. Bat Res. Cons. 2017, 10, 1. [Google Scholar] [CrossRef]
- Williams-Guillén, K.; Olimpi, E.; Maas, B.; Taylor, P.; Arlettaz, R. Bats in the anthropogenic matrix: Challenges and opportunities for the conservation of Chiroptera and their ecosystem services in agricultural landscapes. In Bats in the Anthropocene: Conservation of Bats in a Changing World; Voigt, C., Kingston, T., Eds.; Springer: New York, NY, USA, 2016; pp. 151–186. [Google Scholar]
- Kiesecker, J.M.; Evans, J.S.; Fargione, J.; Doherty, K.; Foresman, K.R.; Kunz, T.H.; Naugle, D.; Nibbelink, N.P.; Niemuth, N.D. Win-win for wind and wildlife: A vision to facilitate sustainable development. PLoS ONE 2011, 6, e17566. [Google Scholar] [CrossRef] [Green Version]
- Allison, T.D.; Diffendorfer, J.E.; Baerwald, E.F.; Beston, J.A.; Drake, D.; Hale, A.M.; Hein, C.D.; Huso, M.M.; Loss, S.R.; Lovich, J.E.; et al. Impacts to wildlife of wind energy siting and operation in the United States. Issues Ecol. 2019, 21, 1–24. [Google Scholar]
- Kunz, T.H.; Arnett, E.B.; Erickson, W.P.; Hoar, A.R.; Johnson, G.D.; Larkin, R.P.; Strickland, M.D.; Thresher, R.W.; Tuttle, M.D.M. Ecological impacts of wind energy development on bats: Questions, research needs, and hypotheses. Fron. Ecol. Environ. 2007, 5, 315–324. [Google Scholar] [CrossRef] [Green Version]
- Erickson, R.A.; Thogmartin, W.E.; Diffendorfer, J.E.; Russell, R.E.; Szymanski, J.A. Effects of wind energy generation and white-nose syndrome on the viability of the Indiana bat. PeerJ. 2016, 4, e2830. [Google Scholar] [CrossRef]
- Frick, W.F.; Baerwald, E.F.; Pollock, J.F.; Barclay, R.M.R.; Szymanski, J.A.; Weller, T.J.; Russell, A.L.; Loeb, S.C.; Medellin, R.A.; McGuire, L.P. Fatalities at wind turbines may threaten population viability of a migratory bat. Biol. Cons. 2017, 209, 172–177. [Google Scholar] [CrossRef]
- Rodhouse, T.J.; Rodriguez, R.M.; Banner, K.M.; Ormsbee, P.C.; Barnett, J.; Irvine, K.M. Evidence of region-wide bat population decline from long-term monitoring and Bayesian occupancy models with empirically informed priors. Ecol. Evo. 2019, 9, 11078–11088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnett, E.B.; Huso, M.M.; Schirmacher, M.R.; Hayes, J.P. Altering turbine speed reduces bat mortality at wind-energy facilities. Fron. Ecol. Environ. 2011, 9, 209–214. [Google Scholar] [CrossRef] [Green Version]
- Martin, C.M.; Arnett, E.B.; Stevens, R.D.; Wallace, M.C. Reducing bat fatalities at wind facilities while improving the economic efficiency of operational mitigation. J. Mamm. 2017, 98, 378–385. [Google Scholar] [CrossRef]
- Hayes, M.A.; Hooton, L.A.; Gilland, K.L.; Grandgent, C.; Smith, R.L.; Lindsay, S.R.; Collins, J.D.; Schumacher, S.M.; Rabie, P.A.; Gruver, J.C.; et al. A smart curtailment approach for reducing bat fatalities and curtailment time at wind energy facilities. Ecol. Appl. 2019, 29, e01881. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.P.; Hein, C.D.; Simpson, T.R.; Evans, J.W.; Castro-Arellano, I. Ultrasonic acoustic deterrents significantly reduce bat fatalities at wind turbines. Global. Ecol. Cons. 2020, e01099. [Google Scholar] [CrossRef]
- Korstian, J.M.; Hale, A.M.; Bennett, V.J.; Williams, D.A. Using DNA barcoding to improve bat carcass identification at wind farms in the United States. Cons. Gen. 2016, 8, 27–34. [Google Scholar] [CrossRef]
- Nelson, D.M.; Nagel, J.; Trott, R.; Campbell, C.J.; Pruitt, L.; Good, R.E.; Iskali, G.; Gugger, P.F. Carcass age and searcher identity affect morphological assessment of sex of bats. J. Wild Man 2018, 82, 1582–1587. [Google Scholar] [CrossRef]
- Korstian, J.M.; Hale, A.M.; Bennett, V.J.; Williams, D.A. Advances in sex determination in bats and its utility in wind-wildlife studies. Mol. Ecol. Res. 2013, 13, 776–780. [Google Scholar] [CrossRef]
- American Wind Wildlife Institute (AWWI). AWWI Technical Report: A Summary of Bat Fatality Data in a Nationwide Database; AWWI: Washington, DC, USA, 2018; Available online: www.awwi.org (accessed on 12 November 2019).
- Arnett, E.; Baerwald, E.F. Impacts of wind energy development on bats: Implications for conservation. In Bat Evolution, Ecology, and Conservation; Adams, R., Pederson, C., Eds.; Springer: New York, NY, USA, 2013; pp. 435–456. [Google Scholar]
- Zimmerling, J.R.; Francis, C.M. Bat mortality due to wind turbines in Canada. J. Wild Man 2016, 80, 1360–1369. [Google Scholar] [CrossRef]
- Baird, A.B.; Braun, J.K.; Mares, M.A.; Morales, J.C.; Patton, J.C.; Tran, C.Q.; Bickham, J.W. Molecular systematic revision of tree bats (Lasiurini): Doubling the native mammals of the Hawaiian Islands. J. Mamm. 2015, 96, 1255–1274. [Google Scholar] [CrossRef] [Green Version]
- Baird, A.B.; Braun, J.K.; Engstrom, M.D.; Holbert, A.C.; Huerta, M.G.; Lim, B.K.; Mares, M.A.; Patton, J.C.; Bickham, J.W. Nuclear and mtDNA phylogenetic analyses clarify the evolutionary history of two species of native Hawaiian bats and the taxonomy of Lasiurini (Mammalia: Chiroptera). PLoS ONE 2017, 12, e0186085. [Google Scholar] [CrossRef] [PubMed]
- Ammerman, L.K.; Hice, C.L.; Schmidly, D.J. Bats of Texas; Texas A&M University Press: College Station, TX, USA, 2012. [Google Scholar]
- Decker, S.K.; Krejsa, D.M.; Lindsey, L.L.; Amoateng, R.P.; Ammerman, L.K. Updated distributions of three species of yellow bat (Dasypterus) in Texas based on specimen records. W Wildl. 2020, 7, 2–8. [Google Scholar]
- Griffith, G.; Bryce, S.; Omernik, J.; Rogers, A. Ecoregions of Texas; Texas Commission on Environmental Quality: Austin, TX, USA,, 2007. [Google Scholar]
- Ceballos, G. Mammals of Mexico; Johns Hopkins University Press: Baltimore, MD, USA, 2014. [Google Scholar]
- Weaver, S.P. Understanding wind energy impacts on bats and testing reduction strategies in south Texas. Ph.D. Thesis, Texas State University, San Marcos, TX, USA, 2019. [Google Scholar]
- Brunet-Rossinni, A.K.; Wilkinson, G.S. Methods for age estimation and the study of senescence in bats. In Ecological and Behavioral Methods for the Study of Bats; Kunz, T.J., Parsons, S., Eds.; Johns Hopkins University Press: Baltimore, MD, USA, 2009; pp. 315–328. [Google Scholar]
- Kunz, T.H.; Whitaker, J.O.; Wadanoli, M.D. Dietary energetics of the insectivorous Mexican free-tailed bat (Tadarida brasiliensis) during pregnancy and lactation. Oecologia 1995, 101, 407–415. [Google Scholar] [CrossRef] [PubMed]
- McCracken, G.F.; Gillam, E.H.; Westbrook, J.K.; Lee, Y.F.; Jensen, M.L.; Balsley, B.B. Brazilian free-tailed bats (Tadarida brasiliensis: Molossidae, Chiroptera) at high altitude: Links to migratory insect populations. Integr. Comp. Biol. 2008, 48, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, A.L.; Cox, M.P.; Brown, V.A.; McCracken, G.F. Population growth of Mexican free-tailed bats (Tadarida brasiliensis mexicana) predates human agricultural activity. BMC Evol. Biol. 2011, 11, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Clare, E.L.; Lim, B.K.; Engstrom, M.D.; Eger, J.L.; Hebert, P.D.N. DNA barcoding of Neotropical bats: Species identification and discovery within Guyana. Mol. Ecol. Notes 2007, 7, 184–190. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Gonzalez, E.; Barquez, R.; Miller, B. Lasiurus blossevillii. The IUCN Red List of Threatened Species 2016: E.T88151055A22120040. Available online: https://dx.doi.org/10.2305/IUCN.UK.2016-1.RLTS.T88151055A22120040.en (accessed on 20 January 2020).
- Miller, B.; Rodriguez, B. Lasiurus intermedius (errata version published in 2017). The IUCN Red List of Threatened Species 2016: E.T11352A115101697. Available online: https://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T11352A22119630.en (accessed on 20 January 2020).
- Barquez, R.; Diaz, M. Lasiurus ega. The IUCN Red List of Threatened Species 2016: E.T11350A22119259. Available online: https://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T11350A22119259.en (accessed on 20 January 2020).
- Arroyo-Cabrales, J.; Álvarez-Castañeda, S.T. Lasiurus xanthinus. The IUCN Red List of Threatened Species 2017: E.T41532A22004260. Available online: https://dx.doi.org/10.2305/IUCN.UK.2017-2.RLTS.T41532A22004260.en. (accessed on 20 January 2020).
- Decker, S.K. Phylogeographic analysis of northern yellow bats, Dasypterus intermedius, by molecular analysis. Honors Thesis, Angelo State University, San Angelo, TX USA, 2019. Available online: https://hdl.handle.net/2346.1/30937.en (accessed on 21 May 2019).
- Zabriskie, J.E.; Cutler, P.L.; Stuart, J.N. Range extension of the western yellow bat (Dasypterus xanthinus) in New Mexico. W Wildl. 2019, 6, 1–4. [Google Scholar]
- Geluso, K.; Valdez, E.W. First records of the eastern red bat (Lasiurus borealis) in Arizona, Utah, and western New Mexico; Occasional Papers, Number 361; Museum of Texas Tech University: Lubbock, TX, USA, 2019. [Google Scholar]
- Solick, D.I.; Barclay, R.M.R.; Bishop-Boros, L.; Hays, Q.R.; Lausen, C.L. Distributions of eastern and western red bats in western North America. West. NA Nat. 2020, 80, 90–97. [Google Scholar] [CrossRef]
- Johnson, G.D.; Erickson, W.P.; Strickland, M.D.; Shepherd, M.F.; Shepherd, D.A.; Sarappo, S.A. Mortality of bats at a large-scale wind power development at Buffalo Ridge, Minnesota. Am. Mid. Nat. 2003, 150, 332–342. [Google Scholar] [CrossRef]
- Arnett, E.B.; Brown, W.K.; Erickson, W.P.; Fiedler, J.K.; Hamilton, B.L.; Henry, T.H.; Jain, A.; Johnson, G.D.; Kerns, J.; Koford, R.R.; et al. Patterns of bat fatalities at wind energy facilities in North America. J. Wild Man 2008, 72, 61–78. [Google Scholar] [CrossRef]
- Grüebler, M.U.; Schuler, H.; Müller, M.; Sparr, R.; Horch, P.; Naef-Daenzer, B. Female biased mortality caused by anthropogenic nest loss contributes to population decline and adult sex ratio of a meadow bird. Biol. Cons. 2008, 141, 3030–3049. [Google Scholar] [CrossRef]
- Cryan, P.M.; Jameson, J.W.; Baerwald, E.F.; Willis, C.K.R.; Barclay, R.M.R.; Snider, E.A.; Crichton, E.G. Evidence of late-summer mating readiness and early sexual maturation in migratory tree-roosting bats found dead at wind turbines. PLoS ONE 2012, 7, e47586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickett, E.J.; Stockwell, M.P.; Pollard, C.J.; Garnham, J.I.; Clulow, J.; Mahony, M.J. Estimates of sex ratio require the incorporation of unequal catchability between sexes. Wild. Res. 2012, 39, 350–354. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chipps, A.S.; Hale, A.M.; Weaver, S.P.; Williams, D.A. Genetic Approaches Are Necessary to Accurately Understand Bat-Wind Turbine Impacts. Diversity 2020, 12, 236. https://doi.org/10.3390/d12060236
Chipps AS, Hale AM, Weaver SP, Williams DA. Genetic Approaches Are Necessary to Accurately Understand Bat-Wind Turbine Impacts. Diversity. 2020; 12(6):236. https://doi.org/10.3390/d12060236
Chicago/Turabian StyleChipps, Austin S., Amanda M. Hale, Sara P. Weaver, and Dean A. Williams. 2020. "Genetic Approaches Are Necessary to Accurately Understand Bat-Wind Turbine Impacts" Diversity 12, no. 6: 236. https://doi.org/10.3390/d12060236
APA StyleChipps, A. S., Hale, A. M., Weaver, S. P., & Williams, D. A. (2020). Genetic Approaches Are Necessary to Accurately Understand Bat-Wind Turbine Impacts. Diversity, 12(6), 236. https://doi.org/10.3390/d12060236