16S rRNA Gene Amplicon Sequencing Data of the Iron Quadrangle Ferruginous Caves (Brazil) Shows the Importance of Conserving This Singular and Threatened Geosystem
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection Sites
2.2. Total Genomic DNA Extraction
2.3. Partial Amplification of the 16S Ribosomal Gene
2.4. 16S rRNA Amplicon Sequencing Using the Ion Torrent Platform
2.5. 16S rRNA Amplicon Sequencing Data Analyses
2.6. Functional Metabolism Prediction from 16S rRNA Amplicon Sequences
3. Results
3.1. Sample Quality and the Number of Sequences Obtained
3.2. Alpha Diversity Characterization
3.3. Beta-Diversity Characterization
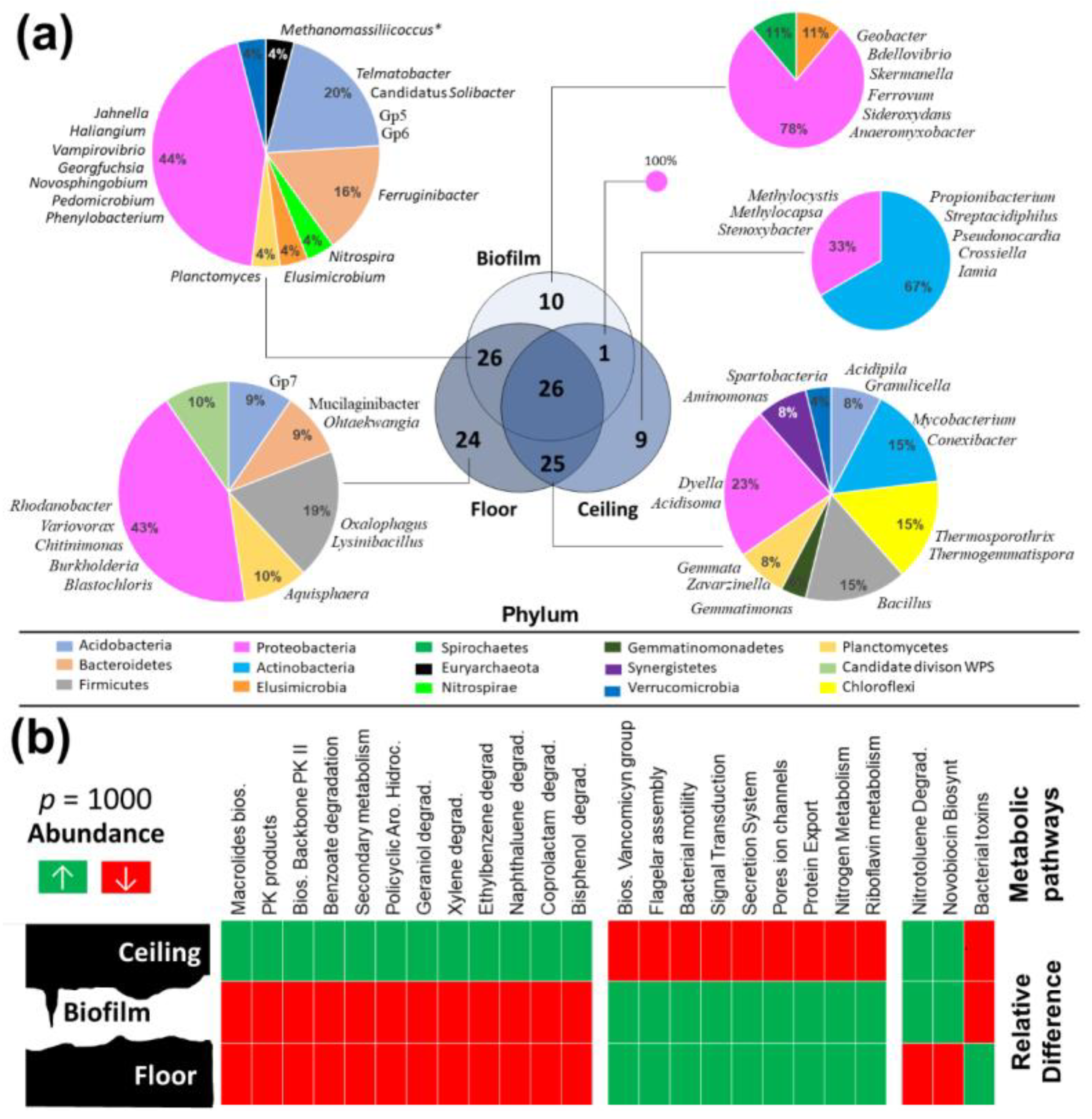
3.4. Comparison of Prokaryotic Community Structures among Caves
3.5. Metabolic Analysis
3.6. Ferruginous Caves Associated with Chapada de Canga
3.7. Inference of Ecological Relations in Ferruginous Caves
4. Discussion
Ferruginous Caves in IQ
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethics Approval and Consent to Participate
References
- Auler, A.; Farrant, A. A brief introduction to karst and caves in Brazil. Proc. Univ. Bristol Spelaeol. Soc. 1996, 20, 187–200. [Google Scholar]
- Piccini, L. Karst in siliceous rocks—Karst landforms and caves in the Auyan-Tepui Massif (Est. Bolivar, Venezuela). Int. J. Speleol. 1995, 24, 41–54. [Google Scholar] [CrossRef]
- Silveira, O.; Maurity, C.; Pinheiro, R.; Henriques, A.; Kern, D.; Souza, S. Estudo das cavernas da província espeleológica arenítica de Monte Alegre—PA. Cad. Geociênc. 1995, 15, 57–63. [Google Scholar]
- Hardt, R.; Pinto, S.d.A.F. Carste Em Litologias Não Carbonáticas. Rev. Bras. De Geomorfol. 2009, 10, 99–105. [Google Scholar] [CrossRef] [Green Version]
- Piccini, L.; Mecchia, M. Solution weathering rate and origin of karst landforms and caves in the quartzite of Auyan-tepui (Gran Sabana, Venezuela). Geomorphology 2009, 106, 15–25. [Google Scholar] [CrossRef]
- Gomes, M.; Azevedo, Ú.; Ferreira, R.; Goulart, F. Landscape Fragmentation around Ferruginous Caves of the Iron Quadrangle, Minas Gerais, Brazil. Cuad. Geogr. Rev. Colomb. Geogr. 2019, 28. [Google Scholar] [CrossRef]
- Simmons, G.C. Canga Caves in the Quadrilátero Ferrífero, Minas Gerais, Brazil. Bull. Natl. Speleol. Soc. 1963, 22, 66–72. [Google Scholar]
- Pereira, M.; Stávale, Y.; Salgado, A. Estudo da gênese das cavidades e depressões em minério de ferro—Quadrilátero Ferrífero/MG: Serras do Rola Moça e do Gandarela. Rev. Bras. Geomorfol. 2013, 13. [Google Scholar] [CrossRef] [Green Version]
- Pereira, M.; Rodet, J.; Salgado, A. Aspectos genéticos e morfológicos das cavidades naturais da Serra da Piedade, Quadrilatero Ferrifero/MG (Brasil). Rev. Bras. Geomorfol. 2012, 13, 465–476. [Google Scholar] [CrossRef] [Green Version]
- Calux, A.S.; Cassimiro, J.R.; Salgado, A.A.R. Caves in iron formations in the Quadrilátero Ferrífero, Minas Gerais, southeastern Brazil: Lithological, morphological and hydrological settings and speleogenesis. Z. Geomorphol. 2019, 62, 125–144. [Google Scholar] [CrossRef]
- Dorr, J.V.N. Physiographic, Stratigraphic and Structural Development of the Quadrilátero Ferrífero Minas Gerais, Brazil, 2nd ed.; United States Government Printing Office: Washington, DC, USA, 1969; 110p. [Google Scholar]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Monteiro, H.S.; Vasconcelos, P.M.; Farley, K.A.; Spier, C.A.; Mello, C.L. (U–Th)/He geochronology of goethite and the origin and evolution of cangas. Geochim. Cosmochim. Acta 2014, 131, 267–289. [Google Scholar] [CrossRef]
- Salgado, A.A.R.; Carmo, F.F. Quadrilátero Ferrífero: A Beautiful and Neglected Landscape Between the Gold and Iron Ore Reservoirs. In Landscapes and Landforms of Brazil; Vieira, B.C., Salgado, A.A.R., Santas, L., Eds.; Springer: London, UK, 2015; pp. 319–330. [Google Scholar]
- Jacobi, C.M.; Carmo, F.F.; Campos, I.C. Soaring extinction threats to endemic plants in Brazilian metal-rich regions. Ambio 2011, 40, 540–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carmo, F.F.; Jacobi, C.M. Diversity and plant trait-soil relationships among rock outcrops in the Brazilian Atlantic rainforest. Plant. Soil 2016, 403, 7–20. [Google Scholar] [CrossRef]
- Ferreira, R.L.; Oliveira, M.P.A.; Silva, M.S. Subterranean Biodiversity in Ferruginous Landscapes: Analysis and Synthesis. In Cave Ecology; Moldovan, O., Kováč, Ľ., Halse, S., Eds.; Springer: Cham, Switzerland, 2018; pp. 435–447. [Google Scholar]
- Calux, A.S.; Cassimiro, J.R. Geoespeleologia das cavernas em rochas ferríferas: Aspectos dimensionais, morfológicos, hidrológicos e sedimentares. In Patrimônio Espeleológico em Rochas Ferruginosas: Propostas para Sua Conservação no Quadrilátero Ferrífero, Minas Gerais; Sociedade Brasileira de Espeleologia, 2015; 26p. [Google Scholar]
- Dutra, G. Síntese dos processos de gênese de cavidades em litologias de ferro. In Proceedings of the 32° Congresso Brasileiro de Espeleologia, Barreiras, Brazil, 11–14 July 2013; pp. 415–426. [Google Scholar]
- Auler, A.; Piló, L.; Parker, C.; Senko, J.; Sasowsky, I.D.; Barton, H. Hypogene Cave Patterns in Iron Ore Caves: Convergence of Forms or Processes? In Hypogene Cave Morphologies; Klimchouk, A., Sasowsky, I.D., Mylroie, J., Engel, S.A., Engel, A.S., Eds.; Karst Waters Institute Special Publication: San Salvador Island, Bahamas, 2014; Volume 18, pp. 15–19. [Google Scholar]
- Culver, D.C. Cave Life: Evolution and Ecology; Harvard University Press: Cambridge, MA, USA, 1982; p. 189. [Google Scholar]
- Scherer, R.S.; Piló, L.B.; Souza-Filho, P.W.M.; Barata, A.S.; Scherer, B.S. Ocorrência de espeleotemas e feições morfológicas raras em cavernas ferríferas da Serra dos Carajás, no Pará. In Proceedings of the 34° Congresso Brasileiro de Espeleologia, Ouro Preto, Brazil, 13–18 June 2017; pp. 409–416. [Google Scholar]
- Parker, C.W.; Auler, A.S.; Barton, M.D.; Sasowsky, I.D.; Senko, J.M.; Barton, H.A. Fe(III) Reducing Microorganisms from Iron Ore Caves Demonstrate Fermentative Fe(III) Reduction and Promote Cave Formation. Geomicrobiol. J. 2018, 35, 311–322. [Google Scholar] [CrossRef]
- Gonçalves, D.F.; Sousa, D.L. Aspectos morfológicos de espeleotemas em feições pseudocársticas da região de Carajás – PA. In Proceedings of the 31° Congresso Brasileiro de Espeleologia, Ponta Grossa, Brazil, 21–24 2011; pp. 141–145. [Google Scholar]
- Carmo, F.F.; Kamino, L.H.Y. Geossistemas Ferruginosos do Brasil—Áreas Prioritárias para Conservação da Diversidade Geológica e Biológica, Patrimônio Cultural e Serviços Ambientais, 1st ed.; 3i Editora: Belo Horizonte, Brazil, 2015; Volume 1. [Google Scholar]
- Salles, D.M.; Carmo, F.F.; Jacobi, C.M. Habitat Loss Challenges the Conservation of Endemic Plants in Mining-Targeted Brazilian Mountains. Environ. Conserv. 2019, 46, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Auler, A.S.; Piló, L.B. Caves and Mining in Brazil: The Dilemma of Cave Preservation Within a Mining Context. In Hydrogeological and Environmental Investigations in Karst Systems; Andreo, B., Carrasco, F., Durán, J.J., Jiménez, P., LaMoreaux, J.W., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 487–496. [Google Scholar]
- Kamino, L.H.Y.; Pereira, E.O.; Carmo, F.F. Conservation paradox: Large-scale mining waste in protected areas in two global hotspots, southeastern Brazil. Ambio 2020, 49, 1629–1638. [Google Scholar] [CrossRef]
- Hatje, V.; Pedreira, R.M.A.; Rezende, C.E.; Schettini, C.A.F.; Souza, G.C.; Marin, D.C.; Hackspacher, P.C. The environmental impacts of one of the largest tailing dam failures worldwide. Sci. Rep. 2017, 7, 10706. [Google Scholar] [CrossRef] [PubMed]
- Carmo, F.F.; Kamino, L.H.Y.; Junior, R.T.; Campos, I.C.; Carmo, F.F.; Silvino, G.; Castro, K.J.d.S.X.d.; Mauro, M.L.; Rodrigues, N.U.A.; Miranda, M.P.d.S.; et al. Fundão tailings dam failures: The environment tragedy of the largest technological disaster of Brazilian mining in global context. Perspect. Ecol. Conserv. 2017, 15, 145–151. [Google Scholar] [CrossRef]
- Parker, C.W.; Wolf, J.A.; Auler, A.S.; Barton, H.A.; Senko, J.M. Microbial Reducibility of Fe(III) Phases Associated with the Genesis of Iron Ore Caves in the Iron Quadrangle, Minas Gerais, Brazil. Minerals 2013, 3, 395. [Google Scholar] [CrossRef] [Green Version]
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–221. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, C.M.; Carmo, F.F.; Vincent, R.C.; Stehmann, J.R. Plant communities on ironstone outcrops: A diverse and endangered Brazilian ecosystem. Biodivers. Conserv. 2007, 16, 2185–2200. [Google Scholar] [CrossRef]
- Jacobi, C.M.; Carmo, F.F.; Carmo, F.F.; Campos, I.C. Iron geosystems: Priority areas for conservation in Brazil. In Mining in Ecologically Sensitive Landscapes; Tibbett, M., Ed.; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [Green Version]
- Fouhy, F.; Clooney, A.G.; Stanton, C.; Claesson, M.J.; Cotter, P.D. 16S rRNA gene sequencing of mock microbial populations- impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016, 16, 123. [Google Scholar] [CrossRef]
- Ion Personal Genome Machine™ Performance Overview. Application Note. Ion. Torrent Life Technol. 2011. Available online: http://tools.thermofisher.com/content/sfs/brochures/cms_094272.pdf (accessed on 15 September 2021).
- Song, L.; Huang, W.; Kang, J.; Huang, Y.; Ren, H.; Ding, K. Comparison of error correction algorithms for Ion Torrent PGM data: Application to hepatitis B virus. Sci. Rep. 2017, 7, 8106. [Google Scholar] [CrossRef]
- Feng, W.; Zhao, S.; Xue, D.; Song, F.; Li, Z.; Chen, D.; He, B.; Hao, Y.; Wang, Y.; Liu, Y. Improving alignment accuracy on homopolymer regions for semiconductor-based sequencing technologies. BMC Genom. 2016, 17, 521. [Google Scholar] [CrossRef] [Green Version]
- Golan, D.; Medvedev, P. Using state machines to model the Ion Torrent sequencing process and to improve read error rates. Bioinformatics 2013, 29, i344–i351. [Google Scholar] [CrossRef] [Green Version]
- Pylro, V.S.; Roesch, L.F.W.; Ortega, J.M.; Amaral, A.M.; Tótola, M.R.; Hirsch, P.R.; Rosado, A.S.; Góes-Neto, A.; Costa da Silva, A.L.; Rosa, C.A.; et al. Brazilian Microbiome Project: Revealing the Unexplored Microbial Diversity—Challenges and Prospects. Microb. Ecol. 2014, 67, 237–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Champaign, IL, USA, 1949; 117p. [Google Scholar]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Chao, A. Non-parametric estimation of the classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar] [CrossRef]
- Hamady, M.; Lozupone, C.; Knight, R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010, 4, 17–27. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: New York, NY, USA, 2016; p. 260. [Google Scholar]
- Aitchison, J.; Barceló-Vidal, C.; Martín-Fernández, J.A.; Pawlowsky-Glahn, V. Logratio Analysis and Compositional Distance. Math. Geol. 2000, 32, 271–275. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [Green Version]
- Maurity, C.; Kotschoubey, B. Evolução recente da cobertura de alteração no Platô N1—Serra dos Carajás-PA. Degradação, pseudocarstificação, espeleotemas. Bol. Mus. Para. Emilio Goeldi Sér. Ciênc. Terra 1995, 7, 331–362. [Google Scholar]
- Caneschi, W.L.; Felestrino, É.B.; Fonseca, N.P.; Villa, M.M.; Lemes, C.G.C.; Cordeiro, I.F.; Assis, R.d.A.B.; Sanchez, A.B.; Vieira, I.T.; Kamino, L.H.Y.; et al. Brazilian Ironstone Plant Communities as Reservoirs of Culturable Bacteria With Diverse Biotechnological Potential. Front. Microbiol. 2018, 9, 1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felestrino, É.B.; Vieira, I.T.; Caneschi, W.L.; Cordeiro, I.F.; Assis, R.A.B.; Lemes, C.G.C.; Fonseca, N.P.; Sanchez, A.B.; Cepeda, J.C.C.; Ferro, J.A.; et al. Biotechnological potential of plant growth-promoting bacteria from the roots and rhizospheres of endemic plants in ironstone vegetation in southeastern Brazil. World J. Microbiol. Biotechnol. 2018, 34, 156. [Google Scholar] [CrossRef] [PubMed]
- Drummond, G.M.; Martins, C.S.; Machado, A.B.M.; Sebaio, F.A.; Antonini, Y. Biodiversidade em Minas Gerais: Um Atlas para Sua Conservação, 2nd ed.; Fundação Biodiversitas: Belo Horizonte, Brazil, 2005; 222p. [Google Scholar]
- Giuliett, A.M.; Rapini, A.; Gomes de Andrade, M.; Queiroz, L.; Da Silva, J.M. Plantas Raras do Brasil; Conservação Internacional: Belo Horizonte, Brazil, 2009; 495p. [Google Scholar]
- Carmo, F.F.; Jacobi, C.M. A vegetação de canga no Quadrilátero Ferrífero, Minas Gerais: Caracterização e contexto fitogeográfico. Rodriguésia 2013, 64, 527–541. [Google Scholar] [CrossRef] [Green Version]
- Gastauer, M.; Vera, M.P.O.; de Souza, K.P.; Pires, E.S.; Alves, R.; Caldeira, C.F.; Ramos, S.J.; Oliveira, G. A metagenomic survey of soil microbial communities along a rehabilitation chronosequence after iron ore mining. Sci. Data 2019, 6, 190008. [Google Scholar] [CrossRef] [PubMed]
- Levett, A.; Gagen, E.; Shuster, J.; Rintoul, L.; Tobin, M.; Vongsvivut, J.; Bambery, K.; Vasconcelos, P.; Southam, G. Evidence of biogeochemical processes in iron duricrust formation. J. S. Am. Earth Sci. 2016, 71, 131–142. [Google Scholar] [CrossRef] [Green Version]
- Parker, C.; Auler, A.; Senko, J.; Sasowsky, I.; Piló, L.; Smith, M.; Johnston, M.; Barton, H. Microbial Iron Cycling and Biospeleogenesis: Cave Development in the Carajás Formation, Brazil. In Proceedings of the Proceedings of the 16th International Congress of Speleology, Brno, Czech Republic, 21 July 2013. [Google Scholar]
- Figueira, R.L.; Coimbra Horbe, A.M.; Herrera Aragón, F.F.; Gonçalves, D.F. Exotic sulphate and phosphate speleothems in caves from eastern Amazonia (Carajás, Brazil): Crystallographic and chemical insights. J. S. Am. Earth Sci. 2019, 90, 412–422. [Google Scholar] [CrossRef]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The Role of Soil Microorganisms in Plant Mineral Nutrition-Current Knowledge and Future Directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef] [Green Version]
- Adetutu, E.M.; Thorpe, K.; Shahsavari, E.; Bourne, S.; Cao, X.S.; Fard, R.; Kirby, G.; Ball, A. Bacterial community survey of sediments at Naracoorte Caves, Australia. Int. J. Speleol. 2012, 41, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Barton, H.A.; Taylor, N.M.; Lubbers, B.R.; Pemberton, A.C. DNA extraction from low-biomass carbonate rock: An improved method with reduced contamination and the low-biomass contaminant database. J. Microbiol. Methods 2006, 66, 21–31. [Google Scholar] [CrossRef]
- Ortiz, M.; Neilson, J.W.; Nelson, W.M.; Legatzki, A.; Byrne, A.; Yu, Y.; Wing, R.A.; Soderlund, C.A.; Pryor, B.M.; Pierson, L.S., III; et al. Profiling bacterial diversity and taxonomic composition on speleothem surfaces in Kartchner Caverns, AZ. Microb. Ecol. 2013, 65, 371–383. [Google Scholar] [CrossRef]
- Gulecal-Pektas, Y. Bacterial Diversity and Composition in Oylat Cave (Turkey) with Combined Sanger/Pyrosequencing Approach. Pol. J. Microbiol. 2016, 65, 69–75. [Google Scholar] [CrossRef] [Green Version]
- D’Auria, G.; Artacho, A.; Rojas, R.A.; Bautista, J.S.; Mendez, R.; Gamboa, M.T.; Gamboa, J.R.; Gomez-Cruz, R. Metagenomics of Bacterial Diversity in Villa Luz Caves with Sulfur Water Springs. Genes 2018, 9, 55. [Google Scholar] [CrossRef] [Green Version]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schaberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Nichols, D.; Cahoon, N.; Trakhtenberg, E.M.; Pham, L.; Mehta, A.; Belanger, A.; Kanigan, T.; Lewis, K.; Epstein, S.S. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 2010, 76, 2445–2450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemoto, P.A.; Zhang, X.; Djuric, S.; Ma, Z. Novel 12-membered ring macrolides with activity against erythromycin-resistant organisms. J. Antibiot. 2003, 56, 392–398. [Google Scholar] [CrossRef] [Green Version]
- Vickers, A.A.; Chopra, I.; O’Neill, A.J. Intrinsic novobiocin resistance in Staphylococcus saprophyticus. Antimicrob. Agents Chemother. 2007, 51, 4484–4485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Zhang, J.; He, S.; Yan, X. A Review Study on Macrolides Isolated from Cyanobacteria. Mar. Drugs 2017, 15, 126. [Google Scholar] [CrossRef] [Green Version]
- Singh, G.; Jayanarayan, K.G.; Dey, C.S. Novobiocin induces apoptosis-like cell death in topoisomerase II over-expressing arsenite resistant Leishmania donovani. Mol. Biochem. Parasitol. 2005, 141, 57–69. [Google Scholar] [CrossRef]
- Romero, L.; Huerfano, C.; Grillo-Ardila, C.F. Macrolides for treatment of Haemophilus ducreyi infection in sexually active adults. Cochrane Database Syst. Rev. 2017, 12, CD012492. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.; Chalmers, J.D.; Crossingham, I.; Relph, N.; Felix, L.M.; Evans, D.J.; Milan, S.J.; Spencer, S. Macrolide antibiotics for bronchiectasis. Cochrane Database Syst. Rev. 2018, 3, CD012406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Hagan, D. The polyketide metabolites; E. Horwood: Sawston, UK, 1991; 176p. [Google Scholar]
- Lewin, G.R.; Carlos, C.; Chevrette, M.G.; Horn, H.A.; McDonald, B.R.; Stankey, R.J.; Fox, B.G.; Currie, C.R. Evolution and Ecology of Actinobacteria and Their Bioenergy Applications. Annu Rev. Microbiol. 2016, 70, 235–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palaniyandi, S.A.; Yang, S.H.; Zhang, L.; Suh, J.W. Effects of actinobacteria on plant disease suppression and growth promotion. Appl. Microbiol. Biotechnol. 2013, 97, 9621–9636. [Google Scholar] [CrossRef]
- Harshey, R.M.; Kawagishi, I.; Maddock, J.; Kenney, L.J. Function, Diversity, and Evolution of Signal Transduction in Prokaryotes. Dev. Cell 2003, 4, 459–465. [Google Scholar] [CrossRef] [Green Version]
- Nazir, R.; Mazurier, S.; Yang, P.; Lemanceau, P.; van Elsas, J.D. The Ecological Role of Type Three Secretion Systems in the Interaction of Bacteria with Fungi in Soil and Related Habitats Is Diverse and Context-Dependent. Front. Microbiol. 2017, 8, 38. [Google Scholar] [CrossRef] [Green Version]
- Desai, M.S.; Assig, K.; Dattagupta, S. Nitrogen fixation in distinct microbial niches within a chemoautotrophy-driven cave ecosystem. ISME J. 2013, 7, 2411. [Google Scholar] [CrossRef] [Green Version]
- Carmo, F.F.; Kamino, L.H.Y. Chapada de Canga: Uma introdução In Chapada de Canga: Patrimônio Natural e Cultural de Relevante Interesse para Conservação; Carmo, F.F., Kamino, L.H.Y., Eds.; 3i Editora: Belo Horizonte, Brazil, 2017; pp. 11–24. [Google Scholar]
- Lopes, L.M.N. O rompimento da barragem de Mariana e seus impactos socioambientais. Sinapse Múlt. 2016, 5, 1–14. [Google Scholar]
- Freitas, C.M.; Silva, M.A.; Menezes, F.C. O desastre na barragem de mineração da samarco—Fratura exposta dos limites do Brasil na redução de risco de desasastres. Ciência Cult. 2016, 68, 25–30. [Google Scholar] [CrossRef] [Green Version]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.P.; Clement, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. MMBR 2016, 80, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Qin, S.; Chen, H.H.; Zhao, G.Z.; Li, J.; Zhu, W.Y.; Xu, L.H.; Jiang, J.H.; Li, W.J. Abundant and diverse endophytic actinobacteria associated with medicinal plant Maytenus austroyunnanensis in Xishuangbanna tropical rainforest revealed by culture-dependent and culture-independent methods. Environ. Microbiol. Rep. 2012, 4, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Li, W.J.; Dastager, S.G.; Hozzein, W.N. Actinobacteria in Special and Extreme Habitats: Diversity, Function Roles, and Environmental Adaptations. Front. Microbiol. 2016, 7, 1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamedi, J.; Mohammadipanah, F.; Ventosa, A. Systematic and biotechnological aspects of halophilic and halotolerant actinomycetes. Extrem. Life Under Extrem. Cond. 2013, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef] [PubMed]
- Piló, L.B.; Coelho, A.; Reino, J.C.R. Geoespeleologia em rochas ferríferas: Cenário atual e conservação. In Geossistemas Ferruginosos do Brasil: Áreas Prioritárias para Conservação da Diversidade Geológica e Biológica, Patrimônio Cultural e Serviços Ambientais; Carmo, F.F.d., Kamino, L.H.Y., Eds.; 3i Editora: Belo Horizonte, Brazil, 2015; pp. 125–148. [Google Scholar]
- Sallstedt, T.; Ivarsson, M.; Lundberg, J.; Sjöberg, R.; Romaní, J.R.V. Speleothem and biofilm formation in a granite/dolerite cave, Northern Sweden. Int. J. Speleol. 2014, 43, 305–313. [Google Scholar] [CrossRef] [Green Version]
- Turrini, P.; Tescari, M.; Visaggio, D.; Pirolo, M.; Lugli, G.A.; Ventura, M.; Frangipani, E.; Visca, P. The microbial community of a biofilm lining the wall of a pristine cave in Western New Guinea. Microbiol. Res. 2020, 241, 126584. [Google Scholar] [CrossRef] [PubMed]
- Caccavo, F., Jr.; Lonergan, D.J.; Lovley, D.R.; Davis, M.; Stolz, J.F.; McInerney, M.J. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 1994, 60, 3752–3759. [Google Scholar] [CrossRef] [Green Version]
- Emerson, D.; Moyer, C. Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl. Environ. Microbiol. 1997, 63, 4784–4792. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.B.; Hallberg, K.B.; Hedrich, S. Uncovering a microbial enigma: Isolation and characterization of the streamer-generating, iron-oxidizing, acidophilic bacterium “Ferrovum myxofaciens”. Appl. Environ. Microbiol. 2014, 80, 672–680. [Google Scholar] [CrossRef] [Green Version]
- Kimura, S.; Bryan, C.; B Hallberg, K.; Johnson, D. Biodiversity and Geochemistry of an extremely acidic, low-temperature subterranean environment sustained by chemolithotrophy. Environ. Microbiol. 2011, 13, 2092–2104. [Google Scholar] [CrossRef]
- Northup, D.E.; Barns, S.M.; Yu, L.E.; Spilde, M.N.; Schelble, R.T.; Dano, K.E.; Crossey, L.J.; Connolly, C.A.; Boston, P.J.; Natvig, D.O.; et al. Diverse microbial communities inhabiting ferromanganese deposits in Lechuguilla and Spider Caves. Environ. Microbiol. 2003, 5, 1071–1086. [Google Scholar] [CrossRef]
- Pacton, M.; Breitenbach, S.; Lechleitner, F.; Vaks, A.; Rollion-Bard, C.; Gutareva, O.S.; Osintcev, A.V.; Vasconcelos, C. The role of microorganisms in the formation of a stalactite in Botovskaya Cave, Siberia—Paleoenvironmental implications. Biogeosciences 2013, 10, 6115–6130. [Google Scholar] [CrossRef] [Green Version]
- Malvankar, N.S.; Vargas, M.; Nevin, K.P.; Franks, A.E.; Leang, C.; Kim, B.-C.; Inoue, K.; Mester, T.; Covalla, S.F.; Johnson, J.P.; et al. Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 2011, 6, 573. [Google Scholar] [CrossRef]
- Prakash, D.; Gabani, P.; Chandel, A.K.; Ronen, Z.; Singh, O.V. Bioremediation: A genuine technology to remediate radionuclides from the environment. Microb. Biotechnol. 2013, 6, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Bosch, J.; Meckenstock, R.U. Use of metal-reducing bacteria for bioremediation of soil contaminated with mixed organic and inorganic pollutants. Environ. Geochem. Health 2012, 34, 135–142. [Google Scholar] [CrossRef]
- Newsome, L.; Morris, K.; Lloyd, J.R. The biogeochemistry and bioremediation of uranium and other priority radionuclides. Chem. Geol. 2014, 363, 164–184. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Khmelenina, V.N.; Suzina, N.E.; Trotsenko, Y.A.; Semrau, J.D.; Liesack, W.; Tiedje, J.M. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 2002, 52, 251–261. [Google Scholar] [CrossRef] [Green Version]
- Lind, H.; Jonsson, H.; Schnürer, J. Antifungal effect of dairy propionibacteria—Contribution of organic acids. Int. J. Food Microbiol. 2005, 98, 157–165. [Google Scholar] [CrossRef]
- Le Lay, C.; Coton, E.; Le Blay, G.; Chobert, J.M.; Haertle, T.; Choiset, Y.; van Long, N.N.; Meslet-Cladiere, L.; Mounier, J. Identification and quantification of antifungal compounds produced by lactic acid bacteria and propionibacteria. Int. J. Food Microbiol. 2016, 239, 79–85. [Google Scholar] [CrossRef]
- Williams, S.T.; Khan, M.R. Antibiotics—A soil microbiologist’s viewpoint. Postepy Hig. Med. Dosw. 1974, 28, 395–408. [Google Scholar]
- Sen, R.; Ishak, H.D.; Estrada, D.; Dowd, S.E.; Hong, E.; Mueller, U.G. Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc. Natl. Acad. Sci. USA 2009, 106, 17805–17810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atashgahi, S.; Liebensteiner, M.G.; Janssen, D.B.; Smidt, H.; Stams, A.J.M.; Sipkema, D. Microbial Synthesis and Transformation of Inorganic and Organic Chlorine Compounds. Front. Microbiol. 2018, 9, 3079. [Google Scholar] [CrossRef] [PubMed]
- Schimel, J.; Schaeffer, S. Microbial control over carbon cycling in soil. Front. Microbiol. 2012, 3, 348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takai, K. The Nitrogen Cycle: A Large, Fast, and Mystifying Cycle. Microbes Environ. 2019, 34, 223–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Li, Y.; Zhang, C.; Liu, H.; Liu, J.; Zheng, W.; Kang, X.; Leng, X.; Zhao, K.; Gu, Y.; et al. Culturable Heavy Metal-Resistant and Plant Growth Promoting Bacteria in V-Ti Magnetite Mine Tailing Soil from Panzhihua, China. PLoS ONE 2014, 9, e106618. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, E.; Lu, Y.; Ouyang, J.; Wang, L.; Wang, X. Bacterial community analysis of anoxic/aeration (A/O) system in a combined process for gibberellin wastewater treatment. PLoS ONE 2017, 12, e0186743. [Google Scholar] [CrossRef] [Green Version]
- Daranas, N.; Rosello, G.; Cabrefiga, J.; Donati, I.; Frances, J.; Badosa, E.; Spinelli, F.; Montesinos, E.; Bonaterra, A. Biological control of bacterial plant diseases with Lactobacillus plantarum strains selected for their broad-spectrum activity. Ann. Appl. Biol. 2019, 174, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Koppel, N.; Maini Rekdal, V.; Balskus, E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, eaag2770. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Baldwin, A.; Dowson, C.G. Burkholderia cepacia complex bacteria: Opportunistic pathogens with important natural biology. J. Appl. Microbiol. 2008, 104, 1539–1551. [Google Scholar] [CrossRef] [PubMed]
- Prakash, O.; Green, S.J.; Jasrotia, P.; Overholt, W.A.; Canion, A.; Watson, D.B.; Brooks, S.C.; Kostka, J.E. Rhodanobacter denitrificans sp. nov., isolated from nitrate-rich zones of a contaminated aquifer. Int. J. Syst. Evol. Microbiol. 2012, 62, 2457–2462. [Google Scholar] [CrossRef] [Green Version]
- Sousa, T.; Chung, A.P.; Pereira, A.; Piedade, A.P.; Morais, P.V. Aerobic uranium immobilization by Rhodanobacter A2-61 through formation of intracellular uranium-phosphate complexes. Met. Integr. Biometal Sci. 2013, 5, 390–397. [Google Scholar] [CrossRef]
- Chung, A.; Sousa, T.; Pereira, A.; Morais, P. Microorganisms—Tools for Bioremediation of Uranium Contaminated Environments. Procedia Earth Planet. Sci. 2014, 8, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Satola, B.; Wubbeler, J.H.; Steinbuchel, A. Metabolic characteristics of the species Variovorax paradoxus. Appl. Microbiol. Biotechnol. 2013, 97, 541–560. [Google Scholar] [CrossRef]
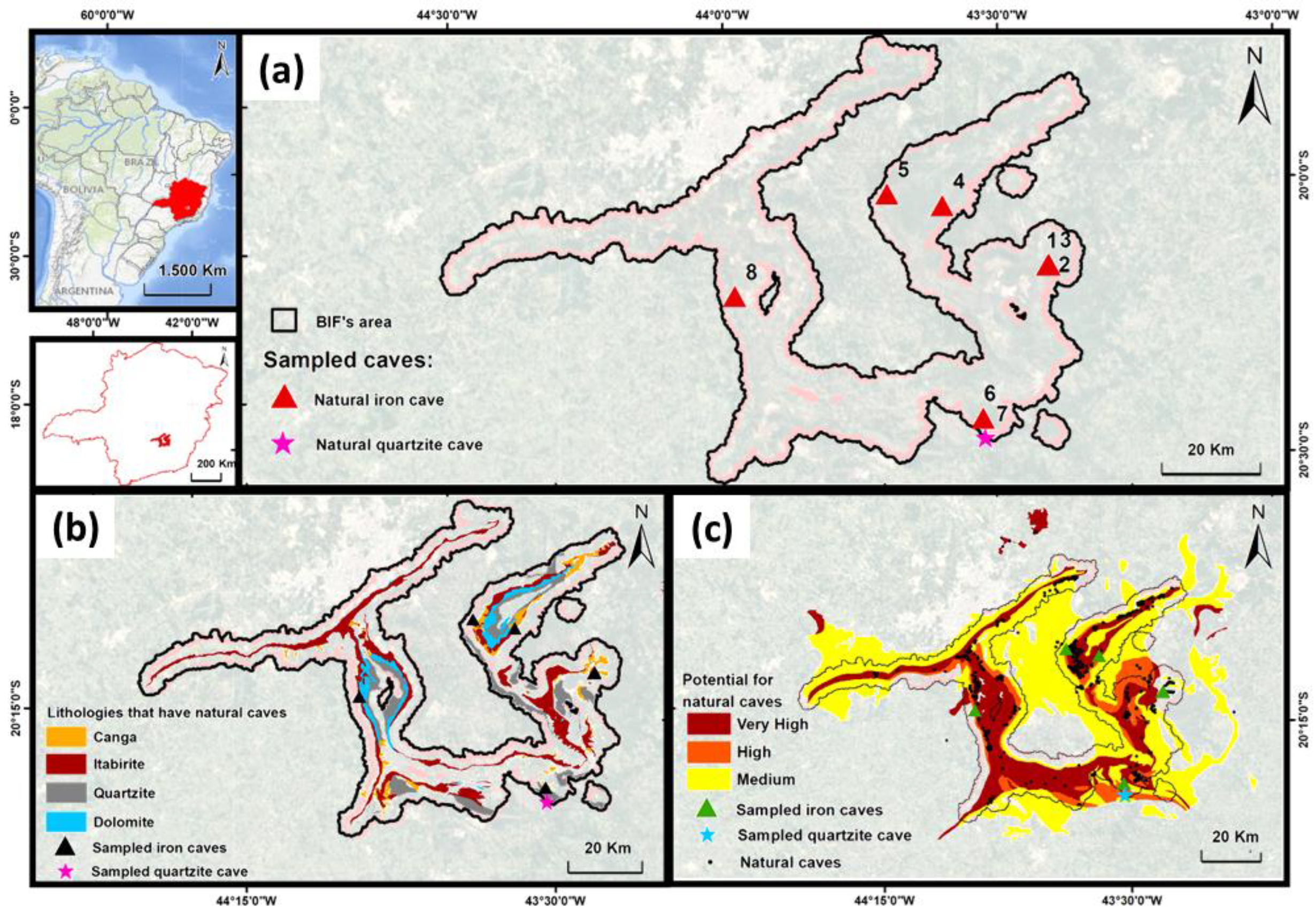

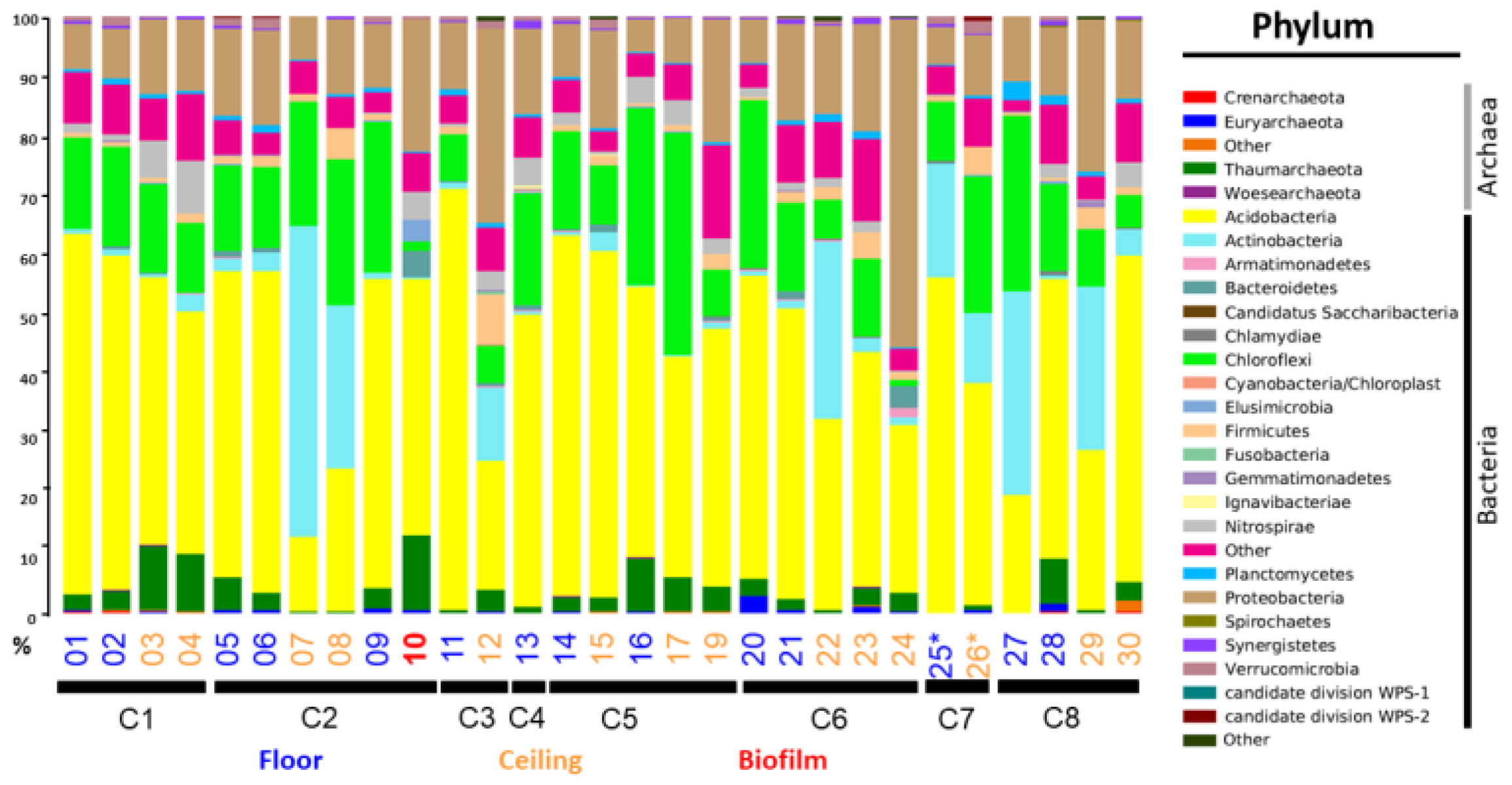
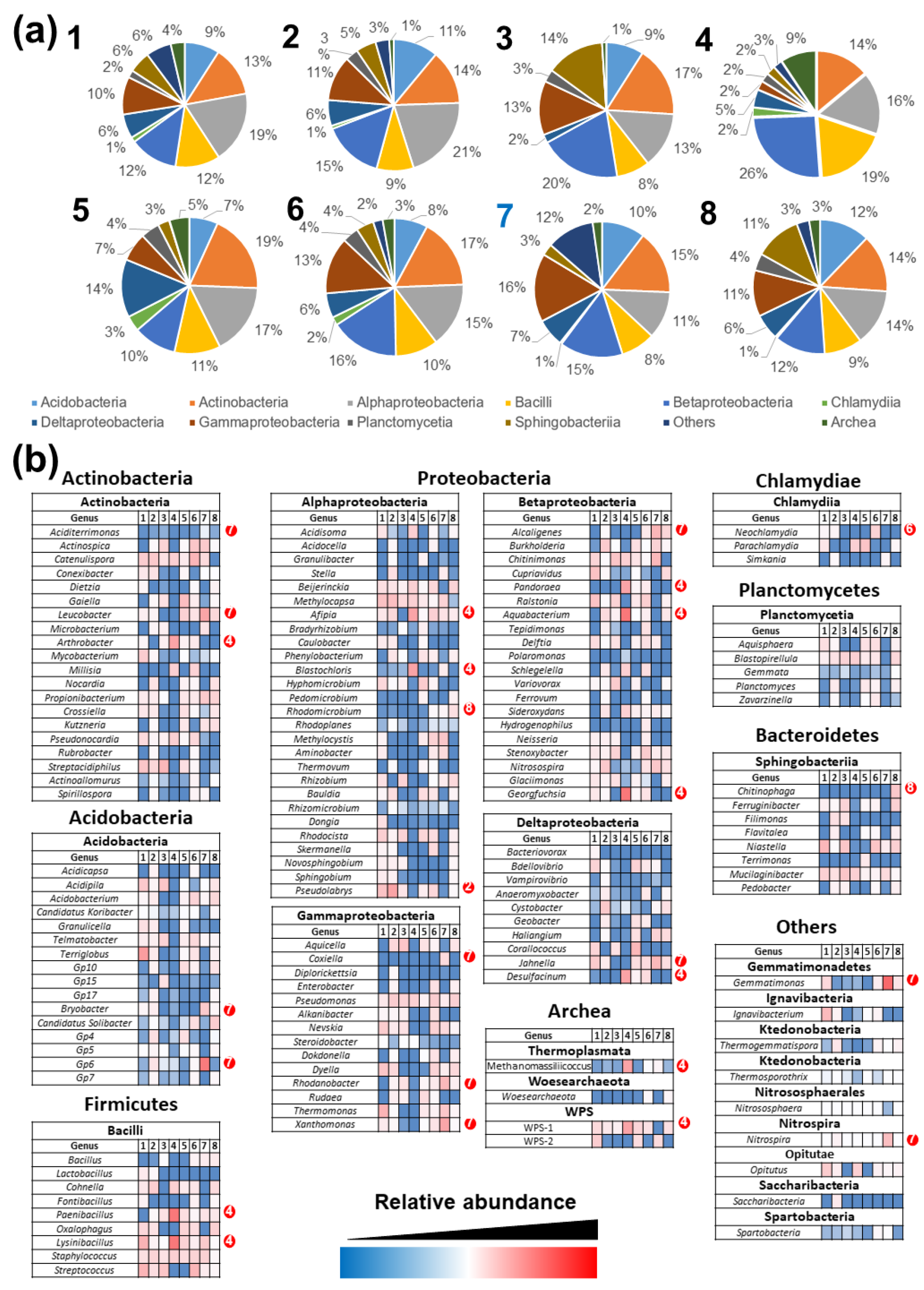

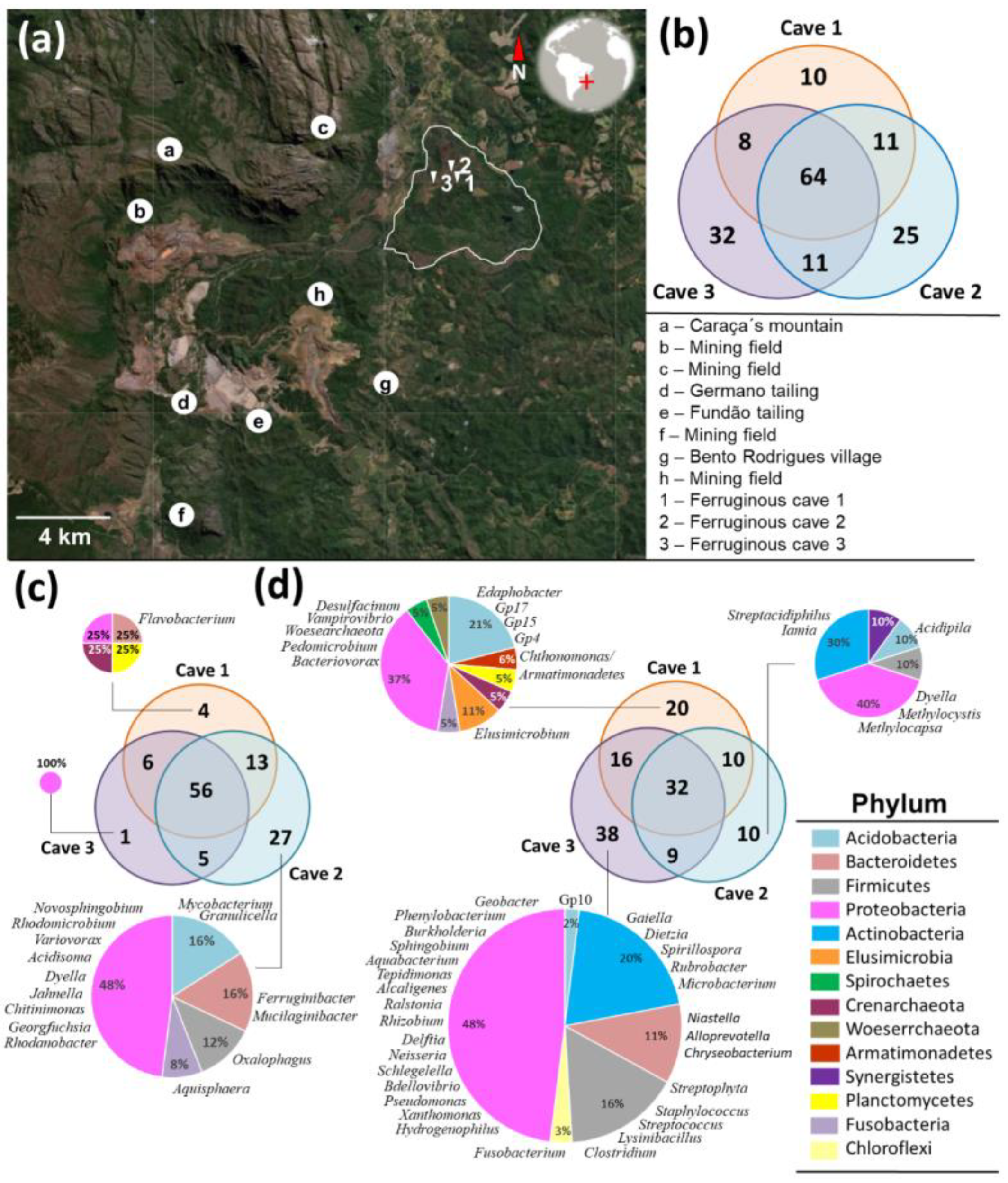
| Cave Number | Localization | Geomorphology | Other Features | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| County | Locality | Biome | Latitude (S) | Longitude (W) | Altitude (m) | Lithology | Ceiling | Floor | Human Activity | Proximity to Mine (up to 500 m) | Protected Area | |
| 1 | Mariana | Chapada | Atlantic Forest | 20°9′53.78″ | 43°24′19.49″ | 879 | Ferruginous | Canga | Canga | No | ~250 m | No |
| 2 | Mariana | Chapada | Atlantic Forest | 20°9′54.03″ | 43°24′29.72″ | 865 | Ferruginous | Canga | Canga | No | ~350 m | No |
| 3 | Mariana | Chapada | Atlantic Forest | 20°9′49.15″ | 43°24′21.85″ | 880 | Ferruginous | Canga | Canga | No | ~350 m | No |
| 4 | Caeté | Gandarela’s Mountain | Atlantic Forest | 20°3′19.58″ | 43°41′42.41″ | 1624 | Ferruginous | Canga | Itabirite | No | No | Yes |
| 5 | Santa Bárbara | Gandarela’s Mountain | Atlantic Forest | 20°3′23.40″ | 43°35′59.94″ | 1236 | Ferruginous | Canga | Itabirite | No | No | Yes |
| 6 | Ouro Preto | Lavras Novas | Atlantic Forest | 20°26′35.76″ | 43°31′31.18″ | 1480 | Ferruginous | Canga | Itabirite | No | No | No |
| 7 | Ouro Preto | Lavras Novas | Atlantic Forest | 20°28′42.20″ | 43°31′15.72″ | 1346 | Quartizite | Quartizite | Quartizite | No | No | No |
| 8 | Nova Lima | Moeda’s Mountain | Atlantic Forest | 20°13′18.26″ | 43°58′38.48″ | 1488 | Ferruginous | Canga | Itabirite | Yes | No | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lemes, C.G.C.; Villa, M.M.; Felestrino, É.B.; Perucci, L.O.; Assis, R.A.B.; Cordeiro, I.F.; Fonseca, N.P.; Guerra, L.C.C.; Caneschi, W.L.; Moraes, L.Â.G.; et al. 16S rRNA Gene Amplicon Sequencing Data of the Iron Quadrangle Ferruginous Caves (Brazil) Shows the Importance of Conserving This Singular and Threatened Geosystem. Diversity 2021, 13, 494. https://doi.org/10.3390/d13100494
Lemes CGC, Villa MM, Felestrino ÉB, Perucci LO, Assis RAB, Cordeiro IF, Fonseca NP, Guerra LCC, Caneschi WL, Moraes LÂG, et al. 16S rRNA Gene Amplicon Sequencing Data of the Iron Quadrangle Ferruginous Caves (Brazil) Shows the Importance of Conserving This Singular and Threatened Geosystem. Diversity. 2021; 13(10):494. https://doi.org/10.3390/d13100494
Chicago/Turabian StyleLemes, Camila G. C., Morghana M. Villa, Érica B. Felestrino, Luiza O. Perucci, Renata A. B. Assis, Isabella F. Cordeiro, Natasha P. Fonseca, Lara C. C. Guerra, Washington L. Caneschi, Lauro Â. G. Moraes, and et al. 2021. "16S rRNA Gene Amplicon Sequencing Data of the Iron Quadrangle Ferruginous Caves (Brazil) Shows the Importance of Conserving This Singular and Threatened Geosystem" Diversity 13, no. 10: 494. https://doi.org/10.3390/d13100494
APA StyleLemes, C. G. C., Villa, M. M., Felestrino, É. B., Perucci, L. O., Assis, R. A. B., Cordeiro, I. F., Fonseca, N. P., Guerra, L. C. C., Caneschi, W. L., Moraes, L. Â. G., do Carmo, F. F., Kamino, L. H. Y., Vale, P. N. C., Guima, S. E. S., Setubal, J. C., Salgado, A. A. R., & Moreira, L. M. (2021). 16S rRNA Gene Amplicon Sequencing Data of the Iron Quadrangle Ferruginous Caves (Brazil) Shows the Importance of Conserving This Singular and Threatened Geosystem. Diversity, 13(10), 494. https://doi.org/10.3390/d13100494






