Abstract
Aegilops tauschii Coss., the D genome donor of hexaploid wheat (Triticum aestivum L.), is the most promising resource used to broaden the genetic diversity of wheat. Taxonomical studies have classified Ae. tauschii into two subspecies, ssp. tauschii and ssp. strangulata. However, molecular analysis revealed three distantly related lineages, TauL1, TauL2 and TauL3. TauL1 and TauL3 includes the only ssp. tauschii, whereas TauL2 includes both subspecies. This study aimed to clarify the phylogeny of Ae. tauschii and to find the traits that can differentiate between TauL1, TauL2 and TauL3, or between ssp. tauschii and ssp. strangulata. We studied the genetic and morpho-physiological diversity in 293 accessions of Ae. tauschii, covering the entire range of the species. A total of 5880 high-quality SNPs derived from DArTseq were used for phylogenetic cluster analyses. As a result, we observed wide morpho-physiological variation in each lineage and subspecies. Despite this variation, no key traits can discriminate lineages or subspecies though some traits were significantly different. Of 124 accessions previously lacking the passport data, 66 were allocated to TauL1, 57 to TauL2, and one to TauL3.
1. Introduction
Wild relatives attract increasing attention because they can provide characters related to adaptation [1]. The genus Aegilops L. (Poaceae) has been intensively studied because of its close relationship with cultivated wheats. The phylogenetic relationship between genera Aegilops and Triticum L. is widely reported [2,3,4,5], and on a world scale, the genus Aegilops includes 23 wild annual species, of which 11 are diploids and 12 are allopolyploids [6,7]. The revision of the genus Aegilops with regards to its genome and taxonomy results in a total of 27 specific and intraspecific taxa [8,9]. Aegilops tauschii Coss. (syn. Ae. squarrosa auct. non L.), a wild diploid self-pollinating species (2n = 2x = 14, DD), is the D genome donor of the hexaploid bread wheat (Triticum aestivum L.; 2n = 6x = 42, AABBDD). This wild species is found mainly at the edges of wheat fields in eastern Turkey, Iraq, Iran, Pakistan, India, China, Afghanistan, Central Asia, Transcaucasia (South Caucasus) and the Caucasus region [10]. About 8000 to 10,000 years ago, the ancestor of the current bread wheat appeared as a result of natural hybridization between cultivated wheat (Triticum turgidum L., 2n = 4x = 28, AABB) and Ae. tauschii [10,11,12]. Inside this last species, two subspecies were first described by Eig (1929) [13] as Ae. squarrosa ssp. eusquarrosa and ssp. strangulata, and their nomenclature was revised by Hammer (1980) [6] as Ae. tauschii ssp. tauschii and ssp. strangulata. Ae. tauschii is genetically and morphologically diverse [13], and the ssp. tauschii has elongated cylindrical spikelets, whereas ssp. strangulata has quadrate spikelets and empty glumes [6,13]. The ssp. tauschii has a wide distribution throughout the species range, whereas ssp. strangulata is limited to the south-eastern Caspian coastal region and the Caucasus [14]. Some of the molecular studies supported the subspecies division [15,16,17], whereas others did not [18,19].
The genetic diversity in Ae. tauschii has been studied at the molecular level by using isozymes [20], random amplified polymorphic DNA (RAPD) [21], chloroplast DNA [14,22] amplified fragment length polymorphisms (AFLPs) [23], simple sequence repeats (SSRs) [24] and DArT-array markers [25]. Most of these studies classified Ae. tauschii into three lineages: TauL1 including only ssp. tauschii, TauL2 including both ssp. tauschii and ssp. strangulata and TauL3 with intermediate forms. However, Arora et al. [26,27] reported that TauL1 is mainly associated with ssp. tauschii and TauL2 with ssp. strangulata. Therefore, this study aims to clarify the phylogeny of Ae. tauschii and to identify morpho-physiological traits that discriminate between the two main lineages (TauL1 and TauL2), ssp. tauschii belonging to TauL1 or TauL2, and the two subspecies (ssp. tauschii and ssp. strangulata).
2. Materials and Methods
2.1. Plant Materials
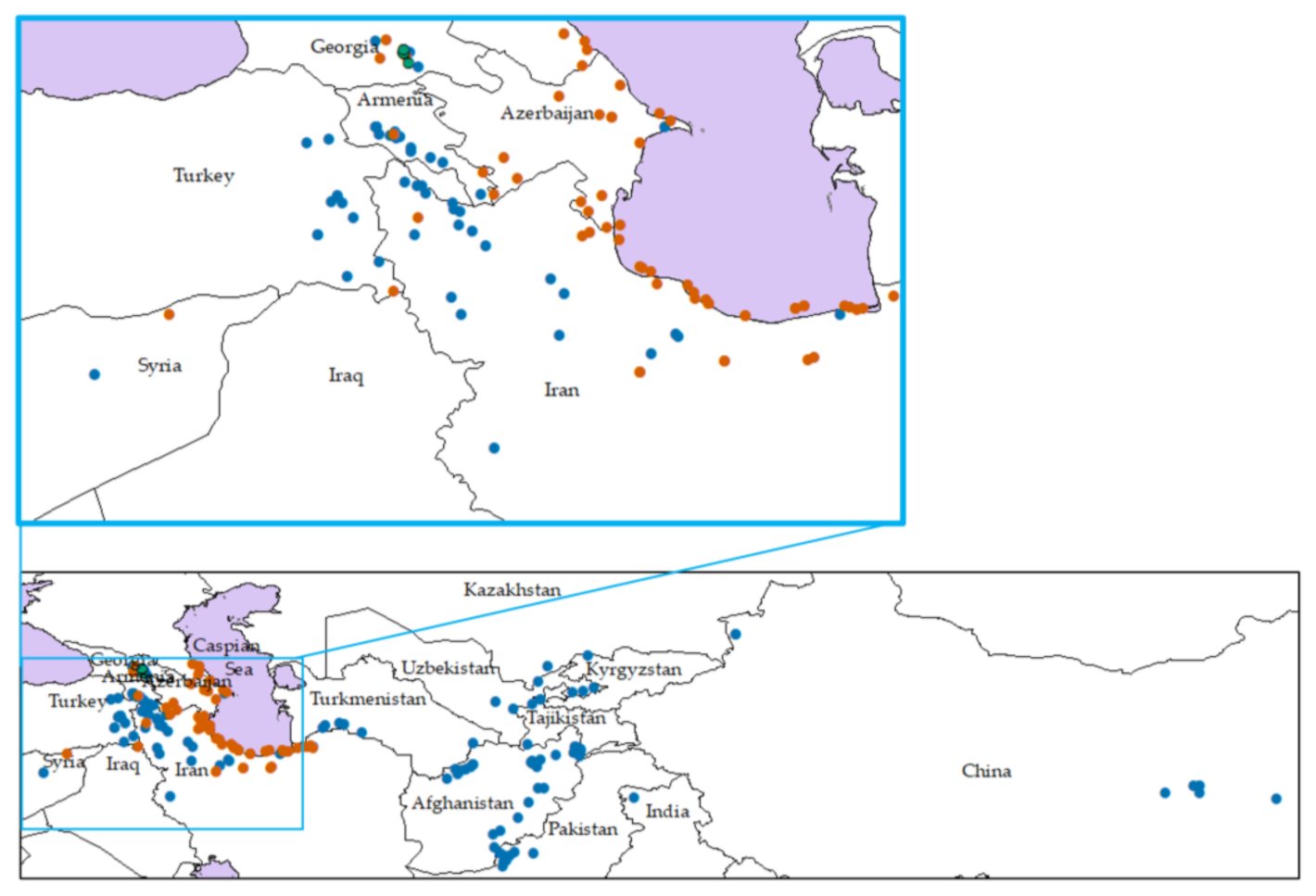
We used 293 Ae. tauschii accessions collected from the entire range of the natural distribution of this species (Table 1, Figure 1). Of these accessions, 201 have full passport data, including geographical coordinates, lineages and subspecies classification [14] (Figure 1). Five of the 201 accessions (AT 55, AT 60, AT 76, PI 499262 and PI 508262) represent adventive populations in the Shaanxi and Henan provinces of China. Among the 201 accessions, 132 belong to TauL1, 64 to TauL2 and 5 to TauL3 [14]. Based on sensu stricto criteria for subspecies classification, only accessions with distinctly moniliform spikes were classified to Ae. tauschii ssp. strangulata. In contrast, accessions having mildly moniliform and cylindrical spikes were classified to Ae. tauschii ssp. tauschii [14]. Of 293 accessions used in this study, 169 were previously studied by Matsuoka et al. (2009) [14] who classified 110, 55 and 4 to TauL1, TauL2 and TauL3, respectively.

Table 1.
Aegilops tauschii accessions used in this study.
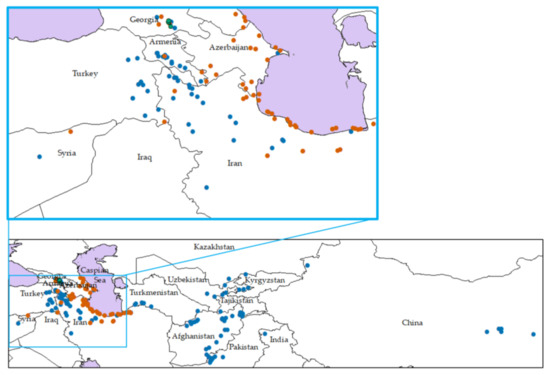
Figure 1.
Geographical distribution of 293 Aegilops tauschii accessions. Blue circles, lineage 1 accessions (TauL1); red circles, lineage 2 accessions (TauL2); and green circles, lineage 3 accessions (TauL3). Western range is enlarged.
2.2. Genomic Analysis and Statistaical Analysis of Molecular Data
Genomic DNA was extracted using the CTAB method [28]. The DNA samples (30 μL; 50–100 ng μL−1) were sent to Diversity Arrays Technology Pty. Ltd, Canberra, Australia (http://www.diversityarrays.com, accessed on 29 January 2018) for a whole-genome scan using the DArTseq platform. Sequencing-based DArT genotyping applies two complexity-reduction methods optimized for several plant species i.e., PstI/HpaII and PstI/HhaI were used to select a subset of the corresponding fragments [29]. At the DArT facility, the DArT soft marker extraction pipeline was used to filter and identify the informative markers. We performed the hierarchical clustering analysis in the statistical software R with the pvclust package [30]. The DArTseq SNPs data of 5880 markers without any missing data for 293 accessions of Ae. tauschii from 16 countries (some accessions are from unknown origin) were used for the analysis. Pvclust package computes the AU (approximately unbiased) p-value and BP (bootstrap probability) value via multiscale bootstrap resampling. These values can show how strong the clustering result is supported by the data. The dendrogram was generated by using the Euclidean distance matrix and complete method. The summary of SNP data sequences used for constructing phylogenetic tree was provided in Table S1.
2.3. Morpho-Physiological Evaluation
The morphological and physiological traits of all the accessions were measured at the research field of the Arid Land Research Center, Tottori University (Tottori, Japan; 35°32′N 134°13′E) during the winter and spring seasons of 2016/17 and 2017/18 by using an augmented complete block design with three randomly selected accessions as checks (GE12-14-O-1, GE12-28-O-2 and KU-20-2), and five plants were grown per accession. To estimate the phenotypic variation, we measured two leaf parameters (flag leaf length, FLL; flag leaf width, FLW), four spike parameters (spike length, SPL; spike width, SPW; seed number per spike, SN/SP; spike weight, SPWg), days to heading (DH), biomass weight (Bio) and three physiological traits (Normalized Difference Vegetative Index, NDVI; canopy temperature, CT; and chlorophyll content, SPAD). To measure SPWg, we covered the spikes with a transparent envelope before physiological maturity to avoid shattering. The measurement methods are summarized in Table 2.

Table 2.
Phenotypic traits analyzed.
2.4. Statistical Analysis of Morpho-Physiological Data
Analyses of the phenotypic data, including mean, standard deviation, range distribution and analysis of variance (F and p-values in one-way ANOVA) for the morpho-physiological variations were calculated using Plant Breeding Tools (PBTools) version 1.4 (International Rice Research Institute, http://bbi.irri.org/products, 15 February 2020). Due to the significant genotype × season interaction, best linear unbiased predictions (BLUPs) were estimated for each trait.
3. Results
3.1. Phylogenetical Allocation of Uncertain Accessions by Molecular Markers
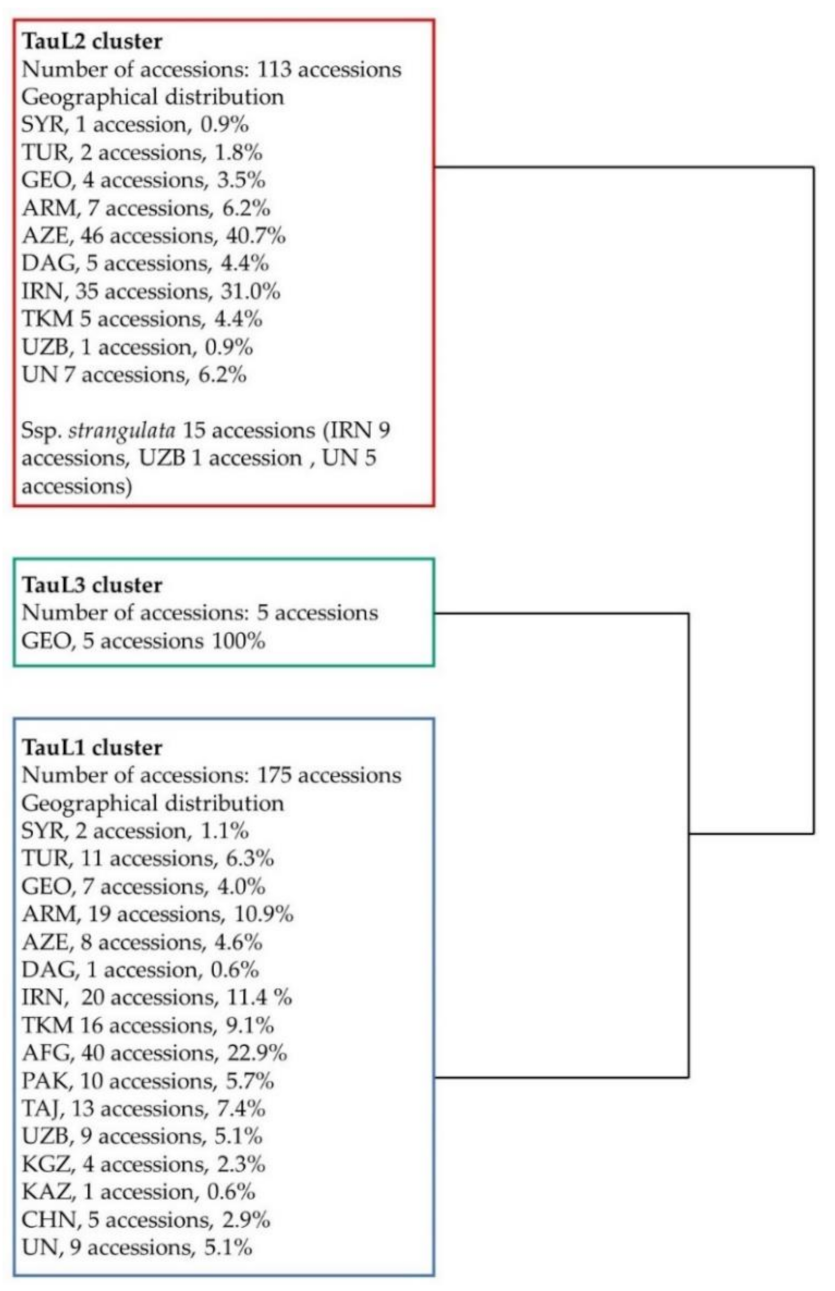
Following Matsouka et al. (2009) [14], we carefully observed the key morphological traits of the 124 accessions that lacked taxonomical information and identified 7 accessions as ssp. strangulata and the remaining 117 as ssp. tauschii. Among the seven accessions identified as ssp. strangulata, AE 525 was collected from Iran, AE 692 from Uzbekistan and AE 426, AE 428, AE 429, AE 430 and AE 434 from unknown regions. To know the lineages (TauL1, TauL2 or TauL3) of all 124 accessions, we conducted cluster analysis using 5880 DArTseq markers. As a result, 66, 57 and 1 were clustered in TauL1, TauL2 and TauL3, respectively (Figure 2, Figure S1). All the accessions in TauL1 were ssp. tauschii, whereas in TauL2, 50 were ssp. tauschii and 7 were ssp. strangulata. The accessions in the TauL3 were ssp. tauschii. These findings supported previous results that the ssp. strangulata is present only in TauL2.
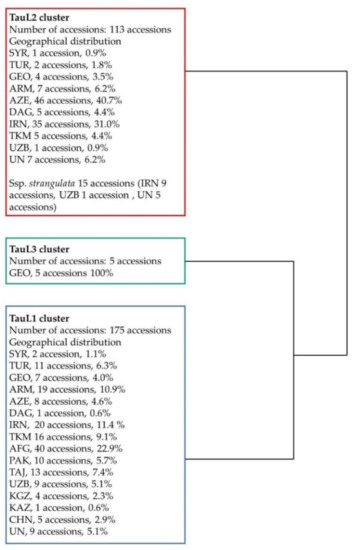
Figure 2.
Schematic form of hierarchical clustering of 293 Ae. tauschii accessions showing the classification of TauL1, TauL2 and TauL3 based on high-quality SNPs derived from 5880 DArTseq markers. Origin of accessions: SYR, Syria; TUR, Turkey; GEO, Georgia; ARM, Armenia; AZE, Azerbaijan; DAG, Dagestan; IRN, Iran; TKM, Turkmenistan; AFG, Afghanistan; PAK, Pakistan; TAJ, Tajikistan; UZB, Uzbekistan; KGZ, Kyrgyzstan; KAS, Kazakhstan; CHN, China and UN, unknown country.
Previously, Matsuoka et al. (2009) [14] classified Ae. tauschii accessions into TauL1, TauL2 and TauL3 based on the chloroplast DNA. To confirm their result, we analyzed the 169 accessions used in Matsuoka et al. (2009) [14] using DArTseq markers. Most of the accessions were clustered as expected with 5 exceptions: KU-2109 and KU-2158 were in TauL1, whereas PI 486274, IG 127015 and IG 120735 were in TauL2.
From these studies, we found that all 293 accessions of Ae. tauschii were classified as 175 TauL1, 113 TauL2 and 5 TauL3. In TauL2, 15 accessions were ssp. strangulata and others including accessions in TauL1 and TauL3 were ssp. tauschii.
The TauL1 cluster contained accessions from Syria, Turkey, Georgia, Armenia, Azerbaijan, Dagestan, Iran, Turkmenistan, Afghanistan, Pakistan, Tajikistan, Uzbekistan, Kyrgyzstan, Kazakhstan, China and unknown countries. The TauL2 cluster contained accessions from Syria, Turkey, Georgia, Armenia, Azerbaijan, Dagestan, Iran, Turkmenistan, Uzbekistan and unknown countries (Table 1, Figure 2, Figure S1). The ssp. strangulata accessions were clustered in one clade in TauL2, and most of the accessions were from Iran.
3.2. Morpho-Physiological Differences between TauL1 and TauL2
A large variation was observed for all the morpho-physiological traits in TauL1 and TauL2 (Table 3). Statistical analyses showed a significant difference between these two lineages in SPW, SPWg, DH and Bio. The means in these traits were larger in TauL2 than in TauL1, indicating that the accessions in TauL2 tend to be higher than TauL1. On the other hand, the means of the physiological traits (NDVI, CT and SPAD), and leaf traits (FLL and FLW) were not significantly different between them. The ranges of these traits overlapped between the two lineages, and thus we cannot discriminate the two groups with these traits (Table 3).

Table 3.
Morpho-physiological variation in two Aegilops tauschii lineages, TauL1 (175 accessions) and TauL2 (113 accessions).
3.3. Morpho-Physiological Variation between ssp. tauschii Belonging to TauL1 and TauL2
We designated ssp. tauschii in TauL1 and TauL2 as ‘TauL1T’ and ‘TauL2T’, respectively, and compared accessions in these groups. A large variation was observed for all the morpho-physiological traits in TauL1T and TauL2T (Table 4). Statistical analyses showed significant differences between the two groups in FLL, DH and Bio. The mean of FLL was higher in TauL1T, whereas those of DH and Bio were higher in TauL2T. On the other hand, the means of the physiological traits (NDVI, CT and SPAD), and spike traits (SPL, SPW, SN/SP and SPWg) were not significantly different between them. The ranges of these traits overlapped between TauL1T and TauL2T, and thus we cannot discriminate the two groups with these traits (Table 4).

Table 4.
Morpho-physiological variation in ssp. tauschii in TauL1 (TauL1T, 175 accessions) and TauL2 (TauL2T, 98 accessions).
3.4. Morpho-Physiological Variation between ssp. tauschii and ssp. strangulata
A large variation was observed for all the morpho-physiological traits in ssp. tauschii and ssp. strangulata (Table 5). Statistical analyses showed significant difference between these two subspecies in SPL, SN/SP, SPWg and DH. The means of SPL and SN/SP were higher in ssp. tauschii than in ssp. strangulata, whereas those of SPWg and DH were higher in ssp. strangulata than in ssp. tauschii. On the other hand, the means of the leaf traits (FLL and FLW), SPW and physiological traits (NDVI, CT and SPAD) were not significantly different between them. The ranges of these traits overlapped between the two subspecies (Table 5).

Table 5.
Morpho-physiological variation in ssp. tauschii (273 accessions) and spp. strangulata (15 accessions) of Aegilops tauschii.
3.5. Morpho-Physiological Variation of Accessions in TauL3
In this study, only five accessions (AE 454, AE 929, AE 929a, KU-2829A and KU-2832) belong to TauL3. Therefore, we did not compare them with TauL1 and TauL2. All the accessions originated from Georgia and showed a similar plant morphology to ssp. tauschii with an intermediate spike shape between TauL1 and TauL2. Genomic analysis revealed that these accessions are clearly differentiated from both TauL1 and TauL2.
4. Discussion
4.1. Geographical Clines of Morphological Variation in Subspecies and Lineage Classification
The main putative area of origin of Ae. tauschii is the Transcaucasus, from which it has spread to the east and south [10] (Figure 1). While ssp. tauschii has cylindrical spike forms and ssp. strangulata moniliform spike forms, some Ae. tauschii accessions have mildly moniliform spike forms (TauL3) which suggest a hybrid origin. Overall, spikelet morphology is the main trait not only for discriminating the two subspecies but also for intraspecific diversification in Ae. tauschii, even though the genetic basis of spikelet morphology divergence has not yet been studied. Nishijima et al. (2017) [31] divided Ae. tauschii into two main lineages TauL1 and TauL2, and a minor lineage (TauL3) by Bayesian population structure analysis with genome-wide marker genotyping. Using DArTseq genotyping of a large number of accessions, we confirmed their results (Figure 2, Figure S1). The TauL1 accessions are spread from the western geographical range (Transcaucasus, northern regions of Iran) to the eastern geographical range (Pakistan and Afghanistan), whereas TauL2 is limited only to the western range, and ssp. strangulata is included only in TauL2.
This result is consistent with Mizuno et al. (2010) [23] using AFLPs. Thus, the differentiation of the ssp. strangulata is believed to have occurred in TauL2. Furthermore, we found that the most probable origin of ssp. strangulata is Iran and that this subspecies clusters in one clade within TauL2 (Figure 2, Figure S1). This finding strongly indicates that speciation had occurred in the ssp. tauschii included in TauL2, resulting in appearance of ssp. strangulata-type spike morphology. The D genome of ssp. strangulata is involved in the D genome of bread wheat. This was revealed by sequencing [32], single nucleotide polymorphisms [33], variation in the AP2 homoeologs, the genes underlying lodicule development [34], SSR markers [35], NADP-dependent aromatic alcohol dehydrogenase [36] and aspartate aminotransferase and alcohol dehydrogenase isoenzymes [37]. Overall, using the DArTseq genotyping platform, we have allocated 124 accessions with no previous lineage description into TauL1, TauL2 or TauL3. Furthermore, based on this data, we have reclassified 5 accessions: 2 accessions from Iran (KU-2109 and KU-2158) formerly classified in TauL2 by chloroplast DNA [14] were now placed in TauL1, and 3 accessions (PI 486274 from Turkey, IG 127015 from Armenia and IG 120735 from Turkmenistan) formerly classified in TauL1 were now placed in TauL2. The inconsistency of the nucleus and cytoplasmic genomes may be attributable to the cytoplasmic substitution origin by hybrids between the two lineages and the backcrossing in the evolution of these accessions. Furthermore, previous studies reported that accessions in TauL2 were distributed in the regions near the Caspian Sea. However, here we found that five accessions (AE 192, AE 213, AE 250, CGN10733 and IG 120735) which originated from Turkmenistan and AE 692 from Uzbekistan were clustered in TauL2 (Table 1). These accessions may have been transferred to the regions naturally or by human activity.
4.2. Potential for Adaptive Convergence in Ae. tauschii Evolution
Molecular evolutionary studies have explained the origin of crops more clearly than before [38,39], especially for the main crops that were domesticated without ploidy modification. Phylogeographic analyses based on nuclear and chloroplast DNA sequences have shown multiple evolutionary origins of cultivated rice in East Asia [40] and barley in the Fertile Crescent and Central Asia [41,42], whereas phylogenetic analysis based on multilocus microsatellite genotyping has shown a single domestication event for maize ca. 9000 years ago [43]. One of the fundamental problems in understanding the evolution of Ae. tauschii is the relationship between the different lineages and subspecies. In the current study, although some traits examined differed significantly between the lineages and subspecies, the range of the diversity was overlapped (Table 3, Table 4 and Table 5). The phenotypes convergence may have originated through either divergent genetic solutions [44,45] or the same pathways, genes or even nucleotide positions in independent lineages [46,47]. Convergence at the genetic level can in turn result from (i) mutations arising independently in separate populations or organisms (parallel genetic evolution); (ii) evolution of a polymorphic allele in a common ancestral population or species (trans-specific polymorphism); and (iii) evolution of an allele introduced by hybridization (introgression) from one population to another (e.g., TauL1 and TauL2). Another possibility that can explain the phenotypic similarities between the different Ae. tauschii lineages is the occurrence of genetic differentiation after the geographical isolation under similar environmental condition without morphological or physiological differentiation. Local standing genetic diversity combined with spatial population structure restricting dispersal in an ecologically patchy area promotes rapid convergence [48].
4.3. Implications of Ae. tauschii Diversity in Wheat Breeding
Among the species in genus Aegilops, only Ae. tauschii can be used efficiently for wheat improvement owing to the mostly regular pairing of its chromosomes with the D genome chromosomes of bread wheat [49]. It is believed that Ae. tauschii is an excellent source to widen the narrow genetic base of bread wheat. Currently, with the new advances in plant science and the rapid development of sequencing and genome-editing tools, identification and characterization of genes of interest in wheat are in progress and can be expected to become easier and more straightforward in the coming decades. Once the gene in question is identified and characterized, it is easy to transfer and utilize the gene in breeding programs. This will pave the way to utilize the genes from Ae. tauschii as it will help to overcome the limitations related to the irregular chromosome pairing.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/d13050217/s1, Figure S1: Hierarchical clustering of 293 Ae. tauschii accessions showing the classification of TauL1, TauL2, and TauL3 based on high-quality SNP markers derived from 5880 DArTseq markers. Values at branches are AU values (upper, red), BP values (down, blue), and cluster labels (medium, gray). Ssp. strangulata is indicated, and others belongs to ssp. tauschii. UN, unknown lineages or country. Origin of accessions: SYR, Syria; TUR, Turkey; GEO, Georgia; ARM, Armenia; AZE, Azerbaijan; DAG, Dagestan; IRN, Iran; TKM, Turkmenistan; AFG, Afghanistan; PAK, Pakistan; TAJ, Tajikistan; UZB, Uzbekistan; KGZ, Kyrgyzstan; KAS, Kazakhstan and CHN, China. The two black circles indicate where these two trees are connected, Table S1: The summary of SNP data sequences used for constructing phylogenetic tree.
Author Contributions
H.T. conceived the project. H.T., T.-S.C., H.I., Y.Y. and M.M.M.M. designed the research. Y.M. and H.T. provided plant materials. M.M.M.M. conducted the experiments and analyzed the data. Y.S.A.G., N.M.K. and M.A. guided the data analyses. M.M.M.M. prepared the first draft of the manuscript. Y.S.A.G., N.M.K., I.S.A.T. and H.T. critically reviewed and improved the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partly supported by the SATREPS Project (JPMJSA1805) funded by Japan Science and Technology Agency (JST), KAKENHI (16H04858) from Japan Society for the Promotion of Science, and by the MRA Project of Tottori University.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Non applicable.
Data Availability Statement
This study did not report any data.
Acknowledgments
We appreciate Michael O. Itam for critical reading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hu, X.G.; Wu, B.H.; Yan, Z.H.; Dai, S.F.; Zhang, L.Q.; Liu, D.C.; Zheng, Y.L. Characteristics and polymorphism of NAM gene from Aegilops section sitopsis species. Afr. J. Agric. Res. 2012, 7, 5252–5258. [Google Scholar] [CrossRef]
- Kimber, G.; Zhao, Y.H. The D genome of the Triticeae. Can. J. Genet. Cytol. 1983, 25, 581–589. [Google Scholar] [CrossRef]
- Kellogg, E.A.; Appels, R.; Mason-Gamer, R.J. When Genes Tell Different Stories: The Diploid Genera of Triticeae (Gramineae). Syst. Bot. 1996, 21, 321. [Google Scholar] [CrossRef]
- Petersen, G.; Seberg, O.; Yde, M.; Berthelsen, K. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol. Phylogenet. Evol. 2006, 39, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Alnaddaf, L.M.; Moualla, M.Y.; Haider, N. Resolving genetic relationships among Aegilops L. and Triticum L. species using analysis of chloroplast DNA by cleaved amplified polymorphic sequence (CAPS). Asian J. Agric. Sci. 2012, 4, 270–279. [Google Scholar]
- Hammer, K. Vorarbeiten zur monographischen Darstellung von Wildpflanzensortimenten: Aegilops L. Kulturpflanze 1980, 28, 33–180. [Google Scholar] [CrossRef]
- Kilian, B.; Mammen, K.; Millet, E.; Sharma, R.; Graner, A.; Salamini, F.; Hammer, K.; Özkan, H. Aegilops. In Wild Crop Relatives: Genomic and Breeding Resources; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–76. [Google Scholar]
- Van Slageren, M.W. Wild Wheats: A monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae); Agricultural University: Wageningen, The Netherlands, 1994. [Google Scholar]
- Čerepanov, S.K. Vascular Plants of Russia and Adjacent Countries; Cambridge University Press: Cambridge, UK, 1996. [Google Scholar]
- Feldman, M. Origin of cultivated wheat. In The World Wheat Book: A history of Wheat Breeding; Bonjean, A.P., Angus, W.J., Eds.; Lavoisier Publishing: Paris, France, 2001; pp. 3–56. [Google Scholar]
- McFadden, E.S.; Sears, E.R. The artificial synthesis of Triticum spelta. Proc. Records Genet. Soc. Am. 1944, 13, 26–27. [Google Scholar]
- Kihara, H. Discovery of the DD-analyzer, one of the ancestors of Triticum vulgare. Agric. Hortic. 1944, 19, 13–14. [Google Scholar]
- Eig, A. Monographisch-kritische Übersicht der Gattung Aegilops. Rep. Spec. Nov. Reg. Veget. Berh. 1929, 55, 1–228. [Google Scholar]
- Matsuoka, Y.; Nishioka, E.; Kawahara, T.; Takumi, S. Genealogical analysis of subspecies divergence and spikelet-shape diversification in central Eurasian wild wheat Aegilops tauschii Coss. Plant Syst. Evol. 2009, 279, 233–244. [Google Scholar] [CrossRef]
- Dvorak, J.; Luo, M.C.; Yang, Z.L.; Zhang, H.B. The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 1998, 97, 657–670. [Google Scholar] [CrossRef]
- Gill, K.S.; Lubbers, E.L.; Gill, B.S.; Raupp, W.J.; Cox, T.S. A genetic linkage map of Triticum tauschii (DD) and its relationship to the D genome of bread wheat (AABBDD). Genome 1991, 34, 362–374. [Google Scholar] [CrossRef]
- Pestsova, E.; Korzun, V.; Goncharov, N.P.; Hammer, K.; Ganal, M.W.; Röder, M.S. Microsatellite analysis of Aegilops tauschii germplasm. Theor. Appl. Genet. 2000, 101, 100–106. [Google Scholar] [CrossRef]
- Lelley, T.; Stachel, M.; Grausgruber, H.; Vollmann, J. Analysis of relationships between Aegilops tauschii and the D genome of wheat utilizing microsatellites. Genome 2000, 43, 661–668. [Google Scholar] [CrossRef]
- Saeidi, H.; Rahiminejad, M.R.; Vallian, S.; Heslop-Harrison, J.S. Biodiversity of diploid D-genome Aegilops tauschii Coss. in Iran measured using microsatellites. Genet. Resour. Crop Evol. 2006, 53, 1477–1484. [Google Scholar] [CrossRef]
- Dudnikov, A.J.; Kawahara, T. Aegilops tauschii: Genetic variation in Iran. Genet. Resour. Crop Evol. 2006, 53, 579–586. [Google Scholar] [CrossRef]
- Okuno, A.; Tamemoto, H.; Tobe, K.; Ueki, K.; Mori, Y.; Iwamoto, K.; Umesono, K.; Akanuma, Y.; Fujiwara, T.; Horikoshi, H.; et al. Troglitazone increases the number of small adipocytes without the change of white adipose tissue mass in obese Zucker rats. J. Clin. Invest. 1998, 101, 1354–1361. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Mori, N.; Kawahara, T. Genealogical use of chloroplast DNA variation for intraspecific studies of Aegilops tauschii Coss. Theor. Appl. Genet. 2005, 111, 265–271. [Google Scholar] [CrossRef]
- Mizuno, N.; Yamasaki, M.; Matsuoka, Y.; Kawahara, T.; Takumi, S. Population structure of wild wheat D-genome progenitor Aegilops tauschii Coss.: Implications for intraspecific lineage diversification and evolution of common wheat. Mol. Ecol. 2010, 19, 999–1013. [Google Scholar] [CrossRef]
- Naghavi, M.R.; Mardi, M. Characterization of genetic variation among accessions of Aegilops tauschii. Asia Pac. J. Mol. Biol. Biotechnol. 2010, 18, 91–94. [Google Scholar]
- Sohail, Q.; Shehzad, T.; Kilian, A.; Eltayeb, A.E.; Tanaka, H.; Tsujimoto, H. Development of diversity array technology (DArT) markers for assessment of population structure and diversity in Aegilops tauschii. Breed. Sci. 2012, 62, 38–45. [Google Scholar] [CrossRef]
- Arora, S.; Singh, N.; Kaur, S.; Bains, N.S.; Uauy, C.; Poland, J.; Chhuneja, P. Genome-wide association study of grain architecture in wild wheat Aegilops tauschii. Front. Plant Sci. 2017, 8, 886. [Google Scholar] [CrossRef]
- Arora, S.; Steuernagel, B.; Gaurav, K.; Chandramohan, S.; Long, Y.; Matny, O.; Johnson, R.; Enk, J.; Periyannan, S.; Singh, N.; et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 2019, 37, 139–143. [Google Scholar] [CrossRef]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef]
- Sansaloni, C.; Petroli, C.; Jaccoud, D.; Carling, J.; Detering, F.; Grattapaglia, D.; Kilian, A. Diversity Arrays Technology (DArT) and next-generation sequencing combined: Genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011, 5, P54. [Google Scholar] [CrossRef]
- Suzuki, R.; Shimodaira, H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef]
- Nishijima, R.; Okamoto, Y.; Hatano, H.; Takumi, S. Quantitative trait locus analysis for spikelet shape-related traits in wild wheat progenitor Aegilops tauschii: Implications for intraspecific diversification and subspecies differentiation. PLoS ONE 2017, 12, e0173210. [Google Scholar] [CrossRef]
- Ling, H.Q.; Ma, B.; Shi, X.; Liu, H.; Dong, L.; Sun, H.; Cao, Y.; Gao, Q.; Zheng, S.; Li, Y.; et al. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature 2018, 557, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Luo, M.C.; Chen, Z.; You, F.M.; Wei, Y.; Zheng, Y.; Dvorak, J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013, 198, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Wang, N.; Sakuma, S.; Pourkheirandish, M.; Koba, T.; Komatsuda, T. Variation in the wheat AP2 homoeologs, the genes underlying lodicule development. Breed. Sci. 2013, 63, 255–266. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Naghavi, M.R.; Aghaei, M.J.; Taleei, A.R.; Omidi, M.; Mozafari, J.; Hassani, M.E. Genetic diversity of the D-genome in T. aestivum and Aegilops species using SSR markers. Genet. Resour. Crop Evol. 2009, 56, 499–506. [Google Scholar] [CrossRef]
- Jaaska, V. NADP-dependent aromatic alcohol dehydrogenase in polyploid wheats and their diploid relatives. On the origin and phylogeny of polyploid wheats. Theor. Appl. Genet. 1978, 53, 209–217. [Google Scholar] [CrossRef]
- Jaaska, V. Aspartate aminotransferase and alcohol dehydrogenase isoenzymes: Intraspecific differentiation in Aegilops tauschii and the origin of the D genome polyploids in the wheat group. Plant Syst. Evol. 1981, 137, 259–273. [Google Scholar] [CrossRef]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef]
- Purugganan, M.D.; Fuller, D.Q. The nature of selection during plant domestication. Nature 2009, 457, 843–848. [Google Scholar] [CrossRef]
- Londo, J.P.; Chiang, Y.C.; Hung, K.H.; Chiang, T.Y.; Schaal, B.A. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. USA 2006, 103, 9578–9583. [Google Scholar] [CrossRef]
- Morrell, P.L.; Clegg, M.T. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc. Natl. Acad. Sci. USA 2007, 104, 3289–3294. [Google Scholar] [CrossRef]
- Saisho, D.; Purugganan, M.D. Molecular phylogeography of domesticated barley traces expansion of agriculture in the old world. Genetics 2007, 177, 1765–1776. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Vigouroux, Y.; Goodman, M.M.; Sanchez, J.G.; Buckler, E.; Doebley, J. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 2002, 99, 6080–6084. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Williams, B.L.; Selegue, J.E.; Carroll, S.B. Drosophila pigmentation evolution: Divergent genotypes underlying convergent phenotypes. Proc. Natl. Acad. Sci. USA 2003, 100, 1808–1813. [Google Scholar] [CrossRef]
- Pascoal, S.; Cezard, T.; Eik-Nes, A.; Gharbi, K.; Majewska, J.; Payne, E.; Ritchie, M.G.; Zuk, M.; Bailey, N.W. Rapid convergent evolution in wild crickets. Curr. Biol. 2014, 24, 1369–1374. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Aardema, M.L.; Medina, E.M.; Schumer, M.; Andolfatto, P. Parallel molecular evolution in an herbivore community. Science 2012, 337, 1634–1637. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Orgogozo, V. The loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution 2013, 67, 1235–1250. [Google Scholar] [CrossRef] [PubMed]
- Ralph, P.L.; Coop, G. Convergent evolution during local adaptation to patchy landscapes. PLoS Genet. 2015, 11, 1–31. [Google Scholar] [CrossRef]
- Kishii, M. An update of recent use of Aegilops species in wheat breeding. Front. Plant Sci. 2019, 10, 585. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).