Genetic Structure and Gene Flow in Eastern Grey Kangaroos in an Isolated Conservation Reserve
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Species
2.2. Study Area
2.3. Sample Collection
2.4. DNA Extraction and Genotyping
2.5. Data Analysis
3. Results
3.1. Genetic Diversity
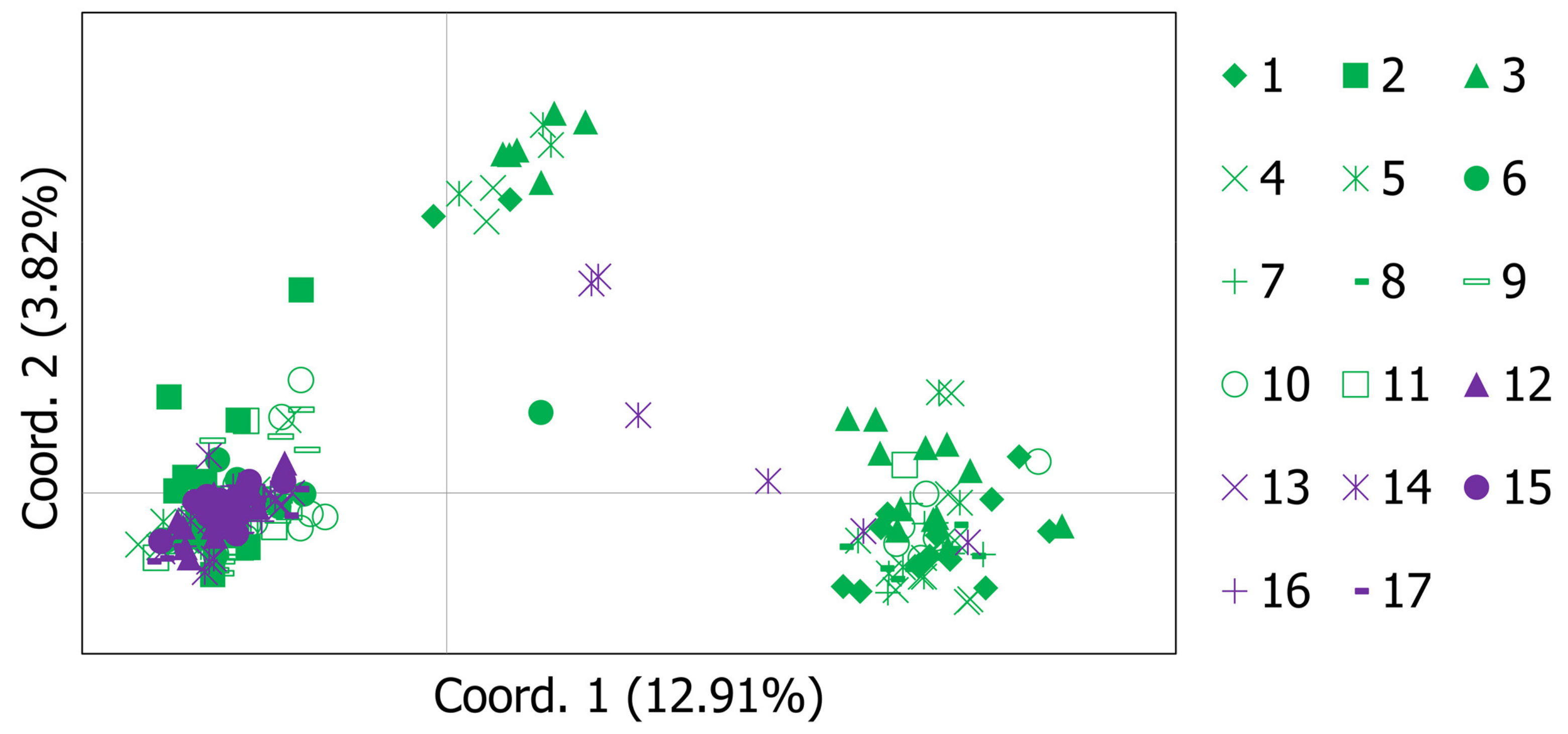
3.2. Genetic Structure and Gene Flow
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fahrig, L. Relative effects of habitat loss and fragmentation on population extinction. J. Wildl. Manag. 1997, 61, 603–610. [Google Scholar] [CrossRef]
- Crooks, K.R.; Burdett, C.L.; Theobald, D.M.; King, S.R.B.; Di Marco, M.; Rondinini, C.; Boitani, L. Quantification of habitat fragmentation reveals extinction risk in terrestrial mammals. Proc. Natl. Acad. Sci. USA 2017, 114, 7635–7640. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Ralls, K.; Eldridge, M.; Dudash, M.R.; Fenster, C.B.; Lacy, R.C.; Sunnucks, P. Genetic Management of Fragmented Animal and Plant Populations; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Shepard, D.B.; Kuhns, A.R.; Dreslik, M.J.; Phillips, C.A. Roads as barriers to animal movement in fragmented landscapes. Anim. Conserv. 2008, 11, 288–296. [Google Scholar] [CrossRef]
- Cayuela, H.; Rougemont, Q.; Prunier, J.G.; Moore, J.S.; Clobert, J.; Besnard, A.; Bernatchez, L. Demographic and genetic approaches to study dispersal in wild animal populations: A methodological review. Mol. Ecol. 2018, 27, 3976–4010. [Google Scholar] [CrossRef]
- Westley, P.A.H.; Berdahl, A.M.; Torney, C.J.; Biro, D. Collective movement in ecology: From emerging technologies to conservation and management. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170004. [Google Scholar] [CrossRef] [PubMed]
- Zemanova, M.A. Towards more compassionate wildlife research through the 3Rs principles: Moving from invasive to non-invasive methods. Wildl. Biol. 2020, 1, wlb.00607. [Google Scholar] [CrossRef]
- Flagstad, O.; Pradhan, N.M.B.; Kvernstuen, L.G.; Wegge, P. Conserving small and fragmented populations of large mammals: Non-invasive genetic sampling in an isolated population of Asian elephants in Nepal. J. Nat. Conserv. 2012, 20, 181–190. [Google Scholar] [CrossRef]
- Davidson, A.; Carmel, Y.; Bar-David, S. Characterizing wild ass pathways using a non-invasive approach: Applying least-cost path modelling to guide field surveys and a model selection analysis. Landsc. Ecol. 2013, 28, 1465–1478. [Google Scholar] [CrossRef]
- Manel, S.; Holderegger, R. Ten years of landscape genetics. Trends Ecol. Evol. 2013, 28, 614–621. [Google Scholar] [CrossRef]
- Schwartz, M.K.; Luikart, G.; Waples, R.S. Genetic monitoring as a promising tool for conservation and management. Trends Ecol. Evol. 2007, 22, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Zemanova, M.A.; Knop, E.; Heckel, G. Introgressive replacement of natives by invading Arion pest slugs. Sci. Rep. 2017, 7, 14908. [Google Scholar] [CrossRef] [PubMed]
- Allendorf, F.W.; Luikart, G. Conservation and the Genetics of Populations; John Wiley & Sons: Oxford, UK, 2009. [Google Scholar]
- Calaby, J.; Grigg, G. Changes in macropoid communities and populations in the past 200 years, and the future. In Kangaroos, Wallabies, and Rat-Kangaroos; Grigg, G., Jarman, P., Hume, I., Eds.; Surrey Beatty: Sydney, Australia, 1989. [Google Scholar]
- Hrdina, F. Marsupial destruction in Queensland 1877-1930. Aust. Zool. 2014, 30, 272–286. [Google Scholar] [CrossRef]
- Ben-Ami, D.; Boom, K.; Boronyak, L.; Townend, C.; Ramp, D.; Croft, D.B.; Bekoff, M. The welfare ethics of the commercial killing of free-ranging kangaroos: An evaluation of the benefits and costs of the industry. Anim. Welfare 2014, 23, 1–10. [Google Scholar] [CrossRef][Green Version]
- Ramp, D. Bringing compassion to the ethical dilemma in killing kangaroos for conservation. J. Bioeth. Inq. 2013, 10, 267–272. [Google Scholar] [CrossRef]
- Boom, K.; Ben-Ami, D.; Croft, D.B.; Cushing, N.; Ramp, D.; Boronyak, L. ’Pest’ and resource: A legal history of Australia’s kangaroos. Anim. Stud. J. 2012, 1, 17–40. [Google Scholar]
- Descovich, K.A.; McDonald, I.J.; Tribe, A.; Phillips, C.J.C. A welfare assessment of methods used for harvesting, hunting and population control of kangaroos and wallabies. Anim. Welfare 2015, 24, 255–265. [Google Scholar] [CrossRef]
- Australian Government. Kangaroo and Wallaby Statistics Archive. Available online: https://www.environment.gov.au/biodiversity/wildlife-trade/natives/kangaroo-wallaby-statistics (accessed on 7 October 2021).
- McKnight, T.L. Barrier fencing for vermin control in Australia. Geogr. Rev. 1969, 59, 330–347. [Google Scholar] [CrossRef]
- Department of Primary Industries and Fisheries. History of Barrier Fences in Queensland; Land Protection: Brisbane, Australia, 2007. [Google Scholar]
- QGIS.org. QGIS Geographic Information System. QGIS Association. 2021. Available online: https://www.qgis.org (accessed on 3 March 2021).
- Kaufmann, J.H. Field observations of the social behaviour of the eastern grey kangaroo, Macropus giganteus. Anim. Behav. 1975, 23, 214–221. [Google Scholar] [CrossRef]
- Miller, E.J.; Eldridge, M.D.B.; Cooper, D.W.; Herbert, C.A. Dominance, body size and internal relatedness influence male reproductive success in eastern grey kangaroos (Macropus giganteus). Reprod. Fertil. Dev. 2010, 22, 539–549. [Google Scholar] [CrossRef]
- Neaves, L.E.; Zenger, K.R.; Cooper, D.W.; Eldridge, M.D.B. Molecular detection of hybridization between sympatric kangaroo species in south-eastern Australia. Heredity 2010, 104, 502–512. [Google Scholar] [CrossRef] [PubMed]
- Dawson, T.J. Kangaroos, 2nd ed.; CSIRO Publishing: Melbourne, 2012. [Google Scholar]
- Neaves, L.E.; Roberts, M.W.; Herbert, C.A.; Eldridge, M.D.B. Limited sex bias in the fine-scale spatial genetic structure of the eastern grey kangaroo and its relationship to habitat. Aust. J. Zool. 2017, 65, 33–44. [Google Scholar] [CrossRef]
- Best, E.C.; Seddon, J.M.; Dwyer, R.G.; Goldizen, A.W. Social preference influences female community structure in a population of wild eastern grey kangaroos. Anim. Behav. 2013, 86, 1031–1040. [Google Scholar] [CrossRef]
- Australia Zoo. Mourachan. Available online: https://www.australiazoo.com.au/experiences-australia-zoo/australia-zoo-expeditions/mourachan/ (accessed on 3 August 2021).
- Paulo, A.A.; Rosa, R.D.; Pereira, L.S. Climate trends and behaviour of drought indices based on precipitation and evapotranspiration in Portugal. Nat. Hazards Earth Syst. Sci. 2012, 12, 1481–1491. [Google Scholar] [CrossRef]
- Begueria, S.; Latorre, B.; Reig, F.; Vicente-Serrano, S.M. Global Drought Monitor. Available online: https://spei.csic.es/map/maps.html#months=1#month=8#year=2021 (accessed on 20 October 2021).
- Zemanova, M.A. Poor implementation of non-invasive sampling in wildlife genetics studies. Rethink. Ecol. 2019, 4, 119–132. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (Version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Gilbert, K.J.; Andrew, R.L.; Bock, D.G.; Franklin, M.T.; Kane, N.C.; Moore, J.S.; Moyers, B.T.; Renaut, S.; Rennison, D.J.; Veen, T. Recommendations for utilizing and reporting population genetic analyses: The reproducibility of genetic clustering using the program STRUCTURE. Mol. Ecol. 2012, 21, 4925–4930. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J.M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S.; Do, C. LDNE: A program for estimating effective population size from data on linkage disequilibrium. Mol. Ecol. Res. 2008, 8, 753–756. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Res. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Sigg, D.P.; Goldizen, A.W.; Pople, A.R. The importance of mating system in translocation programs: Reproductive success of released male bridled nailtail wallabies. Biol. Conserv. 2005, 123, 289–300. [Google Scholar] [CrossRef]
- Zenger, K.R.; Cooper, D.W. A set of highly polymorphic microsatellite markers developed for the eastern grey kangaroo (Macropus giganteus). Mol. Ecol. Notes 2001, 1, 98–100. [Google Scholar] [CrossRef]
- Taylor, A.C.; Cooper, D.W. A set of tammar wallaby (Macropus eugenii) microsatellites tested for genetic linkage. Mol. Ecol. 1998, 7, 925–926. [Google Scholar]
- Zenger, K.R.; Cooper, D.W. Characterization of 14 macropod microsatellite genetic markers. Anim. Genet. 2001, 32, 166–167. [Google Scholar] [CrossRef]
- Green-Barber, J.M.; Old, J.M. The genetic relatedness of a peri-urban population of eastern grey kangaroos. BMC Res. Notes 2018, 11, 856. [Google Scholar] [CrossRef]
- Luikart, G.; Cornuet, J.M. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv. Biol. 1998, 12, 228–237. [Google Scholar] [CrossRef]
- Girod, C.; Vitalis, R.; Leblois, R.; Fréville, H. Inferring population decline and expansion from microsatellite data: A simulation-based evaluation of the Msvar method. Genetics 2011, 188, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.; Sharma, S.; Maldonado, J.E.; Wood, T.C.; Panwar, H.S.; Seidensticker, J. Gene flow and demographic history of leopards (Panthera pardus) in the central Indian highlands. Evolut. Appl. 2013, 6, 949–959. [Google Scholar] [CrossRef]
- Hill, G.J.E. Seasonal movement patterns of the eastern grey kangaroo in southern Queensland. Wildl. Res. 1982, 9, 373–387. [Google Scholar] [CrossRef]
- Bradby, K.; Fitzsimons, J.A.; Del Marco, A.; Driscoll, D.A.; Ritchie, E.G.; Lau, J.; Bradshaw, C.J.A.; Hobbs, R.J. Ecological connectivity or barrier fence? Critical choices on the agricultural margins of Western Australia. Ecol. Manage. Restor. 2014, 15, 180–190. [Google Scholar] [CrossRef]
- Wang, J.; Santiago, E.; Caballero, A. Prediction and estimation of effective population size. Heredity 2016, 117, 193–206. [Google Scholar] [CrossRef]
- Spencer, P.B.S.; Bain, K.; Hayward, M.W.; Hillyer, M.; Friend, J.A.T. Persistence of remnant patches and genetic loss at the distribution periphery in island and mainland populations of the quokka. Aust. J. Zool. 2019, 67, 38–50. [Google Scholar] [CrossRef]
- Vucetich, J.A.; Waite, T.A.; Nunney, L. Fluctuating population size and the ratio of effective to census population size. Evolution 1997, 51, 2017–2021. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Inbreeding and variance effective numbers in populations with overlapping generations. Genetics 1971, 68, 581. [Google Scholar] [CrossRef] [PubMed]
- Crow, J.F.; Denniston, C. Inbreeding and variance effective population numbers. Evolution 1988, 42, 482–495. [Google Scholar] [CrossRef]
- Zenger, K.R.; Eldridge, M.D.B.; Cooper, D.W. Intraspecific variation, sex-biased dispersal and phylogeography of the eastern grey kangaroo (Macropus giganteus). Heredity 2003, 91, 153–162. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zecherle, L.J.; Bar-David, S.; Nichols, H.J.; Templeton, A.R.; Hipperson, H.; Horsburgh, G.J.; Brown, R.P. Landscape resistance affects individual habitat selection but not genetic relatedness in a reintroduced desert ungulate. Biol. Conserv. 2020, 252, 108845. [Google Scholar] [CrossRef]
- Hagenlund, M.; Linløkken, A.; Østbye, K.; Walton, Z.; Odden, M.; Samelius, G.; Willebrand, T.; Wilson, R. Genetic structure and gene flow in red foxes (Vulpes vulpes) in Scandinavia: Implications for the potential future spread of Echinococcus multilocularis tapeworm. Appl. Sci. 2019, 9, 5289. [Google Scholar] [CrossRef]
- Coghlan, B.A.; Seddon, J.M.; Best, E.C.; Thomson, V.A.; Goldizen, A.W. Evidence of male-biased dispersal in eastern grey kangaroos (Macropus giganteus). Aust. J. Zool. 2016, 64, 360–369. [Google Scholar] [CrossRef]
- Smith, D.; Waddell, K.; Allen, B.L. Expansion of vertebrate pest exclusion fencing and its potential benefits for threatened fauna recovery in Australia. Animals 2020, 10, 1550. [Google Scholar] [CrossRef] [PubMed]
- Epps, C.W.; Palsbøll, P.J.; Wehausen, J.D.; Roderick, G.K.; Ramey, R.R.; McCullough, D.R. Highways block gene flow and cause a rapid decline in genetic diversity of desert bighorn sheep. Ecol. Lett. 2005, 8, 1029–1038. [Google Scholar] [CrossRef]
- Smith, D.; King, R.; Allen, B.L. Impacts of exclusion fencing on target and non-target fauna: A global review. Biol. Rev. 2020, 95, 1590–1606. [Google Scholar] [CrossRef]
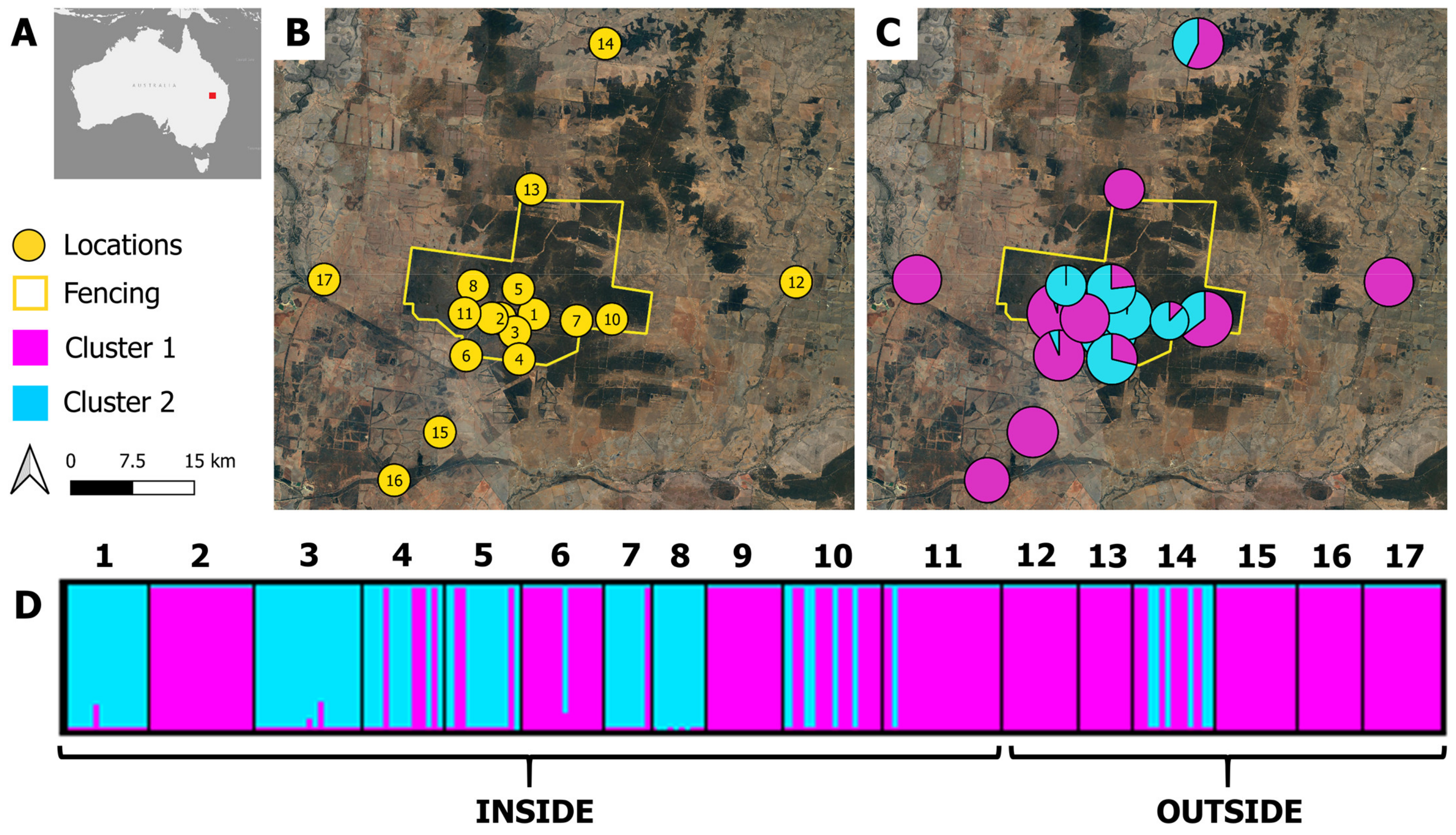
| ID | Location Name | Latitude | Longitude | Position | N | AR | HO | HE | FIS | Q1 | Q2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1000 Acre Dam | −27.779 | 149.067 | Inside | 14 | 4.858 | 0.631 | 0.746 | 0.191 | 0.012 | 0.988 |
| 2 | Black Swan Dam | −27.784 | 149.021 | Inside | 18 | 4.708 | 0.770 | 0.777 | 0.039 | 1.000 | 0.000 |
| 3 | Bob’s Billy Dam | −27.801 | 149.041 | Inside | 18 | 5.136 | 0.635 | 0.796 | 0.240 | 0.016 | 0.984 |
| 4 | Bonza Dam | −27.834 | 149.047 | Inside | 14 | 5.241 | 0.710 | 0.811 | 0.165 | 0.288 | 0.712 |
| 5 | Dead Cow Dam | −27.748 | 149.046 | Inside | 13 | 5.732 | 0.725 | 0.843 | 0.182 | 0.232 | 0.768 |
| 6 | End Dam | −27.829 | 148.974 | Inside | 14 | 5.413 | 0.820 | 0.830 | 0.050 | 0.936 | 0.065 |
| 7 | Far East Dam | −27.787 | 149.125 | Inside | 8 | 4.885 | 0.673 | 0.731 | 0.147 | 0.125 | 0.875 |
| 8 | Nr 11 Dam | −27.744 | 148.983 | Inside | 9 | 4.677 | 0.741 | 0.703 | 0.007 | 0.000 | 1.000 |
| 9 | Shed | −27.784 | 149.009 | Inside | 13 | 4.441 | 0.811 | 0.754 | −0.034 | 1.000 | 0.000 |
| 10 | Sucker Pig Dam | −27.785 | 149.174 | Inside | 17 | 5.609 | 0.749 | 0.853 | 0.154 | 0.647 | 0.353 |
| 11 | Thomby Dam | −27.778 | 148.972 | Inside | 20 | 5.294 | 0.800 | 0.830 | 0.064 | 0.950 | 0.050 |
| 12 | East Road | −27.740 | 149.427 | Outside | 13 | 4.773 | 0.807 | 0.782 | 0.012 | 1.000 | 0.000 |
| 13 | North Road | −27.627 | 149.063 | Outside | 9 | 4.863 | 0.776 | 0.777 | 0.060 | 1.000 | 0.000 |
| 14 | North Road II | −27.449 | 149.165 | Outside | 14 | 5.430 | 0.687 | 0.838 | 0.219 | 0.573 | 0.427 |
| 15 | South Road | −27.922 | 148.938 | Outside | 14 | 5.098 | 0.811 | 0.805 | 0.030 | 1.000 | 0.000 |
| 16 | Warrie Road Dam | −27.981 | 148.875 | Outside | 11 | 5.115 | 0.812 | 0.803 | 0.038 | 1.000 | 0.000 |
| 17 | West Road | −27.737 | 148.779 | Outside | 13 | 5.091 | 0.792 | 0.810 | 0.062 | 1.000 | 0.000 |
| Locus | Reference | Developed in Species | Ngenotyped | NA | HO | HE | HWE | Fnull |
|---|---|---|---|---|---|---|---|---|
| B123 | [45] | Onychogalea fraenata | 232 | 13 | 0.772 | 0.824 | yes | 0.030 |
| B151 | [45] | Onychogalea fraenata | 225 | 29 | 0.631 | 0.898 | no | 0.193 |
| B90 | [45] | Onychogalea fraenata | 225 | 20 | 0.662 | 0.889 | no | 0.148 |
| G12-6 | [46] | Macropus giganteus | 230 | 19 | 0.830 | 0.903 | yes | 0.039 |
| G15-4 | [46] | Macropus giganteus | 221 | 39 | 0.842 | 0.939 | ND | 0.054 |
| G16-1 | [46] | Macropus giganteus | 229 | 25 | 0.664 | 0.850 | no | 0.132 |
| G16-2 | [46] | Macropus giganteus | 232 | 13 | 0.526 | 0.823 | no | 0.232 |
| G20-2 | [46] | Macropus giganteus | 230 | 13 | 0.857 | 0.869 | yes | 0.005 |
| G31-1 | [46] | Macropus giganteus | 212 | 13 | 0.774 | 0.814 | yes | 0.026 |
| G31-3 | [46] | Macropus giganteus | 229 | 29 | 0.738 | 0.878 | no | 0.089 |
| Me15 | [47] | Notamacropus eugenii | 189 | 30 | 0.614 | 0.894 | no | 0.190 |
| Me16 | [47] | Notamacropus eugenii | 211 | 31 | 0.711 | 0.928 | yes | 0.130 |
| Me17 | [47] | Notamacropus eugenii | 218 | 19 | 0.908 | 0.908 | yes | −0.002 |
| Me28 | [47] | Notamacropus eugenii | 202 | 57 | 0.916 | 0.944 | ND | 0.014 |
| T15-1 | [48] | Notamacropus eugenii | 231 | 16 | 0.831 | 0.901 | yes | 0.039 |
| T17-2 | [48] | Notamacropus eugenii | 225 | 43 | 0.747 | 0.915 | yes | 0.100 |
| T19-1 | [48] | Notamacropus eugenii | 215 | 31 | 0.847 | 0.940 | ND | 0.050 |
| T30-1 | [48] | Notamacropus eugenii | 204 | 26 | 0.559 | 0.928 | no | 0.249 |
| T31-1 | [48] | Notamacropus eugenii | 230 | 23 | 0.770 | 0.924 | yes | 0.090 |
| T32-1 | [48] | Notamacropus eugenii | 230 | 34 | 0.843 | 0.937 | ND | 0.050 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zemanova, M.A.; Ramp, D. Genetic Structure and Gene Flow in Eastern Grey Kangaroos in an Isolated Conservation Reserve. Diversity 2021, 13, 570. https://doi.org/10.3390/d13110570
Zemanova MA, Ramp D. Genetic Structure and Gene Flow in Eastern Grey Kangaroos in an Isolated Conservation Reserve. Diversity. 2021; 13(11):570. https://doi.org/10.3390/d13110570
Chicago/Turabian StyleZemanova, Miriam A., and Daniel Ramp. 2021. "Genetic Structure and Gene Flow in Eastern Grey Kangaroos in an Isolated Conservation Reserve" Diversity 13, no. 11: 570. https://doi.org/10.3390/d13110570
APA StyleZemanova, M. A., & Ramp, D. (2021). Genetic Structure and Gene Flow in Eastern Grey Kangaroos in an Isolated Conservation Reserve. Diversity, 13(11), 570. https://doi.org/10.3390/d13110570







