Two New Benthic Diatoms of the Genus Achnanthidium (Bacillariophyceae) from the Hangang River, Korea
Abstract
:1. Introduction
2. Materials and Methods
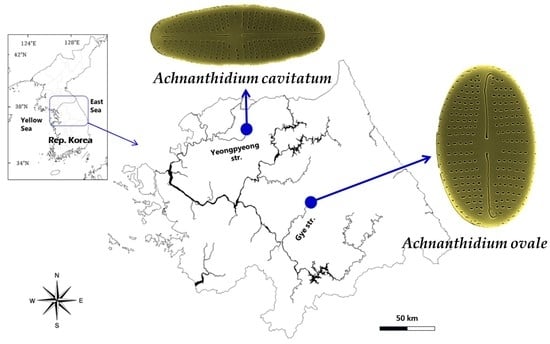
2.1. Sample Collection, Isolation, and Culture
2.2. Light Microscopy (LM)
2.3. Scanning Electron Microscopy (SEM)
2.4. DNA Extraction, PCR Amplification, and Sequencing
2.5. Phylogenetic Analyses
3. Results
3.1. Species Description
3.1.1. Achnanthidium ovale M. Miao & B.-H. Kim, sp. nov.
3.1.2. Achnanthidium cavitatum M. Miao & B.-H. Kim, sp. nov.
3.2. Molecular Phylogeny
4. Discussion
4.1. Achnanthidium ovale as a New Species
4.2. Achnanthidium cavitatum as a New Species
4.3. Areolae Occlusions and Openings
4.4. Ecological Characteristics of Two Achnanthidium Species
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Patrick, R.; Reimer, C.W. The Diatoms of the United States Exclusive of Alaska and Hawaii; Academy of the Natural Sciences: Philadelphia, PA, USA, 1966; p. 688. [Google Scholar]
- Round, F.E.; Crawford, R.M.; Mann, D.G. Diatoms: Biology and Morphology of the Genera; Cambridge University Press: Cambridge, UK, 1990. [Google Scholar]
- Czarnecki, D.B. The freshwater diatoms culture collection at Loras College, Dubuque, Iowa. Mem. Calif. Acad. Sci. 1994, 17, 155–174. [Google Scholar]
- Round, F.E.; Bukhtiyarova, L. Four new genera based on Achnanthes (Achnanthidium) together with a re-definition of Achnanthidium. Diatom Res. 1996, 11, 345–361. [Google Scholar] [CrossRef]
- Kobayashi, H. Comparative Studies among Four Linear-Lanceolate Achnanthidium Species (Bacillariophyceae) with curved terminal raphe endings. Nova Hedwigia 1997, 65, 147–163. [Google Scholar] [CrossRef]
- Potapova, M.; Hamilton, P.B. Morphological and ecological variation within the Achnanthidium minutissimum (Bacillariophyceae) species complex. J. Phycol. 2007, 43, 561–575. [Google Scholar] [CrossRef]
- Yu, P.; Kociolek, J.P.; You, Q.M.; Wang, Q.X. Achnanthidium longissimum sp. nov. (Bacillariophyta), a new diatom species from Jiuzhai Valley, Southwestern China. Diatom Res. 2018, 33, 339–348. [Google Scholar] [CrossRef]
- Witkowski, A.; Kulikovskiy, M.; Riaux-Gobin, C. Achnanthidium sieminskae, a new diatom species from the Kerguelen Archipelago (Austral Islands). In Current Advances in Algal Taxonomy and Its Applications: Phylogenetic, Ecological and Applied Perspective; Wołowski, K., Kaczmarska, I., Ehrman, J.M., Wojtal, A.Z., Eds.; Polish Academy of Sciences: Kraków, Poland, 2012; pp. 61–68. [Google Scholar]
- Pérès, F.; Le Cohu, R.; Delmont, D. Achnanthidium barbei sp. nov. and Achnanthidium costei sp. nov., two new diatom species from French rivers. Diatom Res. 2014, 29, 387–397. [Google Scholar] [CrossRef]
- Potapova, M.G.; Ponader, K.C. Two common North American diatoms, Achnanthidium rivulare sp. nov. and A. deflexum (Reimer) Kingston: Morphology, ecology and comparison with related species. Diatom Res. 2004, 19, 33–57. [Google Scholar] [CrossRef]
- Wojtal, A.Z.; Ector, L.; Van de Vijver, B.; Morales, E.A.; Blanco, S.; Piątek, J.; Smieja, A. The Achnanthidium minutissimum complex (Bacillariophyceae) in southern Poland. Algol. Stud. 2011, 136, 211–238. [Google Scholar] [CrossRef]
- Pérès, F.; Barthès, A.; Ponton, E.; Coste, M.; Ten-Hage, L.; Le Cohu, R. Achnanthidium delmontii sp. nov., a new species from French rivers. Fottea 2012, 12, 189–198. [Google Scholar] [CrossRef] [Green Version]
- Karthick, B.; Taylor, J.C.; Hamilton, P.B. Two new species of Achnanthidium Kützing (Bacillariophyceae) from Kolli Hills, Eastern Ghats, India. Fottea 2017, 17, 65–77. [Google Scholar] [CrossRef] [Green Version]
- Martín, G.; Toja, J.; Sala, S.E.; De los Reyes Fernández, M.; Reyes, I.; Casco, M.A. Application of diatom biotic indices in the Guadalquivir River Basin, a Mediterranean basin. Which one is the most appropriated? Environ. Monit. Assess. 2010, 170, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Ponader, K.C.; Potapova, M.G. Diatoms from the genus Achnanthidium inflowing waters of the Appalachian Mountains (North America): Ecology, distribution and taxonomic notes. Limnologica 2007, 37, 227–241. [Google Scholar] [CrossRef] [Green Version]
- Novais, M.H.; Juttner, I.; Van de Vijver, B.; Morais, M.M.; Hoffmann, L.; Ector, L. Morphological variability within the Achnanthidium minutissimum species complex (Bacillariophyta): Comparison between the type material of Achnanthes minutissima and related taxa, and new freshwater Achnanthidium species from Portugal. Phytotaxa 2015, 224, 101–139. [Google Scholar] [CrossRef]
- Cox, E.J. Ontogeny, homology, and terminology—Wall morphogenesis as an aid to character recognition and character state definition for pennate diatom systematics. J. Phycol. 2012, 48, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Cox, E.J. Symmetry and valve structure in naviculoid diatoms. Nova Hedwigia 1979, 64, 193–206. [Google Scholar]
- Cox, E.J. Variation in patterns of valve morphogenesis between representatives of six biraphid diatom genera (Bacillariophyceae). J. Phycol. 1999, 35, 1297–1312. [Google Scholar] [CrossRef]
- Ross, R.; Cox, E.J.; Karayeva, N.I.; Mann, D.G.; Paddock, T.B.B.; Simonsen, R.; Sims, P.A. An amended terminology for the siliceous components of the diatom cell. Nova Hedwigia 1979, 64, 513–533. [Google Scholar]
- Mann, D.G. Sieves and Flaps: Siliceous Minutiae in the Pores of Raphid Diatoms. In Proceedings of the 6th Symposium on Recent and Fossil Diatoms; Ross, R., Ed.; O. Koeltz: Koenigstein, Germany, 1981; pp. 279–300. [Google Scholar]
- Cox, E.J. Morphogenetic information and the selection of taxonomic characters for raphid diatom systematics. Plant Ecol. Evol. 2010, 143, 271–277. [Google Scholar] [CrossRef]
- Yana, E.; Mayama, S. Two new taxa of Achnanthidium and Encyonema (Bacillariophyceae) from the Yom River, Thailand, with special reference to the areolae occlusions implying ontogenetic relationship. Phycol. Res. 2015, 63, 239–252. [Google Scholar] [CrossRef]
- Hlubikova, D.; Ector, L.; Hoffmann, L. Examination of the type material of some diatom species related to Achnanthidium minutissimum (Kütz.) Czarn. (Bacillariophyceae). Algol. Stud. 2011, 136, 19–43. [Google Scholar] [CrossRef]
- Bruder, K.; Medlin, L.K. Molecular assessment of phylogenetic relationships in selected species/genera in the naviculoid diatoms (Bacillariophyta). I. The genus Placoneis. Nova Hedwigia 2007, 85, 331–352. [Google Scholar] [CrossRef]
- Medlin, L.K.; Kaczmarska, I. Evolution of the diatoms: V. Morphological and cytological support for the major clades and a taxonomic revision. Phycologia 2004, 43, 245–270. [Google Scholar] [CrossRef] [Green Version]
- Medlin, L.K.; Williams, D.M.; Sims, P.A. The evolution of the diatoms (Bacillariophyta). I. Origin of the group and assessment of the monophyly of its major divisions. Eur. J. Phycol. 1993, 28, 261–275. [Google Scholar] [CrossRef] [Green Version]
- Medlin, L.K.; Cooper, A.; Hill, C.; Wrieden, S.; Wellbrock, U. Phylogenetic position of the Chromista plastids based on small subunit rRNA coding regions. Curr. Genet. 1995, 28, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Medlin, L.K.; Kooistra, W.H.; Gersonde, R.; Wellbrock, U. Evolution of the diatoms (Bacillariophyta). II. Nuclear-encoded small-subunit rRNA sequence comparisons confirm a paraphyletic origin for the centric diatoms. Mol. Biol. Evol. 1996, 13, 67–75. [Google Scholar] [CrossRef] [Green Version]
- Sorhannus, U. Diatom phylogenetics inferred based on direct optimization of nuclear-encoded SSU rRNA sequences. Cladistics 2004, 20, 487–497. [Google Scholar] [CrossRef]
- Sato, S.; Mann, D.G.; Matsumoto, S.; Medlin, L.K. Pseudostriatella (Bacillariophyta): A description of a new araphid diatom genus based on observations of frustule and auxospore structure and 18S rDNA phylogeny. Phycologia 2008, 47, 371–391. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Park, B.S.; Kim, J.H.; Kim, J.H.; Lee, H.O.; Han, M.S. Phylogenetic position of eight Amphora sensu lato (Bacillariophyceae) species and comparative analysis of morphological characteristics. Algae 2014, 29, 57–73. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Sui, Z.; Zhang, S.; Ren, Y.; Liu, Y. Comparison of potential diatom ‘barcode’ genes (the 18S rRNA gene and ITS, COI, rbcL) and their effectiveness in discriminating and determining species taxonomy in the Bacillariophyta. Int. J. Syst. Evol. Microbiol. 2015, 65, 1369–1380. [Google Scholar] [CrossRef] [PubMed]
- Mann, D.G.; Simpson, G.E.; Sluiman, H.J.; Möller, M. rbcL gene tree of diatoms: A second large Data-Set for phylogenetic reconstruction. Phycologia 2001, 40, 1–2. [Google Scholar]
- Kulikovskiy, M.S.; Andreeva, S.A.; Gusev, E.S.; Kuznetsova, I.V.; Annenkova, N.V. Molecular phylogeny of monoraphid diatoms and raphe significance in evolution and taxonomy. Biol. Bull. 2016, 43, 398–407. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, P.; Kim, H.K.; Lee, H.; Han, M.S.; Kim, B.H. Lemnicola hungarica (Bacillariophyceae) and the new monoraphid diatom Lemnicola uniseriata sp. nov. (Bacillariophyceae) from Korea. Diatom Res. 2018, 33, 69–87. [Google Scholar] [CrossRef]
- Andersen, R.A. Algal Culturing Techniques; Academic Press: San Diego, CA, USA, 2005; p. 578. [Google Scholar]
- Beakes, G.W.; Canter, H.M.; Jaworski, G.H.M. Zoospore ultrastructure of Zygorhizidium affluens and Z. planktonicum, two chytrids parasitizing the diatom Asterionella formosa. Can. J. Bot. 1988, 66, 1054–1067. [Google Scholar] [CrossRef]
- Katano, T.; Lee, J.; Ki, J.S.; Kang, S.H.; Han, M.S. Effects of temperature and salinity on the growth and the optimum nitrogen to phosphorus ratio for the culture of Diatoma tenue isolated from a temporary Arctic pond in Svalbard, Norway. J. Freshw. Ecol. 2007, 22, 629–635. [Google Scholar] [CrossRef]
- Ki, J.S.; Han, M.S. Molecular analysis of complete SSU to LSU rRNA sequence in the harmful dinoflagellate Alexandrium tamarense (Korean isolate, HY970328M). Ocean Sci. J. 2005, 40, 155–166. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Fredrik, R.; Teslenko, M.; Van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar]
- Kobayasi, H.; Nagumo, T.; Mayama, S. Observation on the two rheophilic species of the genus Achnanthes (Bacillariophyceae), A. convergens H. Kob. And A. japonica H. Kob. Diatom 1986, 2, 83–93. [Google Scholar]
- Thomas, E.W. Exploring Species Boundaries in the Diatom Genus Rhoicosphenia Using Morphology, Phylogeny, Ecology, and Biogeography. Ph.D. Thesis, Department of Ecology & Evolutionary, University of Colorado, Boulder, Co, USA, 2016. [Google Scholar]
- Chessman, B.; Growns, I.; Currey, J.; Plunkett-Cole, N. Predicting diatom communities at the genus level for the rapid biological assessment of rivers. Freshw. Biol. 1999, 41, 317–331. [Google Scholar] [CrossRef]
- Wu, J.T. A generic index of diatom assemblages as bioindicator of pollution in the Keelung River of Taiwan. Hydrobiologia 1999, 397, 79–87. [Google Scholar] [CrossRef]
- Hill, B.H.; Herlihy, A.T.; Kaufmann, P.R.; Stevenson, R.J.; McCormick, F.H.; Johnson, C.B. Use of periphyton assemblage data as an index of biotic integrity. J. N. Am. Benth. Soc. 2000, 19, 50–67. [Google Scholar] [CrossRef]
- NGII (National Geographic Information Institute). Available online: https://www.ngii.go.kr/ (accessed on 11 June 2020).
- Kobayasi, H.; Idei, M.; Mayama, S.; Nagumo, T.; Osada, K. Kobayasi’s Atlas of Japanese Diatoms Based on Electron Microscopy; Uchida Rokakuho: Tokyo, Japan, 2006; p. 531. [Google Scholar]









| Achnanthidium ovale | Achnanthidium cavitatum | |
|---|---|---|
| Coordinates | 37°31′59α″ N, 128°0′42″ E | 38°4′28″ N, 127°24′52″ E |
| pH | 7.07 | 6.72 |
| Dissolved oxygen | 5.93 | 7.93 |
| Temperature (°C) | 11.71 | 11.29 |
| Velocity (cm/sec) | 80 | 20 |
| Conductivity (µS/cm) | 57 | 148 |
| Turbidity (NTU) | 0.0 | 3.1 |
| Gene | Primer | Nucleotide Sequence (5′ to 3′) | Reference |
|---|---|---|---|
| SSU rRNA | AT18F01 | YAC-CTG-GTT-GAT-CCT-GCC-AGT-AG | [40] |
| AT18R02 | GTT-TCA-GCC-TTG-CGA-CCA-TAC-TCC | [40] | |
| AT18F02 | AGA-ACG-AAA-GTT-AAG-GGA-TCG-AAG-ACG | [40] | |
| AT18R01 | GCT-TGA-TCC-TTC-TGC-AGG-TTC-ACC | [40] | |
| rbcL | F3 | GCT-TAC-CGT-GTA-GAT-CCA-GTT-CC | [25] |
| R3 | CCT-TCT-AAT-TTA-CCA-ACA-ACT-G | [25] |
| Species | Strain | Taxonomic Position | Gene Type | Locality | GenBank Accession No. |
|---|---|---|---|---|---|
| Achnanthidium ovale sp. nov. | HYU-D036 | Achnanthales; Achnanthidiaceae | SSU | Korea | MK578710, this study |
| Achnanthidium ovale sp. nov. | HYU-D036 | Achnanthales; Achnanthidiaceae | rbcL | Korea | MK639354, this study |
| Achnanthidium cavitatum sp. nov. | HYU-D037 | Achnanthales; Achnanthidiaceae | SSU | Korea | MK578711, this study |
| Achnanthidium cavitatum sp. nov. | HYU-D037 | Achnanthales; Achnanthidiaceae | rbcL | Korea | MK639355, this study |
| Sequence Name | Achnanthidium ovale MK578710 HYU-D036 | Achnanthidium cavitatum MK578711 HYU-D037 | ||
|---|---|---|---|---|
| Similarity | p-Distance | Similarity | p-Distance | |
| Achnanthidium ovale MK578710 HYU-D036 | - | - | 0.977 | 0.012 |
| Achnanthidium cavitatum MK578711 HYU-D037 | 0.977 | 0.012 | - | - |
| Achnanthidium brevipes AY485476 CCMP100 | 0.921 | 0.044 | 0.913 | 0.046 |
| Achnanthidium catenatum KY863463 TCC849 | 0.971 | 0.016 | 0.99 | 0.004 |
| Achnanthidium cf. longipes AY485500 CCMP101 | 0.912 | 0.050 | 0.904 | 0.052 |
| Achnanthidium coarctatum HQ912594 UTEX FD185 | 0.915 | 0.050 | 0.908 | 0.052 |
| Achnanthidium digitatum KU565386 SPITS-M2A+B-14 | 0.969 | 0.014 | 0.979 | 0.014 |
| Achnanthidium digitatum KU565387 SPITS-M2A+B-38 | 0.967 | 0.014 | 0.977 | 0.014 |
| Achnanthidium digitatum KX946582 SPITS-M2A+B-32 | 0.969 | 0.014 | 0.979 | 0.014 |
| Achnanthidium kranzii KJ658397 A100r | 0.986 | 0.008 | 0.979 | 0.010 |
| Achnanthidium kranzii LC482223 KSA47 | 0.986 | 0.008 | 0.979 | 0.010 |
| Achnanthidium minutissimum AM502032 AT-196Gel02 | 0.967 | 0.018 | 0.975 | 0.018 |
| Achnanthidium minutissimum KF417666 RK6 | 0.935 | 0.030 | 0.933 | 0.030 |
| Achnanthidium minutissimum KF959663 TCC746 | 0.967 | 0.018 | 0.975 | 0.018 |
| Achnanthidium minutissimum KJ658398 AD815 | 0.963 | 0.020 | 0.977 | 0.018 |
| Achnanthidium minutissimum KJ658400 AD819 | 0.963 | 0.020 | 0.977 | 0.018 |
| Achnanthidium minutissimum KJ658401 AM2006 | 0.965 | 0.018 | 0.973 | 0.018 |
| Achnanthidium minutissimum KJ658402 Ashort2 | 0.971 | 0.016 | 0.986 | 0.008 |
| Achnanthidium minutissimum KJ658403 AW2 | 0.971 | 0.016 | 0.986 | 0.008 |
| Achnanthidium minutissimum KJ658404 NJ211 | 0.963 | 0.018 | 0.971 | 0.018 |
| Achnanthidium minutissimum KT072992 TCC688 | 0.967 | 0.018 | 0.975 | 0.018 |
| Achnanthidium minutissimum KY863464 TCC696 | 0.967 | 0.018 | 0.975 | 0.018 |
| Achnanthidium minutissimum LC037436 NIES-407 | 0.967 | 0.018 | 0.975 | 0.018 |
| Achnanthidium minutissimum LC037437 NIES-410 | 0.973 | 0.014 | 0.98 | 0.010 |
| Achnanthidium minutissimum LC037438 NIES-412 | 0.973 | 0.014 | 0.98 | 0.010 |
| Achnanthidium minutissimum LC037439 NIES-414 | 0.967 | 0.018 | 0.975 | 0.018 |
| Achnanthidium minutissimum LC143218 NIES-411 | 0.973 | 0.014 | 0.98 | 0.010 |
| Achnanthidium minutissimum LC143219 NIES-413 | 0.973 | 0.014 | 0.98 | 0.010 |
| Achnanthidium minutissimum MH358459 HYU-D003 | 0.971 | 0.016 | 0.986 | 0.008 |
| Achnanthidium pyrenaicum KY863466 TCC832 | 0.967 | 0.018 | 0.975 | 0.018 |
| Achnanthidium reimeri KJ658405 Arei2 | 0.988 | 0.006 | 0.977 | 0.012 |
| Achnanthidium rivulare KJ658406 Ariv2 | 0.986 | 0.008 | 0.979 | 0.010 |
| Achnanthidium saprophilum LC037434 NIES-372 | 0.971 | 0.016 | 0.986 | 0.008 |
| Achnanthidium saprophilum MN602029 18SCT | 0.971 | 0.014 | 0.979 | 0.010 |
| Achnanthidium saprophilum MN602031 18STP2 | 0.971 | 0.014 | 0.979 | 0.010 |
| Achnanthidium anastasiae KJ658414 Ros1 | 0.988 | 0.006 | 0.975 | 0.010 |
| Achnanthidium sp. KU565379 MIC10 61 | 0.967 | 0.018 | 0.975 | 0.018 |
| Sequence Name | Achnanthidium ovale MK578710 HYU-D036 | Achnanthidium cavitatum MK578711 HYU-D037 | ||
|---|---|---|---|---|
| Similarity | p-Distance | Similarity | p-Distance | |
| Achnanthidium ovale MK639354 HYU-D036 | - | - | 0.924 | 0.075 |
| Achnanthidium cavitatum MK639355 HYU-D037 | 0.924 | 0.075 | - | - |
| Achnanthidium anastasiae KJ658396 Ros1 | 0.956 | 0.045 | 0.946 | 0.052 |
| Achnanthidium catenatum KY799133 TCC849 | 0.925 | 0.066 | 0.961 | 0.033 |
| Achnanthidium cf. lineare KR709273 B397 | 0.930 | 0.068 | 0.957 | 0.043 |
| Achnanthidium cf. lineare KR709274 B398 | 0.930 | 0.068 | 0.957 | 0.043 |
| Achnanthidium daonense KJ658395 PS3 | 0.937 | 0.064 | 0.941 | 0.057 |
| Achnanthidium digitatum KU687471 SPITS-M2A+B-32 | 0.936 | 0.063 | 0.954 | 0.047 |
| Achnanthidium digitatum KU687478 SPITS-M2A+B-14 | 0.936 | 0.063 | 0.954 | 0.047 |
| Achnanthidium digitatum KU687479 SPITS-M2A+B-38 | 0.934 | 0.064 | 0.952 | 0.049 |
| Achnanthidium kranzii KJ658379 A100 | 0.944 | 0.057 | 0.929 | 0.070 |
| Achnanthidium minutissimum AM710499 AT-196Gel02 | 0.936 | 0.063 | 0.959 | 0.042 |
| Achnanthidium minutissimum KF959649 TCC746 | 0.934 | 0.064 | 0.954 | 0.047 |
| Achnanthidium minutissimum KJ658380 AD817 | 0.936 | 0.063 | 0.954 | 0.047 |
| Achnanthidium minutissimum KJ658381 AD819 | 0.936 | 0.063 | 0.954 | 0.047 |
| Achnanthidium minutissimum KJ658382 AD815 | 0.936 | 0.063 | 0.954 | 0.047 |
| Achnanthidium minutissimum KJ658383 AM2006 | 0.934 | 0.064 | 0.954 | 0.047 |
| Achnanthidium minutissimum KJ658384 Ashort2 | 0.929 | 0.070 | 0.966 | 0.035 |
| Achnanthidium minutissimum KJ658385 AW2 | 0.929 | 0.070 | 0.966 | 0.035 |
| Achnanthidium minutissimum KJ658386 NJ211 | 0.937 | 0.061 | 0.956 | 0.045 |
| Achnanthidium minutissimum KR709271 B448 | 0.936 | 0.063 | 0.959 | 0.042 |
| Achnanthidium minutissimum KR709272 B443 | 0.936 | 0.063 | 0.959 | 0.042 |
| Achnanthidium minutissimum KT072938 TCC676 | 0.937 | 0.061 | 0.957 | 0.043 |
| Achnanthidium minutissimum KY799134 TCC748 | 0.932 | 0.064 | 0.951 | 0.047 |
| Achnanthidium minutissimum KY863481 TCC564 | 0.927 | 0.070 | 0.968 | 0.031 |
| Achnanthidium minutissimum KY863482 TCC667 | 0.936 | 0.063 | 0.962 | 0.038 |
| Achnanthidium minutissimum KY863483 TCC688 | 0.934 | 0.064 | 0.959 | 0.042 |
| Achnanthidium minutissimum KY863484 TCC696 | 0.930 | 0.068 | 0.956 | 0.045 |
| Achnanthidium minutissimum MK639350 HYU-D003 | 0.930 | 0.068 | 0.966 | 0.035 |
| Achnanthidium pyrenaicum KY799135 TCC832 | 0.919 | 0.070 | 0.942 | 0.049 |
| Achnanthidium reimeri KJ658387 Arei2 | 0.952 | 0.049 | 0.932 | 0.066 |
| Achnanthidium rivulare KJ658390 Ariv2 | 0.944 | 0.057 | 0.929 | 0.070 |
| Achnanthidium saprophilum KM084941 D06-036 | 0.927 | 0.071 | 0.962 | 0.038 |
| Achnanthidium straubianum KY799136 TCC831 | 0.925 | 0.073 | 0.971 | 0.028 |
| Achnanthidium straubianum KY799137 TCC833 | 0.922 | 0.066 | 0.957 | 0.033 |
| Achnanthidium sp. KU687462 SPITS-M2A+B-26 | 0.934 | 0.064 | 0.957 | 0.043 |
| Achnanthidium sp. KU687463 SPITS-M3-10 | 0.932 | 0.064 | 0.959 | 0.040 |
| Achnanthidium sp. KU687464 MIC10-72 | 0.936 | 0.063 | 0.962 | 0.038 |
| Achnanthidium sp. KU687465 SPITS13 | 0.934 | 0.064 | 0.957 | 0.043 |
| Achnanthidium sp. KU687466 MIC10-61 | 0.936 | 0.063 | 0.962 | 0.038 |
| Achnanthidium sp. KU687467 SPITS-N3-12 | 0.934 | 0.064 | 0.957 | 0.043 |
| Achnanthidium sp. KU687469 SPITS-M2A+B-18a | 0.934 | 0.064 | 0.957 | 0.043 |
| Achnanthidium sp. KU687470 SPITS13 | 0.934 | 0.064 | 0.957 | 0.043 |
| Achnanthidium sp. KU687472 MIC10-52b | 0.936 | 0.063 | 0.962 | 0.038 |
| Achnanthidium sp. KU687474 SPITS-M3-15 | 0.932 | 0.064 | 0.959 | 0.040 |
| Achnanthidium sp. KU687475 MIC10-68a | 0.936 | 0.063 | 0.962 | 0.038 |
| Achnanthidium sp. KU687476 MIC10-53 | 0.936 | 0.063 | 0.962 | 0.038 |
| Achnanthidium sp. KU687477 SPITS-M2A+B-12 | 0.930 | 0.068 | 0.956 | 0.045 |
| Achnanthidium sp. KX783621 CANA109929-J7Run23 | 0.936 | 0.063 | 0.959 | 0.042 |
| Achnanthidium sp. KX783622 CANA109929-J2Run23 | 0.930 | 0.068 | 0.956 | 0.045 |
| A. ovale M. Miao & B.-H. Kim sp. nov. | A. rivulare Potapova & Ponader | A. pyrenaicum (Hustedt) Kobayasi | A. convergens Kobayasi | |
|---|---|---|---|---|
| length (µm) | 6.3–7.7 | 5.4–21.3 | 10.0–16.0 | 10.0–25.0 |
| width (µm) | 3.8–4.1 | 2.6–4.4 | 2.5–4.0 | 4–4.5 |
| valve outline | elliptical | linear-elliptical | linear-lanceolate with slightly drawn-out ends | linear-lanceolate |
| external areolae | elongate and dot-like | small, round, or slightly elongated | elongate or circular (RV) | constricted in various degrees. |
| internal areolae | hymenes partially joined | elliptical internal openings occluded by hymenes | hymenes not joined | hymenes partially joined, linking bars between interstriae partly interrupted |
| areolae in valve mantle | slit-like | slit-like | elongate | elongate |
| raphe valve | ||||
| density of striae (in 10 µm) | 30–35 (up to 55 near apices) | 19–25 (up to 55 near apices) | center: 20–25 apices: 34–40 | center: 18, apices: 36–40 |
| striation pattern | parallel but slightly radiate at apices | parallel but convergent or parallel near apices | parallel or slightly radiate in the central area and slightly convergent at apices | densely convergent striae near the valve ends |
| external raphe endings | laterally expanded | teardrop-shaped | teardrop-shaped | laterally expanded |
| internal raphe endings | deflected in opposite directions | short, hook-shaped | slightly curved to opposite sides | deflected in opposite directions |
| rapheless valve | ||||
| density of striae (in 10 µm) | 30–33 (up to 50 near apices) | 19–28 (up to 43 near apices) | center: 20–28 apices: 32–38 | |
| striation pattern | parallel but slightly radiate near apices | parallel but slightly radiate near apices | parallel or slightly radiate in the central area and slightly convergent at apices | slightly radiate at the ends |
| source | this study | [10] | [13] | [45] |
| A. cavitatum M. Miao & B.-H. Kim sp. Nov. | A. minutissimum (Kützing) Czarnecki | A. saprophilum (Kobayashi & Mayama) Round & Bukhtiyarova | A. eutrophilum (Lange-Bertalot) Lange-Bertalot | A. duriense Novais & Ector | |
|---|---|---|---|---|---|
| length (µm) | 8.8–10.3 | 9.0–14.5 | 9.5–14.5 | 7.5–16.0 | 5.0–9.7 |
| width (µm) | 3.0–3.5 | 2.5–3.177 | 3.0–3.6 | 3.2–4.8 | 2.0–2.7 |
| valve outline | rhombic with slightly drawn-out ends | linear-elliptic to linear-lanceolate | broadly linear | narrowly rhombic | elliptic to linear-elliptic |
| striation pattern | radiate at apices and weakly radiate in central area | radiate, denser toward the apices | radiate at apices and weakly radiate in central area | radiate at apices and weakly radiate in central area | almost parallel near the center; slightly radiate elsewhere |
| areolae in the valve mantle | slit-like or elongate | slit-like | slit-like | elongate or slit-like | elongate or slit-like |
| raphe valve | |||||
| density of striae (in 10 µm) | 30–32 | 30–35 | 28–31 | 25–30 | 35 |
| areola openings | most areolae are rounded or elongate elliptic; some are slit-like in the central area | small and rounded; slit-like near the margin | rounded; slit-like near the margin | rounded to elongated areolae | quadrangular or rounded |
| central area | linear, becoming a little wider in the central area | small and lanceolate to rectangular | linear, becoming a little wider in the central area | small rhombic, almost absent | narrow linear axial area slightly expanded towards the center |
| rapheless valve | |||||
| density of striae (in 10 µm) | 28–40 | 32–35 | 28–31 | 25–30 | 35 |
| areola openings | most areolae are rounded or elongated elliptic, but some are slit-like; slit-like areolae are mostly in the axial central area and more than RV | small and rounded; slit-like near the margin | rounded; slit-like near the margin | rounded to elongated | quadrangular or rounded; sometimes slit-like near the margin |
| central area | broadly lanceolate to linear and narrow | narrowly lanceolate | broadly lanceolate to linear and narrow | narrowly rhombic to lanceolate, almost absent | narrow, linear axial area slightly widening toward the central area |
| Source | this study | [6] | [24] | [24] | [16] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, M.; Li, Z.; Hwang, E.-A.; Kim, H.-K.; Lee, H.; Kim, B.-H. Two New Benthic Diatoms of the Genus Achnanthidium (Bacillariophyceae) from the Hangang River, Korea. Diversity 2020, 12, 285. https://doi.org/10.3390/d12070285
Miao M, Li Z, Hwang E-A, Kim H-K, Lee H, Kim B-H. Two New Benthic Diatoms of the Genus Achnanthidium (Bacillariophyceae) from the Hangang River, Korea. Diversity. 2020; 12(7):285. https://doi.org/10.3390/d12070285
Chicago/Turabian StyleMiao, Minzi, Zhun Li, Eun-A Hwang, Ha-Kyung Kim, Hyuk Lee, and Baik-Ho Kim. 2020. "Two New Benthic Diatoms of the Genus Achnanthidium (Bacillariophyceae) from the Hangang River, Korea" Diversity 12, no. 7: 285. https://doi.org/10.3390/d12070285
APA StyleMiao, M., Li, Z., Hwang, E. -A., Kim, H. -K., Lee, H., & Kim, B. -H. (2020). Two New Benthic Diatoms of the Genus Achnanthidium (Bacillariophyceae) from the Hangang River, Korea. Diversity, 12(7), 285. https://doi.org/10.3390/d12070285