Dimensions of Phyllostomid Bat Diversity and Assemblage Composition in a Tropical Forest-Agricultural Landscape
Abstract
:1. Introduction
2. Materials and Methods
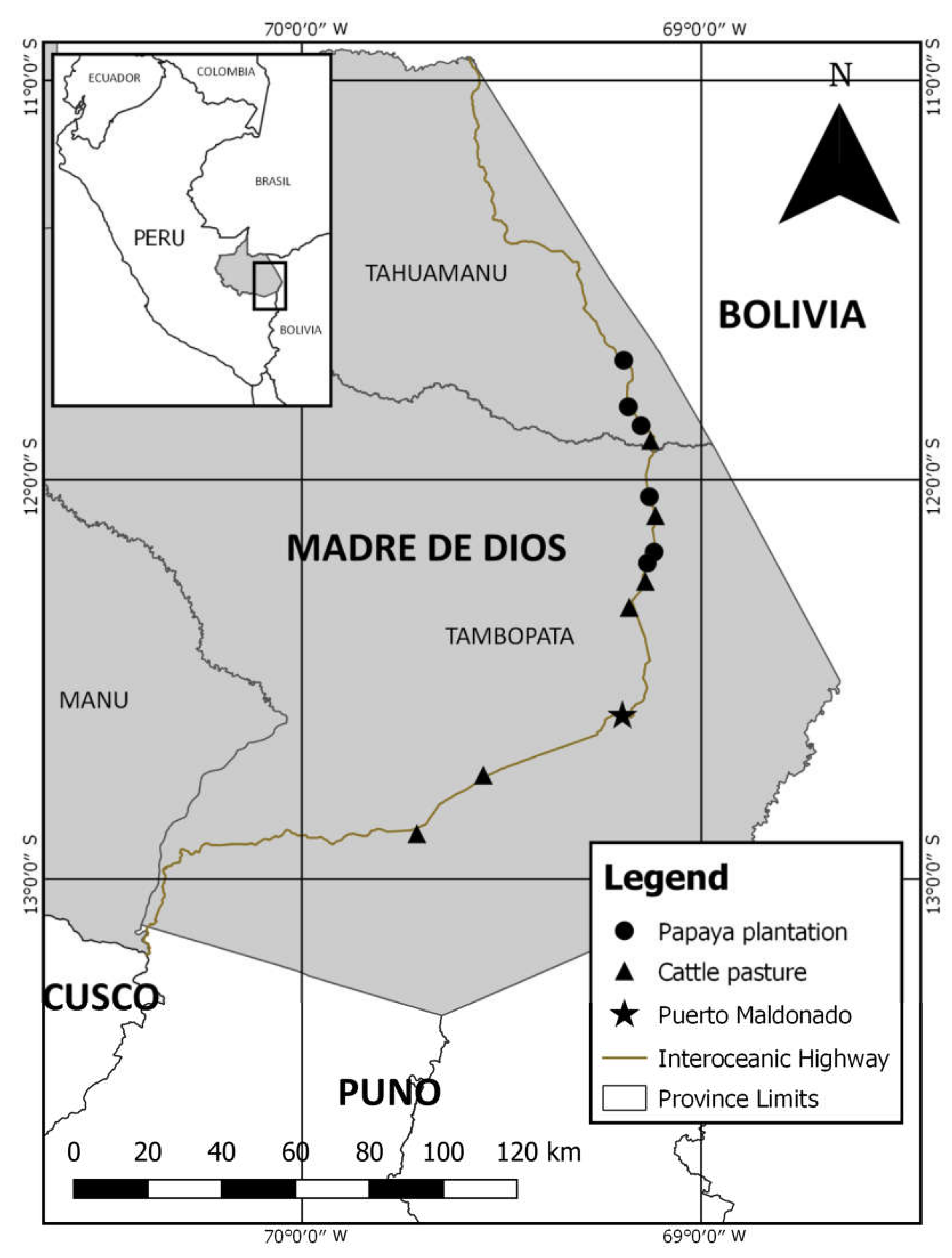
2.1. Sampling Location
2.2. Sampling Design
2.3. Bat Captures
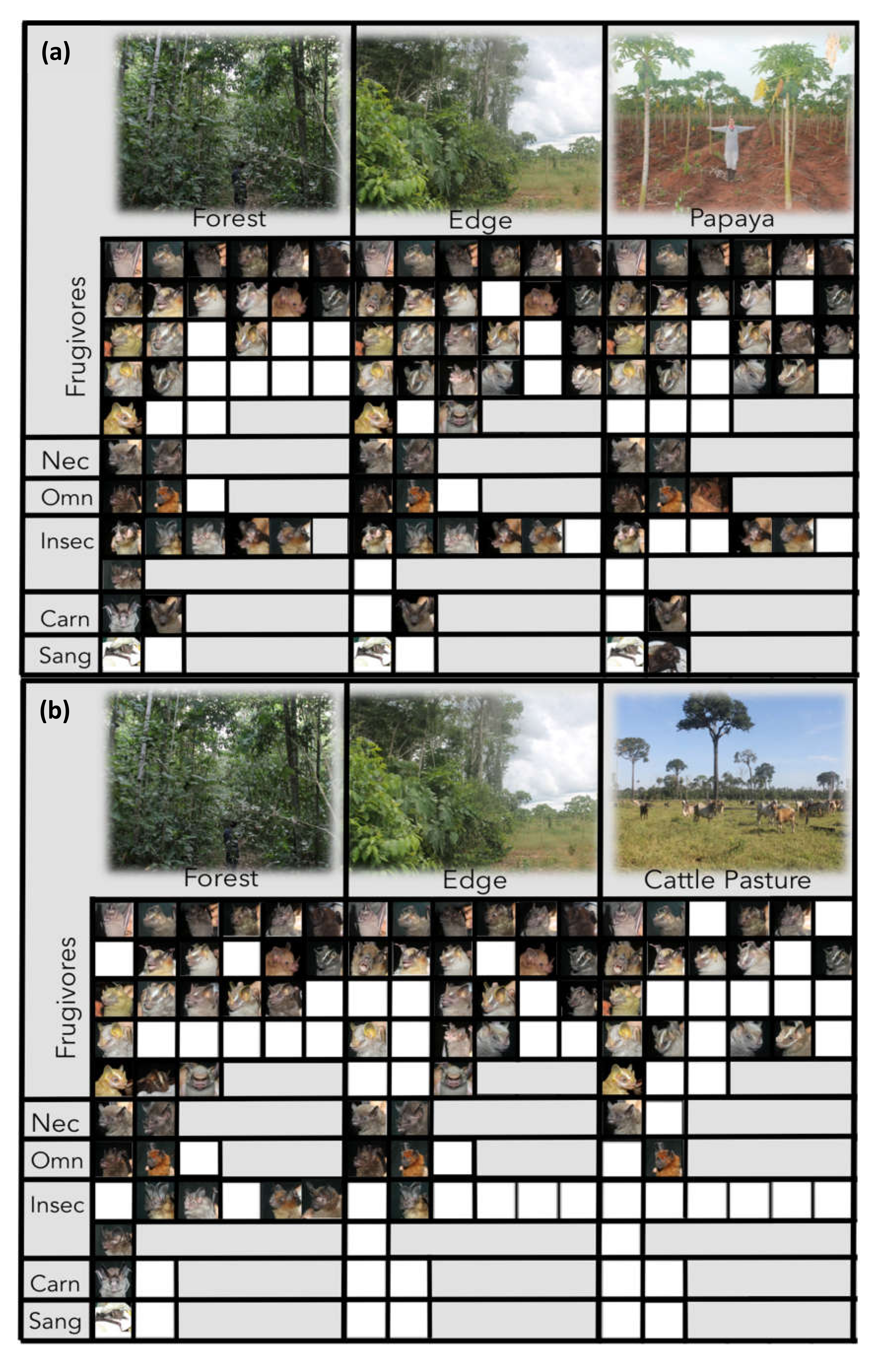
2.4. Traits
2.5. Vegetation Structure and Temperature Variation
2.6. Data Analysis
2.6.1. Effects of Agricultural Land on Composition of Bat Assemblages
- Influence of vegetation structure and temperature variation
2.6.2. Effects of Agricultural Land on Different Dimensions of Diversity
- Taxonomic diversity
- Functional diversity
3. Results
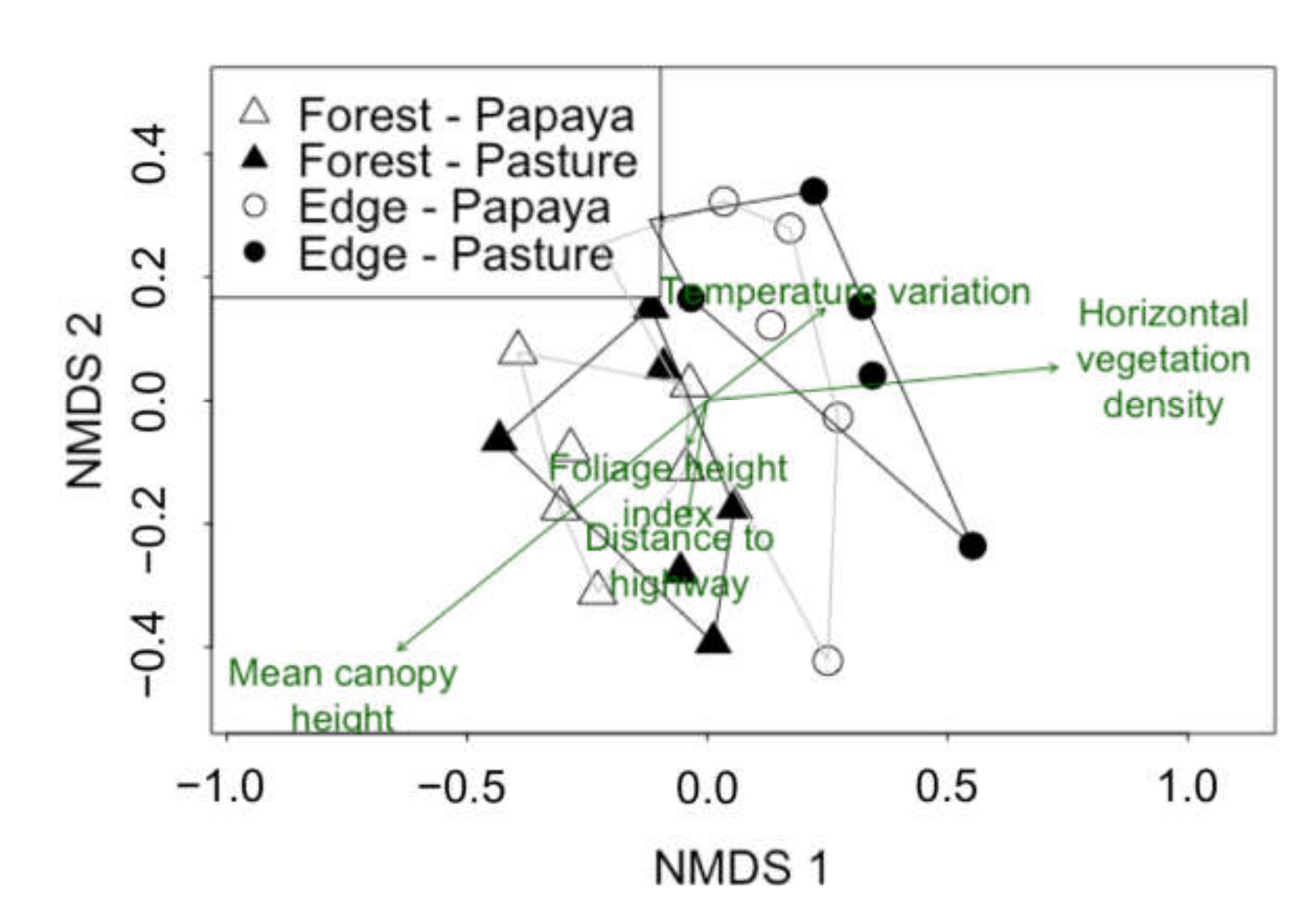
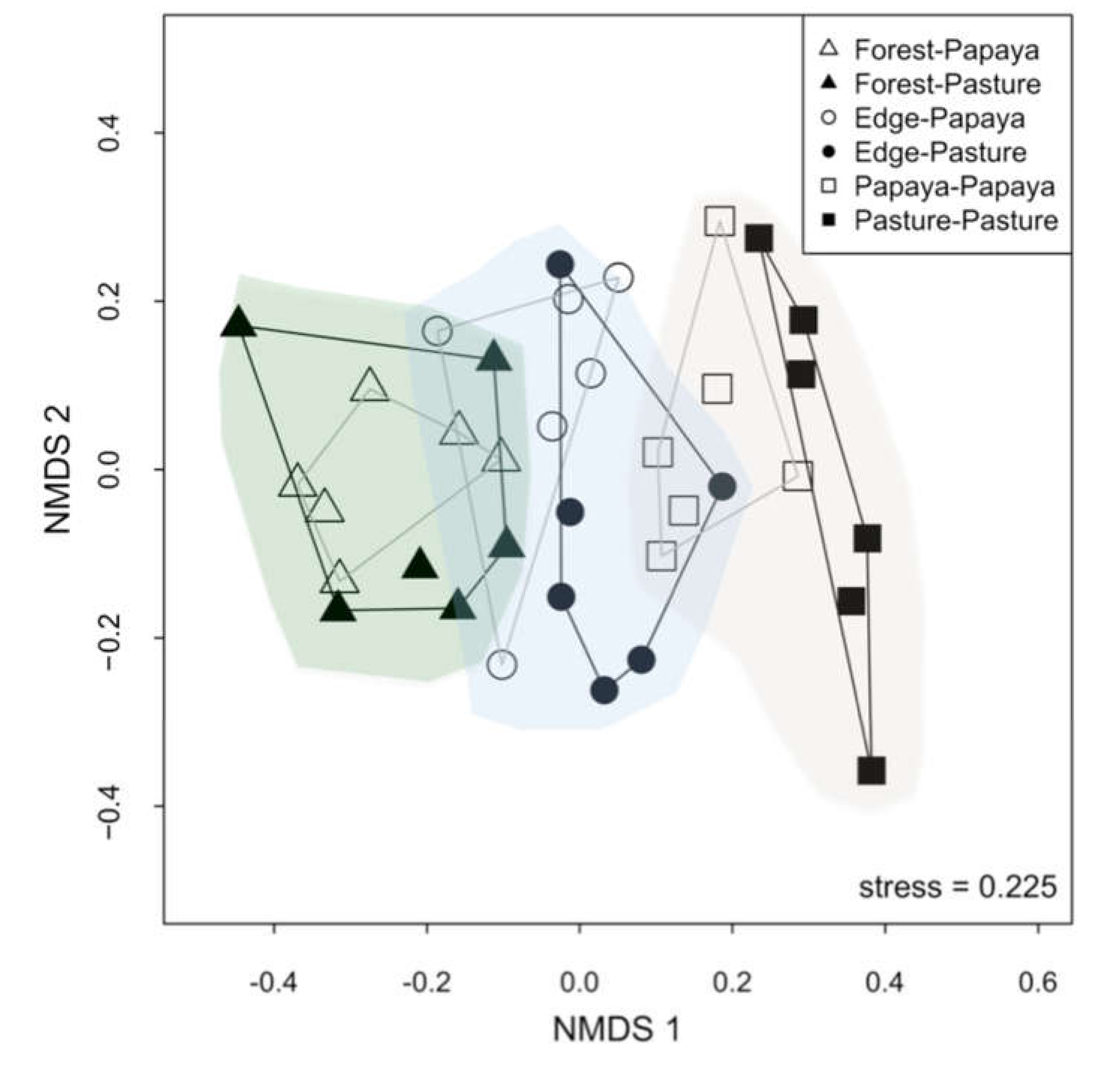
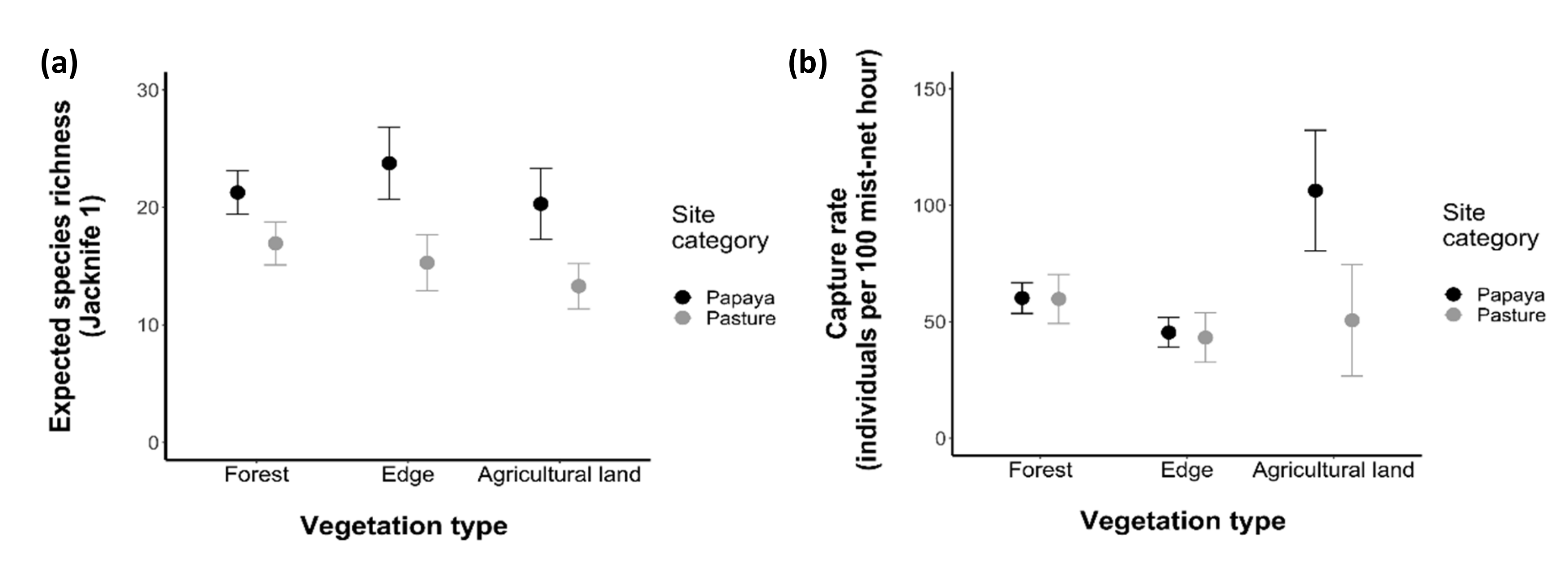
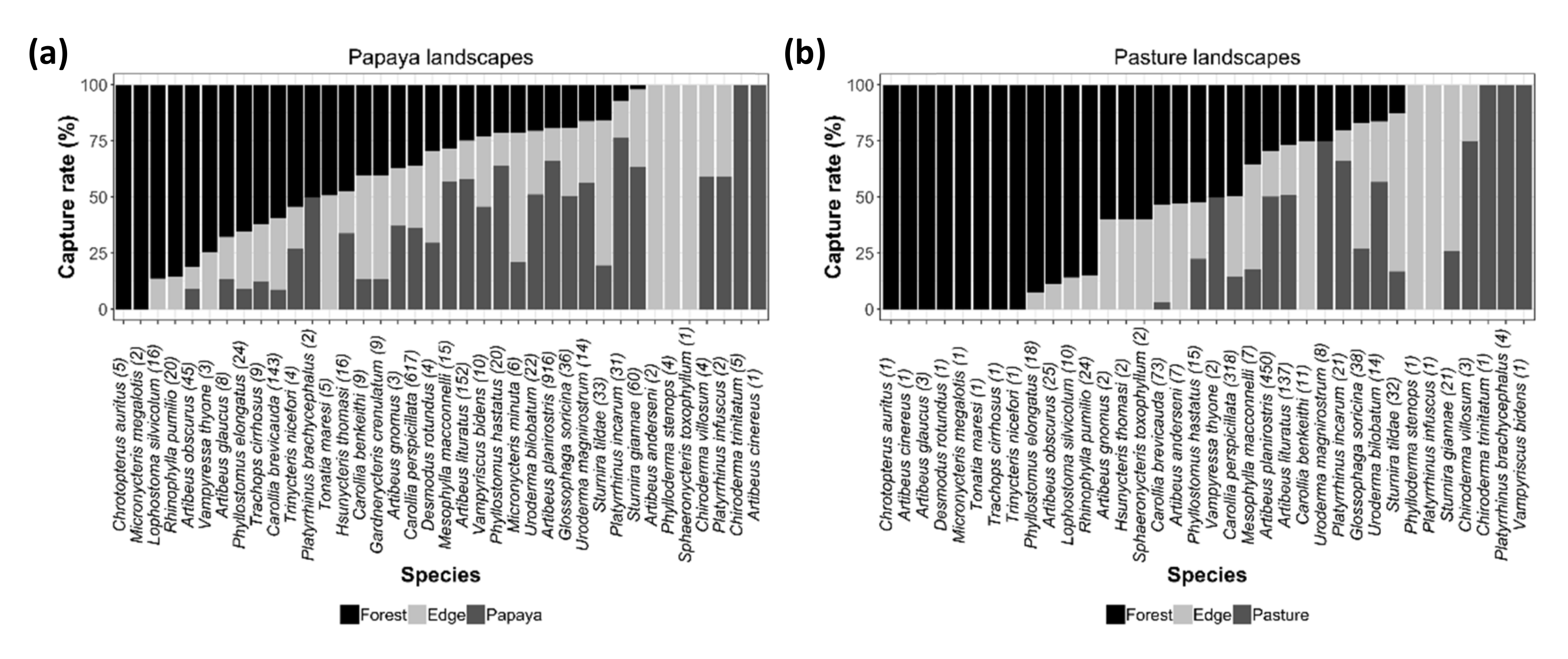
3.1. Effects of Agricultural Land on Composition of Bat Assemblages
3.2. Effects of Agricultural Land on Different Dimensions of Diversity
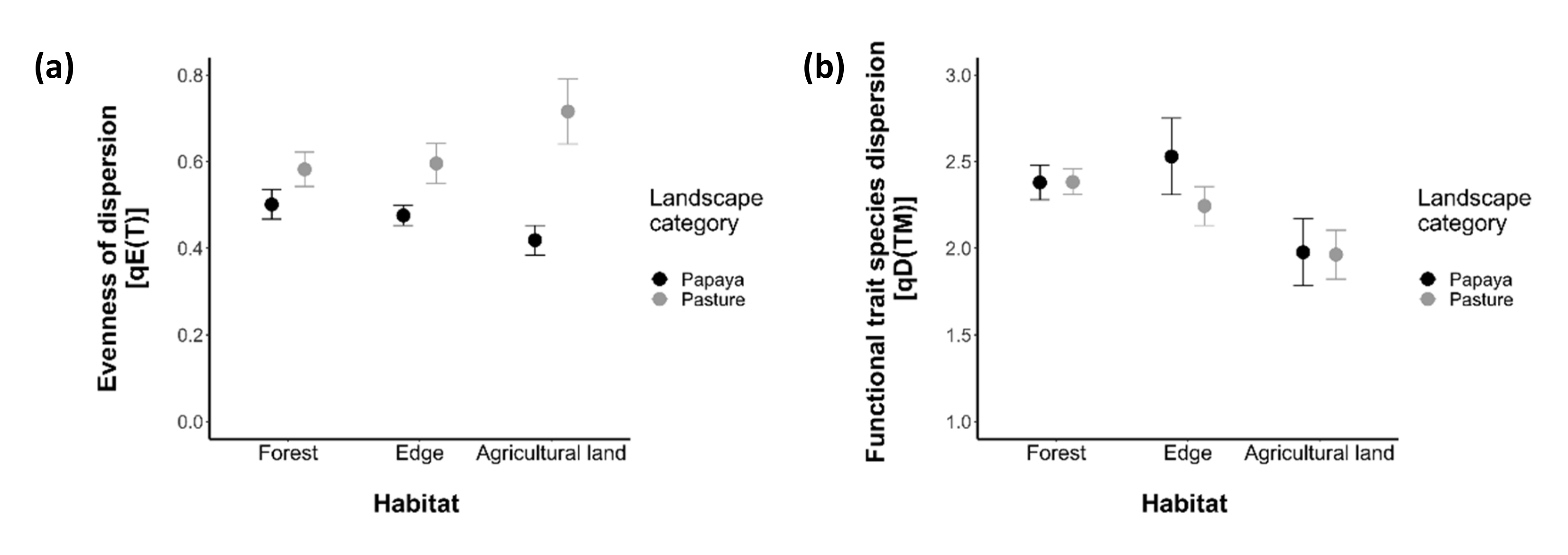
3.2.1. Taxonomic Diversity
3.2.2. Functional Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Taxonomic Diversity Metric | Variable | df | Sum Sq. | Mean Sq. | F Value | Pr(>F) |
|---|---|---|---|---|---|---|
| Capture Rate | Site Category | 1 | 3407 | 3407 | 1.02 | 0.359 |
| Distance to highway | 29 | 42,872 | 1478 | 0.443 | 0.925 | |
| Residuals | 5 | 16,701 | 3340 | |||
| Capture Rate | Vegetation type | 2 | 7021 | 3511 | 0.788 | 0.531 |
| Distance to highway | 30 | 42,595 | 1420 | 0.319 | 0.96 | |
| Residuals | 3 | 13,364 | 4455 | |||
| Species richness (jackknife1) | Site Category | 1 | 393.2 | 393.2 | 5.092 | 0.074 |
| Distance to highway | 29 | 739.2 | 25.5 | 0.33 | 0.974 | |
| Residuals | 5 | 386.1 | 77.2 | |||
| Species richness (jackknife1) | Vegetation type | 30 | 1132.5 | 37.75 | 0.329 | 0.956 |
| Distance to highway | 2 | 42.2 | 21.11 | 0.184 | 0.841 | |
| Residuals | 3 | 343.9 | 114.64 |
| Variable | NMDS1 | NMDS2 | r2 | p |
|---|---|---|---|---|
| Foliage height index | −0.401 | −0.916 | 0.004 | 0.962 |
| Horizontal vegetation density | 0.994 | 0.110 | 0.384 | 0.011 |
| Mean canopy height | −0.836 | −0.548 | 0.424 | 0.002 |
| Temperature variation | 0.831 | 0.556 | 0.052 | 0.564 |
| Distance to highway | −0.169 | −0.986 | 0.025 | 0.776 |
Appendix B
Sampling Effectiveness
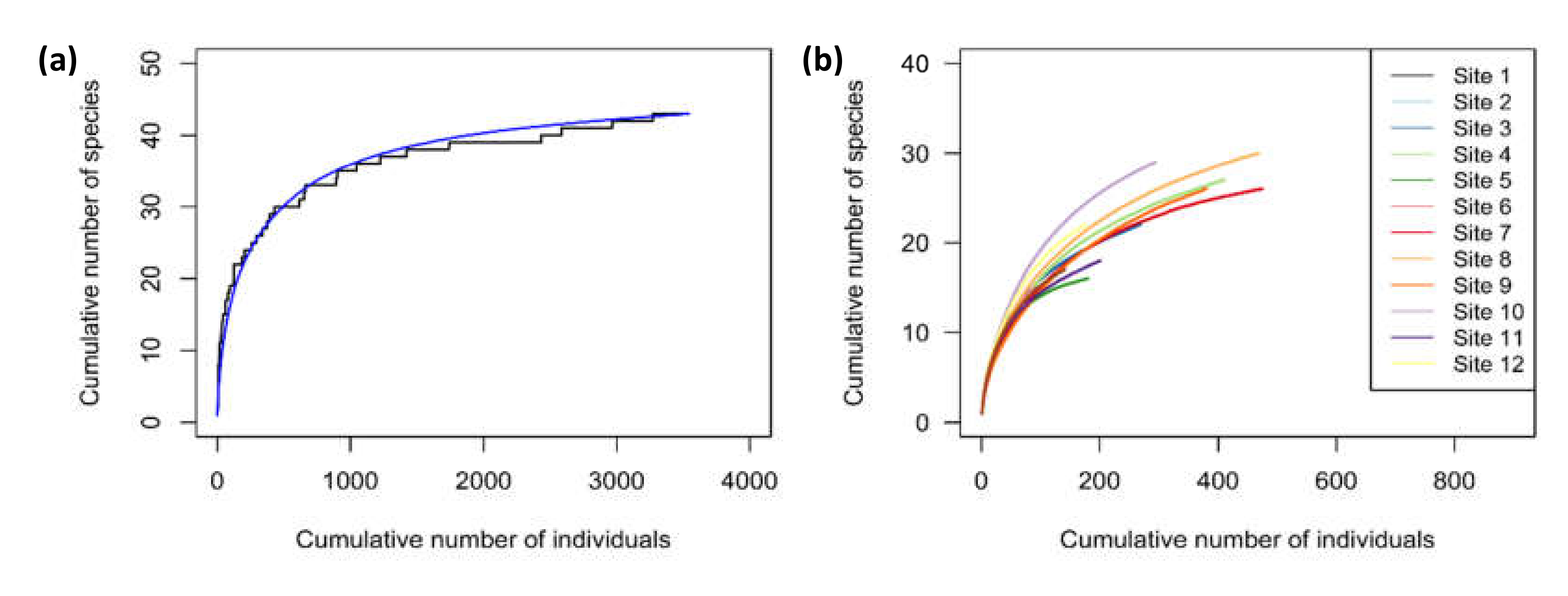
Appendix C
Appendix C.1. Vegetation Structure and Temperature Variation Measurement
Appendix C.1.1. Vegetation Structure
Appendix C.1.2. Temperature
References
- Waters, C.N.; Zalasiewicz, J.; Summerhayes, C.; Barnosky, A.D.; Poirier, C.; Galuszka, A.; Cearreta, A.; Edgeworth, M.; Ellis, E.C.; Ellis, M.; et al. The Anthropocene is functionally and stratigraphically distinct from the Holocene. Science 2016, 351, aad2622. [Google Scholar] [CrossRef] [PubMed]
- McGill, B.J.; Dornelas, M.; Gotelli, N.J.; Magurran, A.E. Fifteen forms of biodiversity trend in the Anthropocene. Trends Ecol. Evol. 2015, 30, 104–113. [Google Scholar] [CrossRef]
- Foley, J.A.; DeFries, R.; Asner, G.P.; Barford, C.; Bonan, G.; Carpenter, S.R.; Chapin, F.S.; Coe, M.T.; Daily, G.C.; Gibbs, H.K.; et al. Global consequences of land use. Science 2005, 309, 570–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mace, G.; Masundire, H.; Baillie, J. Biodiversity. In Ecosystems and Human Well Being, Current State and Trends: Millenium Ecosystem Assessment; Hassan, R., Scholes, R., Ash, N., Eds.; Island Press: New York, NY, USA, 2005; Volume 1, pp. 77–122. [Google Scholar]
- Bello, C.; Galetti, M.; Pizo, M.A.; Magnago, L.F.S.; Rocha, M.F.; Lima, R.A.F.; Peres, C.A.; Ovaskainen, O.; Jordano, P. Defaunation affects carbon storage in tropical forests. Sci. Adv. 2015, 1, e1501105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Mendez, N.; Jordano, P.; Garcia, C.; Valido, A. The signatures of Anthropocene defaunation: Cascading effects of the seed dispersal collapse. Sci. Rep. 2016, 6, 24820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimm, S.L.; Raven, P. Biodiversity—Extinction by numbers. Nature 2000, 403, 843–845. [Google Scholar] [CrossRef]
- Ramankutty, N.; Foley, J.A. Estimating historical changes in global land cover: Croplands from 1700 to 1992. Glob. Biogeochem. Cycles 1992, 13, 997–1027. [Google Scholar] [CrossRef]
- Asner, G.P.; Elmore, A.J.; Olander, L.P.; Martin, R.E.; Harris, A.T. Grazing systems, ecosystem responses, and global change. Annu. Rev. Environ. Resour. 2004, 29, 261–299. [Google Scholar] [CrossRef]
- Dirzo, R.; Raven, P.H. Global state of biodiversity and loss. Annu. Rev. Environ. Resour. 2003, 28, 137–167. [Google Scholar] [CrossRef] [Green Version]
- Frick, W.F.; Kingston, T.; Flanders, J. A review of the major threats and challenges to global bat conservation. Ann. N. Y. Acad. Sci. 2019, 1–21. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Garnett, T. Food security and sustainable intensification. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20120273. [Google Scholar] [CrossRef] [PubMed]
- Laurance, W.F.; Sayer, J.; Cassman, K.G. Agricultural expansion and its impacts on tropical nature. Trends Ecol. Evol. 2014, 29, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Paini, D.R.; Sheppard, A.W.; Cook, D.C.; De Barro, P.J.; Worner, S.P.; Thomas, M.B. Global threat to agriculture from invasive species. Proc. Natl. Acad. Sci. USA 2016, 113, 7575–7579. [Google Scholar] [CrossRef] [Green Version]
- Phalan, B.; Bertzky, M.; Butchart, S.H.M.; Donald, P.F.; Scharlemann, J.P.W.; Stattersfield, A.J.; Balmford, A. Crop expansion and conservation priorities in tropical countries. PLoS ONE 2013, 8, e51759. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, J.L.M.; Pellizari, V.H.; Mueller, R.; Baek, K.; Jesus, E.C.; Paula, F.S.; Mirza, B.; Hamaoui, G.S., Jr.; Siu Mui, T.; Feigl, B.; et al. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2013, 110, 988–993. [Google Scholar] [CrossRef] [Green Version]
- Soares-Filho, B.S.; Nepstad, D.C.; Curran, L.M.; Cerqueira, G.C.; Garcia, R.A.; Ramos, C.A.; Voll, E.; McDonald, A.; Lefebvre, P.; Schlesinger, P. Modelling conservation in the Amazon basin. Nature 2006, 440, 520–523. [Google Scholar] [CrossRef]
- Medellin, R.A.; Equihua, M. Mammal species richness and habitat use in rainforest and abandoned agricultural fields in Chiapas, Mexico. J. Appl. Ecol. 1998, 35, 13–23. [Google Scholar] [CrossRef]
- Mendenhall, C.D.; Sekercioglu, C.H.; Brenes, F.O.; Ehrlich, P.R.; Daily, G.C. Predictive model for sustaining biodiversity in tropical countryside. Proc. Natl. Acad. Sci. USA 2011, 108, 16313–16316. [Google Scholar] [CrossRef] [Green Version]
- Vargas Espinoza, A.; Aguirre, L.F.; Galarza, M.I.; Gareca, E. Ensamble de murciélagos en sitios con diferente grado de perturbación en un bosque montano del Parque Nacional Carrasco, Bolivia. Mastozool Neotrop 2008, 15, 297–308. [Google Scholar]
- Estrada, A.; Coates-Estrada, R. Bats in continuous forest, forest fragments and in an agricultural mosaic habitat-island at Los Tuxtlas, Mexico. Biol. Conserv. 2002, 103, 237–245. [Google Scholar] [CrossRef]
- Harvey, C.A.; Gonzalez Villalobos, J.A. Agroforestry systems conserve species-rich but modified assemblages of tropical birds and bats. Biodivers. Conserv. 2007, 16, 2257–2292. [Google Scholar] [CrossRef]
- Lindenmayer, D.B.; Franklin, J.F. Conserving Forest Biodiversity: A Comprehensive Multiscaled Approach; Island Press: Washington, DC, USA, 2002; p. 351. [Google Scholar]
- Williams-Guillen, K.; Perfecto, I. Effects of Agricultural Intensification on the Assemblage of Leaf-Nosed Bats (Phyllostomidae) in a Coffee Landscape in Chiapas, Mexico. Biotropica 2010, 42, 605–613. [Google Scholar] [CrossRef] [Green Version]
- Dunning, J.B.; Danielson, B.J.; Pulliam, H.R. Ecological processes that affect populations in complex landscapes. Oikos 1992, 65, 169–175. [Google Scholar] [CrossRef] [Green Version]
- Daily, G.C. Ecological forecasts. Nature 2001, 411, 245. [Google Scholar] [CrossRef]
- Presley, S.J.; Cisneros, L.M.; Higgins, C.L.; Klingbeil, B.T.; Scheiner, S.M.; Willig, M.R. Phylogenetic and functional underdispersion in Neotropical phyllostomid bat communities. Biotropica 2018, 50, 135–145. [Google Scholar] [CrossRef]
- Castro-Luna, A.A.; Sosa, V.J.; Castillo-Campos, G. Quantifying phyllostomid bats at different taxonomic levels as ecological indicators in a disturbed tropical forest. Acta Chiropterol. 2007, 9, 219–228. [Google Scholar] [CrossRef]
- Cisneros, L.M.; Fagan, M.E.; Willig, M.R. Environmental and spatial drivers of taxonomic, functional, and phylogenetic characteristics of bat communities in human-modified landscapes. PeerJ 2016, 4, e2551. [Google Scholar] [CrossRef] [Green Version]
- Fenton, M.B.; Acharya, L.; Audet, D.; Hickey, M.B.C.; Merriman, C.; Obrist, M.K.; Syme, D.M.; Adkins, B. Phyllostomid bats (Chiroptera—Phyllostomidae) as indicators of habitat disruption in the Neotropics. Biotropica 1992, 24, 440–446. [Google Scholar] [CrossRef]
- Gorchov, D.L.; Rondon, X.J.; Cornejo, F.; Schaefer, R.L.; Janosko, J.M.; Slutz, G. Edge effects in recruitment of trees, and relationship to seed dispersal patterns, in cleared strips in the Peruvian Amazon. Ecol. Res. 2013, 28, 53–65. [Google Scholar] [CrossRef]
- Lacher, T.E., Jr.; Davidson, A.A.; Fleming, T.H.; Gómez-Ruiz, E.P.; McCraken, G.F.; Owen-Smith, N.; Peres, C.A.; Vander Wall, S.B. The functional roles of mammals in ecosystems. J. Mammal. 2019, 100, 942–964. [Google Scholar] [CrossRef] [Green Version]
- Medellin, R.A.; Equihua, M.; Amin, M.A. Bat diversity and abundance as indicators of disturbance in neotropical rainforests. Conserv. Biol. 2000, 14, 1666–1675. [Google Scholar] [CrossRef]
- Barlow, J.; Gardner, T.A.; Araujo, I.S.; Avila-Pires, T.C.; Bonaldo, A.B.; Costa, J.E.; Esposito, M.C.; Ferreira, L.V.; Hawes, J.; Hernandez, M.M.; et al. Quantifying the biodiversity value of tropical primary, secondary, and plantation forests. Proc. Natl. Acad. Sci. USA 2007, 104, 18555–18560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cisneros, L.M.; Fagan, M.E.; Willig, M.R. Effects of human-modified landscapes on taxonomic, functional and phylogenetic dimensions of bat biodiversity. Divers. Distrib. 2015, 21, 523–533. [Google Scholar] [CrossRef]
- Meyer, C.F.J.; Struebig, M.J.; Willig, M.R. Responses of tropical bats to habitat fragmentation, logging, and deforestation. In Bats in the Anthropocene: Conservation of Bats in a Changing World; Voigt, C.C., Kingston, T., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 63–103. [Google Scholar]
- Meyer, C.F.J.; Kalko, E.K.V. Assemblage-level responses of phyllostomid bats to tropical forest fragmentation: Land-bridge islands as a model system. J. Biogeogr. 2008, 35, 1711–1726. [Google Scholar] [CrossRef]
- Dourojeanni, M.J. Una nueva estrategia para el desarrollo de la Amazonía Peruana. RFP 1976, 6, 1–22. [Google Scholar]
- Papaya–Potential New Driver of Deforestation in Madre de Dios. Available online: https://maaproject.org/2016/papaya-peru/ (accessed on 16 April 2020).
- Klingbeil, B.T.; Willig, M.R. Seasonal differences in population-, ensemble- and community-level responses of bats to landscape structure in Amazonia. Oikos 2010, 119, 1654–1664. [Google Scholar] [CrossRef]
- Mendenhall, C.D.; Karp, D.S.; Meyer, C.F.J.; Hadly, E.A.; Daily, G.C. Predicting biodiversity change and averting collapse in agricultural landscapes. Nature 2014, 509, 213–217. [Google Scholar] [CrossRef]
- Saldanha Bueno, A.; Peres, C.A. The role of baseline suitability in assessing the impacts of land-use change on biodiversity. Biol. Conserv. 2020, 243, 108396. [Google Scholar] [CrossRef]
- Scheiner, S.M.; Kosman, E.; Presley, S.J.; Willig, M.R. Decomposing functional diversity. Methods Ecol. Evol. 2017, 8, 809–820. [Google Scholar] [CrossRef] [Green Version]
- Bovendorp, R.S.; Brum, F.T.; McCleery, R.A.; Baiser, B.; Loyola, R.; Cianciaruso, M.V.; Galetti, M. Defaunation and fragmentation erode small mammal diversity dimensions in tropical forests. Ecography 2019, 42, 23–35. [Google Scholar] [CrossRef]
- Farneda, F.Z.; Rocha, R.; Lopez-Baucells, A.; Groenenberg, M.; Silva, I.; Palmeirim, J.M.; Bobrowiec, P.E.D.; Meyer, C.F.J. Trait-related responses to habitat fragmentation in Amazonian bats. J. Appl. Ecol. 2015, 52, 1381–1391. [Google Scholar] [CrossRef]
- Farneda, F.Z.; Rocha, R.; Lopez-Baucells, A.; Sampaio, E.M.; Palmeirim, J.M.; Bobrowiec, P.E.D.; Grelle, C.E.V.; Meyer, C.F.J. The Road to Functional Recovery: Temporal Effects of Matrix Regeneration on Amazonian Bats. Trop. Conserv. Sci. 2018, 11, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Farneda, F.Z.; Grelle, C.E.V.; Rocha, R.; Ferreira, D.F.; Lopez-Baucells, A.; Meyer, C.F.J. Predicting biodiversity loss in island and countryside ecosystems through the lens of taxonomic and functional biogeography. Ecography 2020, 43, 97–106. [Google Scholar] [CrossRef]
- Petchey, O.L.; Gaston, K.J. Functional diversity: Back to basics and looking forward. Ecol. Lett. 2006, 9, 741–758. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Delgado, N.; Perez-Torres, J. Habitat edge context and the distribution of phyllostomid bats in the Andean forest and anthropogenic matrix in the Central Andes of Colombia. Biodivers. Conserv. 2011, 20, 987–999. [Google Scholar] [CrossRef]
- Garcia-Morales, R.; Badano, E.I.; Moreno, C.E. Response of Neotropical Bat Assemblages to Human Land Use. Conserv. Biol. 2013, 27, 1096–1106. [Google Scholar] [CrossRef]
- Daily, G.C.; Ceballos, G.; Pacheco, J.; Suzan, G.; Sanchez-Azofeifa, A. Countryside biogeography of neotropical mammals: Conservation opportunities in agricultural landscapes of Costa Rica. Conserv. Biol. 2003, 17, 1814–1826. [Google Scholar] [CrossRef]
- Sánchez-Cuervo, A.M.; de Santos Lima, L.; Dallmeier, F.; Garate, P.; Bravo, A.; Vanthomme, H. Twenty years of land cover change in the southeastern Peruvian Amazon: Implications for biodiversity conservation. Reg. Environ. Chang. 2020, 20. [Google Scholar] [CrossRef]
- Presidencia del Consejo de Ministros (PCM); Gobierno Regional de Madre de Dios (GOREMAD). Estudio de Diagnóstico y Zonificación Para el Tratamiento de la Demarcacion Territorial de la Provincia Tambopata; GOREMAD: Puerto Maldonado, Peru, 2010; Volume I, p. 310. Available online: http://www2.congreso.gob.pe/sicr/cendocbib/con4_uibd.nsf/007886A2CDEC740505257B7A0075FC13/%24FILE/doc_edz_tambopata.pdf (accessed on 28 May 2020).
- Murcia, C. Edge effects in fragmented forests—Implications for conservation. Trends Ecol. Evol. 1995, 10, 58–62. [Google Scholar] [CrossRef]
- Galindo-Gonzalez, J.; Sosa, V.J. Frugivorous bats in isolated trees and riparian vegetation associated with human-made pastures in a fragmented tropical landscape. Southwest Nat. 2003, 48, 579–589. [Google Scholar] [CrossRef]
- Saldana-Vazquez, R.A.; Munguia-Rosas, M.A. Lunar phobia in bats and its ecological correlates: A meta-analysis. Mamm. Biol. 2013, 78, 216–219. [Google Scholar] [CrossRef]
- Helbig-Bonitz, M.; Rutten, G.; Kalko, E.K.V. Fruit bats can disperse figs over different land-use types on Mount Kilimanjaro, Tanzania. Afr. J. Ecol. 2014, 52, 122–125. [Google Scholar] [CrossRef]
- Klingbeil, B.T.; Willig, M.R. Guild-specific responses of bats to landscape composition and configuration in fragmented Amazonian rainforest. J. App. Ecol. 2009, 46, 203–213. [Google Scholar] [CrossRef]
- Diaz, M.M.; Solari, S.; Aguirre, L.F.; Aguiar, L.M.S.; Barquez, R.M. Clave de Identificación de los Murciélagos de Sudamérica; Publicación Especial N° 2; PCMA (Programa de Conservación de los Murciélagos de Argentina): Tucumán, Argentina, 2016; p. 160. [Google Scholar]
- Gardner, A.L. Mammals of South America. Marsupials, Xenarthrans, Shrews, and Bats; University of Chicago Press: Chicago, IL, USA; London, UK, 2007; Volume 1, p. 669. [Google Scholar]
- Giannini, N.P.; Barquez, R.M. Sturnira erythromos. Mamm. Species 2003, 729, 1–5. [Google Scholar] [CrossRef]
- López-Baucells, A.; Rocha, R.; Bobrowiec, P.; Bernard, E.; Palmeirim, J.; Meyer, C. Field Guide to Amazonian Bats; INPA: Manaus, Brazil, 2016; p. 168. [Google Scholar]
- Basantes, M.; Tinoco, N.; Velazco, P.M.; Hofmann, M.J.; Rodriguez-Posada, M.E.; Camacho, M.A. Systematics and Taxonomy of Tonatia saurophila Koopman & Williams, 1951 (Chiroptera, Phyllostomidae). Zookeys 2020, 59–86. [Google Scholar] [CrossRef]
- Velazco, P.M.; Patterson, B.D. Small mammals of the Mayo River Basin in Northern Peru, with the description of a new species of Sturnira (Chiroptera: Phyllostomidae). Bull. Am. Mus. Nat. Hist. 2019, 1–67. [Google Scholar] [CrossRef] [Green Version]
- Hurtado, N.; Pacheco, V. Análisis filogenético del género Mimon Gray, 1847 (Mammalia, Chiroptera, Phyllosto midae) con la descripción de un nuevo género. Therya 2014, 5, 751–791. [Google Scholar] [CrossRef]
- Sikes, R.S. The Animal Care and Use Committee of the American Society of Mammalogists. 2016 Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J. Mammal. 2016, 97, 663–668. [Google Scholar]
- AVMA. AVMA Guidelines for the Euthanasia of Animals: 2013 Edition; American Veterinary Medical Association: Schaumbutg, IL, USA, 2013; p. 102. [Google Scholar]
- Kwiecinski, G.G. Phyllostomus discolor. Mamm Species 2006, 801, 1–11. [Google Scholar] [CrossRef]
- García-García, J.L.; Santos-Moreno, A. Efectos de la estructura del paisaje y de la vegetación en la diversidad de murciélagos filostómidos (Chiroptera: Phyllostomidae) de Oaxaca, México. Rev. Biol. Trop. 2014, 62, 226–249. [Google Scholar] [CrossRef] [Green Version]
- Martins, A.C.M.; Willig, M.R.; Presley, S.J.; Marinho-Filho, J. Effects of forest height and vertical complexity on abundance and biodiversity of bats in Amazonia. Forest. Ecol. Manag. 2017, 391, 427–435. [Google Scholar] [CrossRef]
- Kalko, E.K.V. Organisation and diversity of tropical bat communities through space and time. ZACS 1998, 101, 281–297. [Google Scholar]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 326–349. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, R.B. Vegan: Community Ecology Package. Version 1.7-81. 2005. Available online: http://cran.r-project.org/i (accessed on 16 April 2020).
- Nenadic, O.; Greenacre, M. Correspondence Analysis in R, with two- and three-dimensional graphics: The ca package. J. Stat. Softw. 2007, 20, 1–13. [Google Scholar]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002; p. 495. [Google Scholar]
- Dray, S.; Dufour, A.; Chessel, D. The ade4 Package: Implementing the Duality Diagram for Ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Walther, B.A.; Moore, J.L. The concepts of bias, precision and accuracy, and their use in testing the performance of species richness estimators, with a literature review of estimator performance. Ecography 2005, 28, 815–829. [Google Scholar] [CrossRef]
- Burnham, K.P.; Overton, W.S. Robust estimation of population size when capture probabilities vary among animals. Ecology 1979, 60, 927–936. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.r-project.org (accessed on 16 April 2020).
- Paradis, E.; Schliep, K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2018, 35, 526–528. [Google Scholar] [CrossRef]
- Kalko, E.K.V. Diversity in tropical bats. In Tropical Biodiversity and Systematics, Proceedings of the International Symposium on Biodiversity and Systematics in Tropical Ecosystems, Bonn, Germany, 2–7 May 1994; Ulrich, H., Ed.; Zoologisches Forschungsinstitut und Museum Alexander Koenig: Bonn, Germany, 1997; pp. 13–43. [Google Scholar]
- Laurance, W.F.; Lovejoy, T.E.; Vasconcelos, H.L.; Bruna, E.M.; Didham, R.K.; Stouffer, P.C.; Gascon, C.; Bierregaard, R.O.; Laurance, S.G.; Sampaio, E. Ecosystem decay of Amazonian forest fragments: A 22-year investigation. Conserv. Biol. 2002, 16, 605–618. [Google Scholar] [CrossRef] [Green Version]
- Ober, H.K.; Hayes, J.P. Influence of vegetation on bat use of riparian areas at multiple spatial scales. J. Wildl. Manag. 2008, 72, 396–404. [Google Scholar] [CrossRef]
- Cleary, K.A.; Waits, L.; Finegan, B. Agricultural intensification alters bat assemblage composition and abundance in a dynamic Neotropical landscape. Biotropica 2016, 48, 667–676. [Google Scholar] [CrossRef]
- Cadenasso, M.L.; Pickett, S.T.A.; Weathers, K.C.; Jones, C.G. A framework for a theory of ecological boundaries. Bioscience 2003, 53, 750–758. [Google Scholar] [CrossRef]
- Fagan, W.E.; Cantrell, R.S.; Cosner, C. How habitat edges change species interactions. Am. Nat. 1999, 153, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Ries, L.; Fletcher, R.J.; Battin, J.; Sisk, T.D. Ecological responses to habitat edges: Mechanisms, models, and variability explained. Annu. Rev. Ecol. Evol. Syst. 2004, 35, 491–522. [Google Scholar] [CrossRef] [Green Version]
- Arroyo-Rodríguez, V.; Pérez-Elissetche, G.K.; Ordóñez-Gómez, J.D.; González-Zamora, A.; Chavez, O.M.; Sánchez-López, S.; Chapman, C.A.; Moralez-Hernández, K.; Pablo-Rodríguez, M.; Ramos-Fernández, G. Spider monkeys in human-modified landscapes: The importance of the matrix. Trop. Conserv. Sci. 2017, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Syiem, B.L.N.; Goswami, V.R.; Vasudev, D. “In a tree by the brook, there’s a songbird who sings”: Woodlands in an agricultural matrix maintain functionality of a wintering bird community. PLoS ONE 2018, 13, e0201657. [Google Scholar] [CrossRef] [Green Version]
- de la Pena-Cuellar, E.; Benitez-Malvido, J.; Avila-Cabadilla, D.L.; Martinez-Ramos, M.; Estrada, A. Structure and diversity of phyllostomid bat assemblages on riparian corridors in a human-dominated tropical landscape. Ecol. Evol. 2015, 5, 903–913. [Google Scholar] [CrossRef]
- Silveira, J.M.; Louzada, J.; Barlow, J.; Andrade, R.; Mestre, L.; Solar, R.; Lacau, S.; Cochrane, M.A. A multi-taxa assessment of biodiversity change after single and recurrent wildfires in a Brazilian Amazon forest. Biotropica 2015, 48, 170–180. [Google Scholar] [CrossRef]
- Bernard, E.; Fenton, M.B. Bat mobility and roosts in a fragmented landscape in central Amazonia, Brazil. Biotropica 2003, 35, 262–277. [Google Scholar] [CrossRef]
- Gorrensen, P.M.; Willig, M.R. Landscape responses of bats to habitat fragmentation in Atlantic forest of Paraguay. J. Mammal. 2004, 85, 688–697. [Google Scholar] [CrossRef] [Green Version]
- Willig, M.R.; Presley, S.J.; Bloch, C.P.; Hice, C.L.; Yanoviak, S.P.; Diaz, M.M.; Chauca, L.A.; Pacheco, V.; Weaver, S.C. Phyllostomid bats of lowland Amazonia: Effects of habitat alteration on abundance. Biotropica 2007, 39, 737–746. [Google Scholar] [CrossRef]
- Cordero-Schmidt, E.; Medeiros-Guimarāes, M.; Vargas-Mena, J.C.; Carvalho, B.; Lopes Ferreira, R.; Rodriguez-Herrera, B.; Venticinque, E.M. Are leaves good option in Caatinga’s menu? First record of folivory in Artibeus planirostris (Phyllostomidae) in the semiarid forest, Brazil. Acta Chiropterol. 2016, 18, 489–497. [Google Scholar] [CrossRef]
- Faria, D. Phyllostomid bats of a fragmented landscape in the north-eastern Atlantic forest, Brazil. J. Trop. Ecol. 2006, 22, 531–542. [Google Scholar] [CrossRef] [Green Version]
- Remsen, J.V.; Good, D.A. Misuse of data from mist-net captures to assess relative abundance in bird populations. Auk 1986, 113, 381–398. [Google Scholar] [CrossRef]
- Claireau, F.; Bas, Y.; Pauwels, J.; Puechmaille, S.; Julien, J.F.; Machon, N.; Allegrini, B.; Kerbiriou, C. Major roads have important effects on insectivorous bats activity. Biol. Conserv. 2019, 235, 53–62. [Google Scholar] [CrossRef]
- Medinas, D.; Marques, J.T.; Mira, A. Assessing road effects on bats: The role of landscape, road features, and bat activity on road-kills. Ecol. Res. 2013, 28, 227–237. [Google Scholar] [CrossRef] [Green Version]
- Medinas, D.; Ribeiro, V.; Marques, J.T.; Silva, B.; Barbosa, A.M.; Rebelo, H.; Mira, A. Road effects on bat activity depend on surrounding habitat type. Sci. Total Environ. 2019, 660, 340–347. [Google Scholar] [CrossRef]
- Yoh, N.; Syme, P.; Rocha, R.; Meyer, C.F.J.; López-Baucells, A. Echolocation of Central Amazonian ‘whispering’ phyllostomid bats: Call design and interspecific variation. Mamm. Res. 2020. [Google Scholar] [CrossRef]
- Boulinier, T.; Nichols, J.D.; Sauer, J.R.; Hines, J.E.; Pollock, K.H. Estimating species richness: The importance of heterogeneity in species detectability. Ecology 1998, 79, 1018–1028. [Google Scholar] [CrossRef]
- Kéry, M.; Plattner, M. Species richness estimation and determinants of species detectability in butterfly monitoring programmes. Ecol. Entomol. 2007, 32, 53–61. [Google Scholar] [CrossRef]
- Kéry, M.; Royle, J.A. Hierarchical Bayes estimation of species richness and occupancy in spatially replicated surveys. J. Appl. Ecol. 2008, 45, 589–598. [Google Scholar] [CrossRef]
- Meyer, C.F.J.; Aguiar, L.M.S.; Aguirre, L.F.; Baumgarten, J.; Clarke, F.M.; Cosson, J.F.; Estrada Villegas, S.; Fahr, J.; Faria, D.; Furey, N.; et al. Accounting for detectability improves estimates of species richness in tropical bat surveys. J. Appl. Ecol. 2011, 48, 777–787. [Google Scholar] [CrossRef]
- Boyles, J.G.; Cryan, P.M.; McCracken, G.F.; Kunz, T.H. Economic Importance of Bats in Agriculture. Science 2011, 332, 41–42. [Google Scholar] [CrossRef]
- Karp, D.S.; Mendenhall, C.D.; Sandí, R.F.; Chaumont, N.; Ehrlich, P.R.; Hadly, E.A.; Daily, G.C. Forest bolsters bird abundance, pest control and coffee yield. Ecol. Lett. 2013, 16, 1339–1347. [Google Scholar] [CrossRef]
- Maas, B.; Karp, D.S.; Bumrungsri, S.; Darras, K.; Gonthier, D.; Huang, J.C.C.; Lindell, C.A.; Maine, J.J.; Mestre, L.; Mitchel, N.L.; et al. Bird and bat predation services in tropical forest and agroforestry landscapes. Biol. Rev. 2015, 91, 1081–1101. [Google Scholar] [CrossRef] [Green Version]
- Cottam, G.; Curtis, J.T. The use of distance measures in phytosociological sampling. Ecology 1956, 37, 451–460. [Google Scholar] [CrossRef]
- MacArthur, R.; MacArthur, J.W. On bird species-diversity. Ecology 1961, 42, 594–598. [Google Scholar] [CrossRef]
- Zellweger, F.; Baltensweiler, A.; Ginzler, C.; Roth, T.; Braunisch, V.; Bugmann, H.; Bollmann, K. Environmental predictors of species richness in forest landscapes: Abiotic factors versus vegetation structure. J. Biogeogr. 2016, 43, 1080–1090. [Google Scholar] [CrossRef]
- Marsden, S.J.; Fielding, A.H.; Mead, C.; Hussin, M.Z. A technique for measuring the density and complexity of understorey vegetation in tropical forests. For. Ecol. Manag. 2002, 165, 117–123. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef] [PubMed]


| Species | Papaya Plantation Sites | Cattle Pasture Sites | Total | Guild | Collection Number | ||||
|---|---|---|---|---|---|---|---|---|---|
| Edge | Forest | Agricultural Area | Edge | Forest | Agricultural Area | ||||
| Artibeus lituratus | 35 | 35 | 82 | 41 | 33 | 63 | 289 | Fru | CEBIOMAS0497 |
| Artibeus obscurus | 6 | 35 | 4 | 4 | 21 | 0 | 70 | Fru | |
| Artibeus planirostris | 178 | 167 | 571 | 124 | 121 | 205 | 1366 | Fru | CEBIOMAS0505 |
| Artibeus anderseni | 2 | 0 | 0 | 4 | 3 | 0 | 9 | Fru | CEBIOMAS0493, CEBIOMAS0495 |
| Artibeus cinereus | 0 | 0 | 1 | 0 | 1 | 0 | 2 | Fru | |
| Artibeus glaucus | 2 | 5 | 1 | 0 | 3 | 0 | 11 | Fru | CEBIOMAS0485, CEBIOMAS0498 |
| Artibeus gnomus | 1 | 1 | 1 | 1 | 1 | 0 | 5 | Fru | CEBIOMAS0499 |
| Carollia benkeithi | 5 | 3 | 1 | 9 | 2 | 0 | 20 | Fru | |
| Carollia brevicauda | 58 | 74 | 11 | 39 | 32 | 2 | 216 | Fru | CEBIOMAS0484, CEBIOMAS0506 |
| Carollia perspicillata | 221 | 197 | 199 | 145 | 134 | 39 | 935 | Fru | CEBIOMAS0490, CEBIOMAS0491 |
| Chiroderma salvini | 1 | 0 | 0 | 0 | 0 | 0 | 1 | Fru | |
| Chiroderma trinitatum | 0 | 0 | 5 | 0 | 0 | 1 | 6 | Fru | |
| Chiroderma villosum | 2 | 0 | 2 | 1 | 0 | 2 | 7 | Fru | |
| Chrotopterus auritus | 0 | 5 | 0 | 0 | 1 | 0 | 6 | Car | |
| Desmodus rotundus | 2 | 1 | 1 | 0 | 1 | 0 | 5 | San | |
| Diphylla ecaudata | 0 | 0 | 1 | 0 | 0 | 0 | 1 | San | |
| Glossophaga soricina | 14 | 6 | 16 | 25 | 5 | 8 | 74 | Nec | CEBIOMAS0496, CEBIOMAS0504, CEBIOMAS0508, CEBIOMAS0510, CEBIOMAS0512 |
| Hsunycteris thomasi | 4 | 7 | 5 | 1 | 1 | 0 | 18 | Nec | CEBIOMAS0511 |
| Lophostoma brasiliense | 0 | 0 | 0 | 0 | 1 | 0 | 1 | HCIns | CEBIOMAS0503 |
| Lophostoma silvicolum | 3 | 13 | 0 | 2 | 8 | 0 | 26 | HCIns | |
| Mesophylla macconnelli | 3 | 4 | 8 | 4 | 2 | 1 | 22 | Fru | |
| Micronycteris megalotis | 0 | 2 | 0 | 0 | 1 | 0 | 3 | HCIns | CEBIOMAS0502 |
| Micronycteris minuta | 4 | 1 | 1 | 0 | 0 | 0 | 6 | HCIns | CEBIOMAS0500 |
| Gardnerycteris crenulatum | 5 | 3 | 1 | 0 | 0 | 0 | 9 | HCIns | CEBIOMAS0507 |
| Phylloderma stenops | 4 | 0 | 0 | 1 | 0 | 0 | 5 | Fru | |
| Phyllostomus discolor | 0 | 0 | 1 | 0 | 0 | 0 | 1 | Omn | |
| Phyllostomus elongatus | 8 | 14 | 2 | 2 | 16 | 0 | 42 | Omn | |
| Phyllostomus hastatus | 4 | 4 | 12 | 5 | 7 | 3 | 35 | Omn | |
| Platyrrhinus brachycephalus | 0 | 1 | 1 | 0 | 0 | 4 | 6 | Fru | |
| Platyrrhinus incarum | 7 | 2 | 22 | 4 | 4 | 13 | 52 | Fru | CEBIOMAS0509 |
| Platyrrhinus infuscus | 1 | 0 | 1 | 1 | 0 | 0 | 3 | Fru | |
| Rhinophylla pumilio | 4 | 16 | 0 | 5 | 19 | 0 | 44 | Fru | |
| Sphaeronycteris toxophyllum | 1 | 0 | 0 | 1 | 1 | 0 | 3 | Fru | |
| Sturnira giannae | 26 | 1 | 33 | 17 | 0 | 4 | 81 | Fru | CEBIOMAS0487 |
| Sturnira tildae | 24 | 4 | 5 | 25 | 3 | 4 | 65 | Fru | CEBIOMAS0488, CEBIOMAS0494, CEBIOMAS0492 |
| Tonatia maresi | 3 | 2 | 0 | 0 | 1 | 0 | 6 | HCIns | |
| Trachops cirrhosus | 3 | 5 | 1 | 0 | 1 | 0 | 10 | Car | |
| Trinycteris nicefori | 1 | 2 | 1 | 0 | 1 | 0 | 5 | HCIns | |
| Uroderma bilobatum | 8 | 4 | 10 | 5 | 2 | 7 | 36 | Fru | CEBIOMAS0486, CEBIOMAS0489 |
| Uroderma magnirostrum | 5 | 2 | 7 | 0 | 2 | 6 | 22 | Fru | CEBIOMAS0501 |
| Vampyressa thyone | 1 | 2 | 0 | 0 | 1 | 1 | 5 | Fru | |
| Vampyriscus bidens | 4 | 2 | 4 | 0 | 0 | 1 | 11 | Fru | |
| Vampyrodes caracciolli | 0 | 0 | 0 | 0 | 1 | 0 | 1 | Fru | |
| Variable | NMDS1 | NMDS2 | r2 | p |
|---|---|---|---|---|
| Foliage height index | −0.491 | −0.871 | 0.005 | 0.95 |
| Horizontal vegetation density | 0.997 | 0.074 | 0.389 | 0.008 |
| Mean canopy height | −0.846 | −0.533 | 0.422 | 0.005 |
| Temperature variation | 0.855 | 0.519 | 0.06 | 0.501 |
| Site Category | Vegetation Type | Observed Species | Jackknife 1 | Jackknife 1–SE | Individuals Captured |
|---|---|---|---|---|---|
| Papaya plantation | Forest interior | 31 | 35.99 | 2.23 | 620 |
| Papaya plantation | Forest edge | 34 | 39.99 | 2.44 | 650 |
| Papaya plantation | Agricultural land | 31 | 43.99 | 3.60 | 1011 |
| Cattle pasture | Forest interior | 31 | 43.97 | 3.60 | 430 |
| Cattle pasture | Forest edge | 23 | 28.99 | 2.44 | 466 |
| Cattle pasture | Agricultural land | 17 | 20.99 | 1.99 | 364 |
| Taxonomic Diversity Metric | Variable | df | Sum Square | Mean Square | F | p |
|---|---|---|---|---|---|---|
| Estimated species richness | vegetation type | 2 | 51.7 | 25.9 | 0.741 | 0.485 |
| site category | 1 | 393.2 | 393.2 | 11.266 | 0.002 | |
| vegetation type:site category | 2 | 26.5 | 13.2 | 0.379 | 0.688 | |
| Residuals | 30 | 1047.1 | 34.9 | |||
| Capture rate | vegetation type | 2 | 7021 | 3511 | 2.259 | 0.122 |
| site category | 1 | 3407 | 3407 | 2.193 | 0.149 | |
| vegetation type:site category | 2 | 5929 | 2964 | 1.908 | 0.166 | |
| Residuals | 30 | 46,623 | 1554 |
| Functional Diversity Metric | Variable | df | Sum Square | Mean Square | F | p |
|---|---|---|---|---|---|---|
| 1E(T) | vegetation type | 2 | 0.0067 | 0.0034 | 0.275 | 0.761 |
| site category | 1 | 0.2486 | 0.2486 | 20.358 | <0.001 | |
| vegetation type:site category | 2 | 0.0797 | 0.0399 | 3.264 | 0.052 | |
| Residuals | 30 | 0.3663 | 0.0122 | |||
| 1D(TM) | vegetation type | 2 | 1.37 | 0.69 | 5.111 | 0.012 |
| site category | 1 | 0.09 | 0.09 | 0.651 | 0.426 | |
| vegetation type:site category | 2 | 0.16 | 0.08 | 0.589 | 0.561 | |
| Residuals | 30 | 4.02 | 0.13 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrasco-Rueda, F.; Loiselle, B.A. Dimensions of Phyllostomid Bat Diversity and Assemblage Composition in a Tropical Forest-Agricultural Landscape. Diversity 2020, 12, 238. https://doi.org/10.3390/d12060238
Carrasco-Rueda F, Loiselle BA. Dimensions of Phyllostomid Bat Diversity and Assemblage Composition in a Tropical Forest-Agricultural Landscape. Diversity. 2020; 12(6):238. https://doi.org/10.3390/d12060238
Chicago/Turabian StyleCarrasco-Rueda, Farah, and Bette A. Loiselle. 2020. "Dimensions of Phyllostomid Bat Diversity and Assemblage Composition in a Tropical Forest-Agricultural Landscape" Diversity 12, no. 6: 238. https://doi.org/10.3390/d12060238






