Bacterial Number and Genetic Diversity in a Permafrost Peatland (Western Siberia): Testing a Link with Organic Matter Quality and Elementary Composition of a Peat Soil Profile
Abstract
:1. Introduction
2. Study Site and Methods
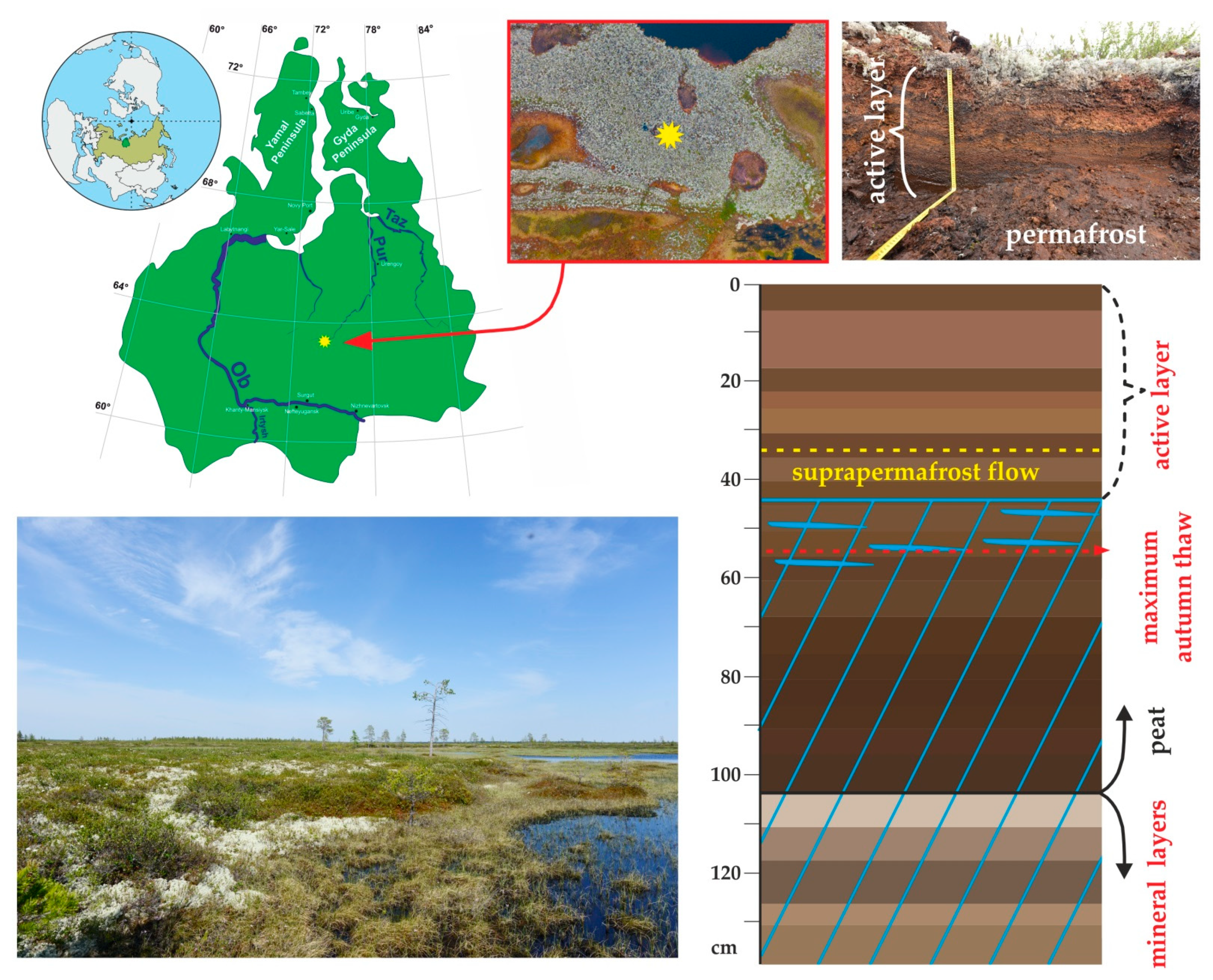
2.1. Peat Core Collection and Temperature Pattern of the WSL Peat Profile
2.2. Total Chemical Analysis
2.3. Organic Matter Isotopic Composition and Quality
2.4. Analysis of Bacterial Number and Genetic Diversity
2.5. Statistical Treatment
3. Results
3.1. Peat Formation History, Thermal Regime, Physical and Elementary Characteristics of the Peat Core and Chemical Composition of the Pore Water
3.2. Organic Matter Isotopic Composition and Functional Group Analysis by NMR
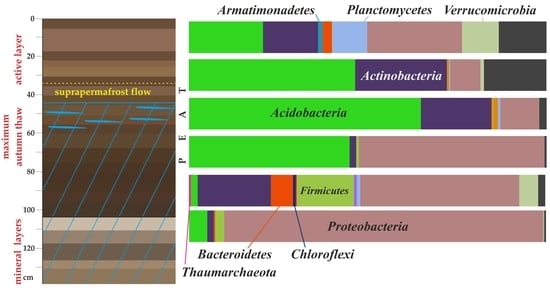
3.3. Cell Number and Microbial Diversity as a Function of Depth
3.4. Pair Correlation and Multiparametric Statistics of the Full Data Set
4. Discussion
4.1. Elementary Peat and Peat Fluid Composition and Organic Matter Quality
4.2. Bacterial Number and Genetic Diversity of Soil Microorganisms
4.3. Multiple Factors Governing the Microbial Diversity in the Peat Core
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilhelm, R.C.; Niederberger, T.D.; Greer, C.; Whyte, L.G. Microbial diversity of active layer and permafrost in an acidic wetland from the Canadian High Arctic. Can. J. Microbiol. 2011, 57, 303–315. [Google Scholar] [CrossRef] [Green Version]
- Steven, B.; Briggs, G.; McKay, C.P.; Pollard, W.H.; Greer, C.W.; Whyte, L.G. Characterization of the microbial diversity in a permafrost sample from the Canadian high Arctic using culture-dependent and culture-independent methods. FEMS Microbiol. Ecol. 2007, 59, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Steven, B.; Pollard, W.H.; Greer, C.W.; Whyte, L.G. Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic. Environ. Microbiol. 2008, 10, 3388–3403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, A.A.; Herbert, R.A.; Mikkelsen, K.; Jensen, L.L.; Kristoffersen, T.; Tiedje, J.M.; Lomstein, B.A.; Finster, K. Viability, diversity and composition of the bacterial community in a high Arctic permafrost soil from Spitsbergen, Northern Norway. Environ. Microbiol. 2007, 9, 2870–2884. [Google Scholar] [CrossRef] [PubMed]
- Vorobyova, E.; Soina, V.; Gorlenko, M.; Minkovskaya, N.; Zalinova, N.; Mamukelashvili, A.; Gilichinsky, D.; Rivkina, E.; Vishnivetskaya, T. The deep cold biosphere: Facts and hypothesis. FEMS Microbiol. Rev. 1997, 20, 277–290. [Google Scholar] [CrossRef]
- Schnecker, J.; Wild, B.; Hofhansl, F.; Alves, R.J.E.; Bárta, J.; Capek, P.; Fuchslueger, L.; Gentsch, N.; Gittel, A.; Guggenberger, G.; et al. Effects of Soil Organic Matter Properties and Microbial Community Composition on Enzyme Activities in Cryoturbated Arctic Soils. PLoS ONE 2014, 9, e94076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wild, B.; Schnecker, J.; Alves, R.J.E.; Barsukov, P.; Bárta, J.; Capek, P.; Gentsch, N.; Gittel, A.; Guggenberger, G.; Lashchinskiy, N.; et al. Input of easily available organic C and N stimulates microbial decomposition of soil organic matter in arctic permafrost soil. Soil Biol. Biochem. 2014, 75, 143–151. [Google Scholar] [CrossRef] [Green Version]
- Wallenstein, M.D.; McMahon, S.; Schimel, J. Bacterial and fungal community structure in Arctic tundra tussock and shrub soils. FEMS Microbiol. Ecol. 2007, 59, 428–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackelprang, R.; Waldrop, M.; DeAngelis, K.; David, M.M.; Chavarria, K.L.; Blazewicz, S.J.; Rubin, E.M.; Jansson, J. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 2011, 480, 368–371. [Google Scholar] [CrossRef] [Green Version]
- Gittel, A.; Bárta, J.; Kohoutová, I.; Mikutta, R.; Owens, S.; Gilbert, J.; Schnecker, J.; Wild, B.; Hannisdal, B.; Maerz, J.; et al. Distinct microbial communities associated with buried soils in the Siberian tundra. ISME J. 2013, 8, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Yergeau, E.; Bokhorst, S.; Huiskes, A.H.L.; Boschker, H.T.S.; Aerts, R.; Kowalchuk, G.A. Size and structure of bacterial, fungal and nematode communities along an Antarctic environmental gradient. FEMS Microbiol. Ecol. 2006, 59, 436–451. [Google Scholar] [CrossRef] [Green Version]
- Yergeau, E.; Kang, S.; He, Z.; Zhou, J.; Kowalchuk, G.A. Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. ISME J. 2007, 1, 163–179. [Google Scholar] [CrossRef] [Green Version]
- Yergeau, E.; Schoondermark-Stolk, S.A.; Brodie, E.L.; Déjean, S.; DeSantis, T.Z.; Gonçalves, O.; Piceno, Y.M.; Andersen, G.L.; Kowalchuk, G.A. Environmental microarray analyses of Antarctic soil microbial communities. ISME J. 2009, 3, 340–351. [Google Scholar] [CrossRef] [Green Version]
- Kremenetski, K.; Velichko, A.; Borisova, O.; MacDonald, G.; Smith, L.; Frey, K.; Orlova, L. Peatlands of the Western Siberian lowlands: Current knowledge on zonation, carbon content and Late Quaternary history. Quat. Sci. Rev. 2003, 22, 703–723. [Google Scholar] [CrossRef]
- Tyrtikov, A.P. Thawing of soils in tundra of western Siberia. In Natural Environment of Western Siberia; Popov, A.I., Ed.; MG Izd-vo: Moscow, Russia, 1973; pp. 160–169. [Google Scholar]
- Tyrtikov, A.P. Dynamics of Vegetation Coverage and Permafrost Development in Western Siberia; Nauka: Moscow, Russia, 1979; 114p. [Google Scholar]
- Vasil’evskaya, V.D.; Ivanov, V.V.; Bogatyrev, L.G. Soils of North of Western Siberia; Moscow University Publ. House: Moscow, Russia, 1986; 228p. [Google Scholar]
- Novikov, S.M.; Moskvin, Y.P.; Trofimov, S.A.; Usova, L.I.; Batuev, V.I.; Tumanovskaya, S.M.; Smirnova, V.P.; Markov, M.L.; Korotkevicth, A.E.; Potapova, T.M. Hydrology of Bog Territories of the Permafrost Zone of Western Siberia; BBM publ. House: St. Petersbourg, Russia, 2009; 535p. [Google Scholar]
- Khrenov, V.Y. Soils of Cryolithozone of Western Siberia: Morphology, Physico-Chemical Properties and Geochemistry; Nauka: Moscow, Russia, 2011; 214p. [Google Scholar]
- Morgalev, Y.N.; Lushchaeva, I.V.; Morgaleva, T.G.; Kolesnichenko, L.G.; Loiko, S.; Krickov, I.V.; Morgalev, S.Y.; Lim, A.; Raudina, T.V.; Volkova, I.I.; et al. Bacteria primarily metabolize at the active layer/permafrost border in the peat core from a permafrost region in western Siberia. Polar Biol. 2017, 40, 1645–1659. [Google Scholar] [CrossRef]
- Yergeau, E.; Hogues, H.; Whyte, L.G.; Greer, C.W. The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses. ISME J. 2010, 4, 1206–1214. [Google Scholar] [CrossRef]
- Gittel, A.; Barta, J.; Lacmanova, I.; Schnecker, J.; Wild, B.; Capek, P.; Kaiser, C.; Torsvik, V.; Richter, A.; Schleper, C.; et al. Site- and horizon-specific patterns of microbial community structure and enzyme activities in permafrost-affected soils of Greenland. Front. Microbiol. 2014, 5, 541. [Google Scholar] [CrossRef]
- Deng, J.; Gu, Y.; Zhang, J.; Xue, K.; Qin, Y.; Yuan, M.; Yin, H.; He, Z.; Wu, L.; Schuur, E.A.G.; et al. Shifts of tundra bacterial and archaeal communities along a permafrost thaw gradient in A laska. Mol. Ecol. 2015, 24, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Johnston, E.; Rodriguez-R, L.M.; Eluo, C.; Yuan, M.M.; Ewu, L.; He, Z.; Konstantinidis, K.T.; Luo, Y.; Tiedje, J.M.; Zhou, J.; et al. Metagenomics Reveals Pervasive Bacterial Populations and Reduced Community Diversity across the Alaska Tundra Ecosystem. Front. Microbiol. 2016, 7, 579. [Google Scholar] [CrossRef] [PubMed]
- Inglese, C.N.; Christiansen, C.; Lamhonwah, D.; Moniz, K.; Montross, S.N.; Lamoureux, S.; Lafrenière, M.; Grogan, P.; Walker, V.K. Examination of Soil Microbial Communities After Permafrost Thaw Subsequent to an Active Layer Detachment in the High Arctic. Arct. Antarct. Alp. Res. 2017, 49, 455–472. [Google Scholar] [CrossRef] [Green Version]
- Malard, L.A.; Pearce, D.A. Microbial diversity and biogeography in Arctic soils. Environ. Microbiol. Rep. 2018, 10, 611–625. [Google Scholar] [CrossRef]
- Monteux, S.; Weedon, J.T.; Blume-Werry, G.; Gavazov, K.; Jassey, V.E.J.; Johansson, M.; Keuper, F.; Olid, C.; Dorrepaal, E. Long-term in situ permafrost thaw effects on bacterial communities and potential aerobic respiration. ISME J. 2018, 12, 2129–2141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malard, L.A.; Anwar, M.Z.; Jacobsen, C.S.; Pearce, D.A. Biogeographical patterns in soil bacterial communities across the Arctic region. FEMS Microbiol. Ecol. 2019, 95, fiz128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.; Jonassen, I.; Øvreås, L.; Taş, N. Metagenome-assembled genome distribution and key functionality highlight importance of aerobic metabolism in Svalbard permafrost. FEMS Microbiol. Ecol. 2020, 96, fiaa057. [Google Scholar] [CrossRef]
- Blaud, A.; Lerch, T.Z.; Phoenix, G.K.; Osborn, A.M. Arctic soil microbial diversity in a changing world. Res. Microbiol. 2015, 166, 796–813. [Google Scholar] [CrossRef]
- Lipson, D.A.; Haggerty, J.M.; Srinivas, A.; Raab, T.K.; Sathe, S.; Dinsdale, E.A. Metagenomic Insights into Anaerobic Metabolism along an Arctic Peat Soil Profile. PLoS ONE 2013, 8, e64659. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, A.A.; Kulichevskaya, I.S.; Merkel, A.Y.; Toshchakov, S.; Dedysh, S.N. High Diversity of Planctomycetes in Soils of Two Lichen-Dominated Sub-Arctic Ecosystems of Northwestern Siberia. Front. Microbiol. 2016, 7, 2065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grodnitskaya, I.D.; Trusova, M.Y.; Syrtsov, S.N.; Koroban, N.V. Structure of microbial communities of peat soils in two bogs in Siberian tundra and forest zones. Microbiology 2018, 87, 89–102. [Google Scholar] [CrossRef]
- Schnecker, J.; Wild, B.; Takriti, M.; Alves, R.J.E.; Gentsch, N.; Gittel, A.; Mikutta, R.; Richter, A. Microbial community composition shapes enzyme patterns in topsoil and subsoil horizons along a latitudinal transect in Western Siberia. Soil Biol. Biochem. 2015, 83, 106–115. [Google Scholar] [CrossRef]
- Kim, H.M.; Jung, J.Y.; Yergeau, E.; Hwang, C.; Hinzman, L.; Nam, S.; Hong, S.G.; Kim, O.-S.; Chun, J.; Lee, Y.K. Bacterial community structure and soil properties of a subarctic tundra soil in Council, Alaska. FEMS Microbiol. Ecol. 2014, 89, 465–475. [Google Scholar] [CrossRef]
- Tveit, A.; Schwacke, R.; Svenning, M.M.; Urich, T. Organic carbon transformations in high-Arctic peat soils: Key functions and microorganisms. ISME J. 2013, 7, 299–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tveit, A.T.; Urich, T.; Frenzel, P.; Svenning, M.M. Metabolic and trophic interactions modulate methane production by Arctic peat microbiota in response to warming. Proc. Natl. Acad. Sci. USA 2015, 112, E2507–E2516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martineau, C.; Pan, Y.; Bodrossy, L.; Yergeau, E.; Whyte, L.G.; Greer, C.W. Atmospheric methane oxidizers are present and active in Canadian high Arctic soils. FEMS Microbiol. Ecol. 2014, 89, 257–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.-H.; Jang, I.; Chae, N.; Choi, T.; Kang, H. Organic Layer Serves as a Hotspot of Microbial Activity and Abundance in Arctic Tundra Soils. Microb. Ecol. 2012, 65, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Taş, N.; Prestat, E.; Wang, S.; Wu, Y.; Ulrich, C.; Kneafsey, T.; Tringe, S.G.; Torn, M.; Hubbard, S.S.; Jansson, J.K. Landscape topography structures the soil microbiome in arctic polygonal tundra. Nat. Commun. 2018, 9, 777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vishnivetskaya, T.A.; Buongiorno, J.; Bird, J.; Krivushin, K.; Spirina, E.V.; Oshurkova, V.; Shcherbakova, V.A.; Wilson, G.; Lloyd, K.G.; Rivkina, E.M. Methanogens in the Antarctic Dry Valley permafrost. FEMS Microbiol. Ecol. 2018, 94, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trotsenko, Y.A.; Khmelenina, V.N. Aerobic methanotrophic bacteria of cold ecosystems. FEMS Microbiol. Ecol. 2005, 53, 15–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shcherbakova, V.; Yoshimura, Y.; Ryzhmanova, Y.; Taguchi, Y.; Segawa, T.; Oshurkova, V.; Rivkina, E. Archaeal communities of Arctic methane-containing permafrost. FEMS Microbiol. Ecol. 2016, 92, fiw135. [Google Scholar] [CrossRef] [Green Version]
- Comte, J.; Monier, A.; Crevecoeur, S.; Lovejoy, C.; Vincent, W.F. Microbial biogeography of permafrost thaw ponds across the changing northern landscape. Ecography 2015, 39, 609–618. [Google Scholar] [CrossRef]
- Roiha, T.; Peura, S.; Cusson, M.; Rautio, M. Allochthonous carbon is a major regulator to bacterial growth and community composition in subarctic freshwaters. Sci. Rep. 2016, 6, 34456. [Google Scholar] [CrossRef]
- Peura, S.; Wauthy, M.; Simone, D.; Eiler, A.; Einarsdóttir, K.; Rautio, M.; Bertilsson, S. Ontogenic succession of thermokarst thaw ponds is linked to dissolved organic matter quality and microbial degradation potential. Limnol. Oceanogr. 2019, 65, S248–S263. [Google Scholar] [CrossRef]
- Przytulska, A.; Comte, J.; Crevecoeur, S.; Lovejoy, C.; Laurion, I.; Vincent, W. Phototrophic pigment diversity and picophytoplankton in permafrost thaw lakes. Biogeosciences 2016, 13, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Steven, B.; Léveillé, R.; Pollard, W.H.; Whyte, L.G. Microbial ecology and biodiversity in permafrost. Extremophiles 2006, 10, 259–267. [Google Scholar] [CrossRef]
- Liebner, S.; Harder, J.; Wagner, D. Bacterial diversity and community structure in polygonal tundra soils from Samoylov Island, Lena Delta, Siberia. Int. J. Microbiol. 2008, 1, 195–202. [Google Scholar]
- Barbier, B.A.; Dziduch, I.; Liebner, S.; Ganzert, L.; Lantuit, H.; Pollard, W.; Wagner, D. Methane-cycling communities in a permafrost-affected soil on Herschel Island, Western Canadian Arctic: Active layer profiling ofmcrAandpmoAgenes. FEMS Microbiol. Ecol. 2012, 82, 287–302. [Google Scholar] [CrossRef] [Green Version]
- Ganzert, L.; Bajerski, F.; Wagner, D. Bacterial community composition and diversity of five different permafrost-affected soils of Northeast Greenland. FEMS Microbiol. Ecol. 2014, 89, 426–441. [Google Scholar] [CrossRef]
- Zhou, L.; Zhou, Y.; Yao, X.; Cai, J.; Liu, X.; Tang, X.; Zhang, Y.; Jang, K.S.; Jeppesen, E. Decreasing diversity of rare bacterial subcommunities relates to dissolved organic matter along permafrost thawing gradients. Environ. Int. 2020, 134, 105330. [Google Scholar] [CrossRef] [PubMed]
- Coolen, M.J.L.; Van De Giessen, J.; Zhu, E.Y.; Wuchter, C. Bioavailability of soil organic matter and microbial community dynamics upon permafrost thaw. Environ. Microbiol. 2011, 13, 2299–2314. [Google Scholar] [CrossRef] [PubMed]
- Reimer, P.J.; Austin, W.E.N.; Bard, E.; Bayliss, A.; Blackwell, P.G.; Ramsey, C.B.; Butzin, M.; Cheng, H.; Edwards, R.L.; Friedrich, M.; et al. The IntCal20 Northern Hemisphere radiocarbon age calibration curve (0–55 cal kBP). Radiocarbon 2020, 62, 1–33. [Google Scholar] [CrossRef]
- Stepanova, V.M.; Pokrovsky, O.S.; Viers, J.; Mironycheva-Tokareva, N.P.; Kosykh, N.P.; Vishnyakova, E.K. Major and trace elements in peat profiles in Western Siberia: Impact of the landscape context, latitude and permafrost coverage. Appl. Geochem. 2015, 53, 53–70. [Google Scholar] [CrossRef]
- Koroleff, F. Determination of phosphorus. In Methods for Seawater Analysis; Grasshoff, K., Ehrhardt, M., Kremling, K., Eds.; Wiley-VCH Verlag GmbH: Weinheim, Germany, 1983; pp. 125–136. [Google Scholar]
- Raudina, T.V.; Loiko, S.V.; Lim, A.; Manasypov, R.M.; Shirokova, L.S.; Istigechev, G.I.; Kuzmina, D.M.; Kulizhsky, S.P.; Vorobyev, S.N.; Pokrovsky, O.S. Permafrost thaw and climate warming may decrease the CO2, carbon, and metal concentration in peat soil waters of the Western Siberia Lowland. Sci. Total. Environ. 2018, 634, 1004–1023. [Google Scholar] [CrossRef]
- Lim, A.G.; Loiko, S.V.; Kuzmina, D.M.; Krickov, I.V.; Shirokova, L.S.; Kulizhsky, S.P.; Vorobyev, S.N.; Pokrovsky, O.S. Dispersed ground ice of permafrost peatlands: Potential unaccounted carbon, nutrient and metal sources. Chemosphere 2020, 266, 128953. [Google Scholar] [CrossRef]
- Massiot, D.; Fayon, F.; Capron, M.; King, I.; Le Calvé, S.; Alonso, B.; Durand, J.O.; Bujoli, B.; Gan, Z.; Hoatson, G. Modelling one- and two-dimensional solid-state NMR spectra. Magn. Reson. Chem. 2002, 40, 70–76. [Google Scholar] [CrossRef]
- Kögel-Knabner, I. 13C and 15N NMR spectroscopy as a tool in soil organic matter studies. Geoderma 1997, 80, 243–270. [Google Scholar] [CrossRef]
- Dymov, A.; Zhangurov, E.; Hagedorn, F. Soil organic matter composition along altitudinal gradients in permafrost affected soils of the Subpolar Ural Mountains. Catena 2015, 131, 140–148. [Google Scholar] [CrossRef]
- Gundelwein, A.; Müller-Lupp, T.; Sommerkorn, M.; Haupt, E.T.; Pfeiffer, E.M.; Wiechmann, H. Carbon in tundra soils in the Lake Labaz region of arctic Siberia. Eur. J. Soil Sci. 2007, 58, 1164–1174. [Google Scholar] [CrossRef] [Green Version]
- Maksimova, V.; Klavina, L.; Bikovens, O.; Zicmanis, A.; Purmalis, O. Structural Characterization and Chemical Classification of Some Bryophytes Found in Latvia. Chem. Biodivers. 2013, 10, 1284–1294. [Google Scholar] [CrossRef] [PubMed]
- Amundson, R.; Austin, A.T.; Schuur, E.A.G.; Yoo, K.; Matzek, V.; Kendall, C.; Uebersax, A.; Brenner, D.; Baisden, W.T. Global patterns of the isotopic composition of soil and plant nitrogen. Glob. Biogeochem. Cycles 2003, 17. [Google Scholar] [CrossRef]
- Craine, J.M.; Brookshire, E.N.J.; Cramer, M.D.; Hasselquist, N.J.; Koba, K.; Marin-Spiotta, E.; Wang, L. Ecological interpretations of nitrogen isotope ratios of terrestrial plants and soils. Plant Soil 2015, 396, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Parinkina, O.M. Microflora of Tundra Soils: Ecologico-Geographical Features and Productivity; Nauka: St. PetersburgLeningrad, Russia, 1989; 159p. [Google Scholar]
- Cowan, D.A.; Russell, N.J.; Mamais, A.; Sheppard, D.M. Antarctic Dry Valley mineral soils contain unexpectedly high levels of microbial biomass. Extremophiles 2002, 6, 431–436. [Google Scholar] [CrossRef] [Green Version]
- Altshuler, I.; Goordial, J.; Whyte, L.G. Microbial life in permafrost. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Schinner, F., Marx, J.C., Gerday, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 153–179. [Google Scholar]
- Jansson, J.; Taş, N. The microbial ecology of permafrost. Nat. Rev. Microbiol. 2014, 12, 414–425. [Google Scholar] [CrossRef]
- Mackelprang, R.; Burkert, A.; Haw, M.; Mahendrarajah, T.; Conaway, C.H.; Douglas, T.A.; Waldrop, M.P. Microbial survival strategies in ancient permafrost: Insights from metagenomics. ISME J. 2017, 11, 2305–2318. [Google Scholar] [CrossRef] [Green Version]
- Frank-Fahle, B.A.; Yergeau, É.; Greer, C.W.; Lantuit, H.; Wagner, D. Microbial Functional Potential and Community Composition in Permafrost-Affected Soils of the NW Canadian Arctic. PLoS ONE 2014, 9, e84761. [Google Scholar] [CrossRef] [Green Version]
- Tuorto, S.J.; Darias, P.; McGuinness, L.R.; Panikov, N.; Zhang, T.; Haggblom, M.; Kerkhof, L.J. Bacterial genome replication at subzero temperatures in permafrost. ISME J. 2013, 8, 139–149. [Google Scholar] [CrossRef] [Green Version]
- Huhe; Chen, X.; Hou, F.; Wu, Y.; Cheng, Y. Bacterial and Fungal Community Structures in Loess Plateau Grasslands with Different Grazing Intensities. Front. Microbiol. 2017, 8, 606. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.M.; Souza, R.C.; Barcellos, F.G.; Hungria, M.; Vasconcelos, A.T.R. Genomic and evolutionary comparisons of diazotrophic and pathogenic bacteria of the order Rhizobiales. BMC Microbiol. 2010, 10, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erlacher, A.; Cernava, T.; Cardinale, M.; Soh, J.; Sensen, C.W.; Grube, M.; Berg, G. Rhizobiales as functional and endosymbiontic members in the lichen symbiosis of Lobaria pulmonaria L. Front. Microbiol. 2015, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Oter, R.; Nakano, R.T.; Dombrowski, N.; Ma, K.-W.; Tea, T.A.; McHardy, A.C.; Schulze-Lefert, P. Modular Traits of the Rhizobiales Root Microbiota and Their Evolutionary Relationship with Symbiotic Rhizobia. Cell Host Microbe 2018, 24, 155–167.e5. [Google Scholar] [CrossRef] [Green Version]
- Cole, J.K.; Morton, B.R.; Cardamone, H.C.; Lake, H.R.R.; Dohnalkova, A.C.; Kim, Y.-M.; Romine, M.F.; Kyle, J.E.; Maezato, Y.; Dana, K.L.; et al. Salinivirga fredricksonii gen. nov., sp. nov., a heterotrophic halophile isolated from a photosynthetic mat, a member of a novel lineage (Salinarimonadaceae fam. nov.) within the order Rhizobiales, and reclassification of the genus Salinarimonas Liu et al. 2010 into Salinarimonadaceae. Int. J. Syst. Evol. Microbiol. 2018, 68, 1591–1598. [Google Scholar] [CrossRef]
- Taylor, B.N.; Simms, E.L.; Komatsu, K.J. More Than a Functional Group: Diversity within the Legume–Rhizobia Mutualism and Its Relationship with Ecosystem Function. Divers. 2020, 12, 50. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Meade, A.; Lam, H.-M.; Luo, H. Evolutionary Timeline and Genomic Plasticity Underlying the Lifestyle Diversity in Rhizobiales. mSystems 2020, 5, e00438-20. [Google Scholar] [CrossRef] [PubMed]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; Van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef] [Green Version]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lladó, S.; Žifčáková, L.; Větrovský, T.; Eichlerová, I.; Baldrian, P. Functional screening of abundant bacteria from acidic forest soil indicates the metabolic potential of Acidobacteria subdivision 1 for polysaccharide decomposition. Biol. Fertil. Soils 2016, 52, 251–260. [Google Scholar] [CrossRef]





| Depth, cm | Substrate | Average Depth, cm | Average T, °C | Minimal T, °C | Maximal T, °C | Peat, Bulk Density, g/cm3 | Degree of Decomposition, % |
|---|---|---|---|---|---|---|---|
| 10–20 | peat | −15 | −0.047 | −8.4 | 11.3 | 0.16 | 5 |
| 30–40 | peat | −35 | −0.63 | −6.1 | 4.4 | 0.29 | 10 |
| 50–60 | peat | −55 | −0.8 | −4.1 | 0.2 | 0.29 | 17 |
| 70–80 | peat | −75 | −0.74 | −2.8 | −0.1 | 0.23 | 30 |
| 90–100 | peat | −95 | −0.65 | −2.3 | −0.1 | 0.23 | 32 |
| 100–110 | mineral | −105 | −0.58 | −1.3 | −0.1 | 1.30 | |
| Depth, cm | substrate | N, % | C, % | C:N | L* by CIE lab | a* by CIE lab | b* by CIE lab |
| 10–20 | peat | 0.79 | 44.6 | 56.1 | 43.7 | 9.9 | 16.5 |
| 30–40 | peat | 0.61 | 46.0 | 75.4 | 39.2 | 9.1 | 17.5 |
| 50–60 | peat | 0.83 | 40.6 | 48.9 | 33.2 | 8.7 | 17.5 |
| Depth, cm | substrate | N, % | C, % | C:N | L* by CIE lab | a* by CIE lab | b* by CIE lab |
| 70–80 | peat | 1.36 | 56.1 | 41.3 | 25.9 | 7.8 | 14.0 |
| 90–100 | peat | 0.84 | 54.41 | 64.8 | 25.3 | 6.9 | 12.3 |
| 100–110 | mineral | 0.03 | 1.905 | 63.5 | 66.2 | 3.9 | 10.6 |
| Depth, cm | substrate | S, % | P-lab, mg/kg | δ13C | δ15N | Alkyl C | O-alkyl C |
| 10–20 | peat | 0.38 | 0.76 | −24.7 | 4.4 | 24.05 | 71.9 |
| 30–40 | peat | 0.38 | 0.34 | −25.9 | 4.8 | 15.3 | 77.3 |
| 50–60 | peat | 0.29 | 0.06 | −26.4 | 4.25 | 26.2 | 63.9 |
| 70–80 | peat | 0.39 | 0.04 | −26.9 | 3.7 | 37.1 | 50.5 |
| 90–100 | peat | 0.22 | 0.03 | −26.2 | 4.1 | 47.1 | 39.3 |
| 100–110 | mineral | 0.05 | 0.00 | −26.9 | |||
| Depth, cm | substrate | aromatic C | carboxylic C | Li peat | B peat | Na peat | Mg peat |
| ppb | |||||||
| 10–20 | peat | 1.6 | 2.3 | 577 | 4518 | 372,422 | 296,970 |
| 30–40 | peat | 5.2 | 2.2 | 294 | 4544 | 103,287 | 327,233 |
| 50–60 | peat | 7.8 | 2.1 | 498 | 4434 | 72,538 | 194,873 |
| 70–80 | peat | 10.4 | 2.0 | 294 | 4892 | 29,929 | 169,643 |
| 90–100 | peat | 10.1 | 3.5 | 424 | 11,608 | 83,755 | 145,879 |
| 100–110 | mineral | 2441 | 3359 | 235,580 | 74,206 | ||
| Depth, cm | substrate | Al peat | P peat | K peat | Ca peat | Ti peat | V peat |
| ppb | |||||||
| 10–20 | peat | 2,302,515 | 448,947 | 938,435 | 1,049,413 | 183,028 | 3059 |
| 30–40 | peat | 1,392,605 | 295,471 | 232,556 | 1,525,343 | 52,626 | 1199 |
| 50–60 | peat | 1,813,247 | 209,406 | 284,074 | 911,927 | 86,250 | 1987 |
| 70–80 | peat | 1,925,618 | 200,513 | 192,876 | 921,297 | 59,999 | 1936 |
| 90–100 | peat | 2,797,844 | 172,804 | 327,006 | 832,506 | 116,491 | 2942 |
| 100–110 | mineral | 1,748,772 | 22,357 | 991,020 | 279,474 | 218,869 | 1677 |
| Depth, cm | substrate | Cr peat | Mn peat | Fe peat | Co peat | Ni peat | Cu peat |
| ppb | |||||||
| 10–20 | peat | 2504 | 10,234 | 1,311,464 | 281 | 1169 | 2138 |
| 30–40 | peat | 1966 | 5228 | 1,433,817 | 426 | 1083 | 594 |
| 50–60 | peat | 2745 | 5699 | 472,019 | 218 | 942 | 1152 |
| 70–80 | peat | 2793 | 2139 | 451,913 | 284 | 1841 | 2718 |
| 90–100 | peat | 3871 | 5225 | 519,426 | 522 | 1573 | 5064 |
| 100–110 | mineral | 1895 | 8516 | 290,073 | 177 | 263 | 914 |
| Depth, cm | substrate | Zn peat | As peat | Rb peat | Sr peat | Y peat | Zr peat |
| ppb | |||||||
| 10–20 | peat | 15,946 | 1354 | 3086 | 9573 | 618 | 3614 |
| 30–40 | peat | 15,957 | 315 | 748 | 13,479 | 345 | 1356 |
| Depth, cm | substrate | Zn peat | As peat | Rb peat | Sr peat | Y peat | Zr peat |
| ppb | |||||||
| 50–60 | peat | 7367 | 261 | 1185 | 8224 | 463 | 2777 |
| 70–80 | peat | 9325 | 391 | 694. | 8406 | 332 | 1843 |
| 90–100 | peat | 17,238 | 380 | 1084 | 8394 | 414 | 5941 |
| 100–110 | mineral | 2355 | 299 | 2854 | 7518 | 913 | 14,104 |
| Depth, cm | substrate | Nb peat | Mo peat | Cd peat | Sb peat | Cs peat | Ba peat |
| ppb | |||||||
| 10–20 | peat | 476 | 155 | 130 | 260 | 189 | 27,243 |
| 30–40 | peat | 462 | 87 | 132 | 39 | 46 | 20,066 |
| 50–60 | peat | 863 | 88 | 71 | 29 | 72 | 22,801 |
| 70–80 | peat | 579 | 211 | 86 | 42 | 41 | 29,938 |
| 90–100 | peat | 887 | 108 | 72 | 37 | 65 | 39,452 |
| 100–110 | mineral | 924 | 48 | 14 | 139 | 83 | 69,278 |
| Depth, cm | substrate | La peat | Ce peat | Dy peat | Yb peat | Pb peat | U peat |
| ppb | |||||||
| 10–20 | peat | 1184 | 2365 | 120 | 69 | 6485 | 80 |
| 30–40 | peat | 537 | 1167 | 72 | 32 | 1009 | 50 |
| 50–60 | peat | 775 | 1559 | 90 | 54 | 647 | 92 |
| 70–80 | peat | 534 | 1090 | 65 | 35 | 532 | 88 |
| 90–100 | peat | 727 | 1413 | 83 | 47 | 837 | 111 |
| 100–110 | mineral | 850 | 1498 | 108 | 86 | 1036 | 162 |
| Depth, cm | substrate | Humidity, % | |||||
| 10–20 | peat | 317 | |||||
| 30–40 | peat | 337 | |||||
| 50–60 | peat | 700 | |||||
| 70–80 | peat | 458 | |||||
| 90–100 | peat | 650 | |||||
| 100–110 | mineral | 82 | |||||
| Depth | δ13C | δ15N | Alkyl C | O-Alkyl C | Aromatic C | Carbonyl C | Alkyl C: O-Alkyl C |
|---|---|---|---|---|---|---|---|
| ‰ | ‰ | % | |||||
| (0–5) | −23.6 | 2.8 | 16.7 | 79.8 | 0.0 | 3.5 | 0.21 |
| (19–21) | −25.8 | 6.0 | 31.4 | 64.1 | 3.3 | 1.2 | 0.49 |
| (30–35) | −25.9 | 4.8 | 15.3 | 77.3 | 5.2 | 2.2 | 0.20 |
| (65–75) | −26.7 | 3.3 | 30.5 | 57.5 | 10.4 | 1.7 | 0.53 |
| (75–85) | −27.1 | 4.1 | 43.7 | 43.5 | 10.4 | 2.4 | 1.00 |
| (85–95) | −26.2 | 4.1 | 47.1 | 39.3 | 10.1 | 3.5 | 1.20 |
| (105–115) | −26.9 | n.m * | n.m * | n.m * | n.m * | n.m * | n.m * |
| Abbreviation | Taxon Unit (Phylum: Class) | Peat Horizon Depth, cm | |||||
|---|---|---|---|---|---|---|---|
| 10–20 | 30–40 | 50–60 | 70–80 | 90–100 | 100–110 | ||
| Share of 16S rRNA Gene Sequences in % of the Number of Obtained Sequences | |||||||
| EuMeth | Euryarchaeota; Methanobacteria | 0.0 | 0.1 | 0.0 | 0.0 | 0.3 | 0.0 |
| ThNit | Thaumarchaeota; Nitrososphaeria | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.1 |
| Ac | Acidobacteria; Acidobacteriia | 20.5 | 46.0 | 64.7 | 44.8 | 1.0 | 4.9 |
| Ac6 | Acidobacteria; Subgroup 6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.4 | 0.1 |
| AcAci | Actinobacteria; Acidimicrobiia | 2.1 | 2.8 | 1.3 | 0.1 | 0.6 | 0.1 |
| AcAct | Actinobacteria; Actinobacteria | 10.6 | 1.8 | 12.1 | 0.4 | 18.4 | 1.0 |
| AcOt | Actinobacteria; Others | 0.6 | 20.7 | 4.6 | 0.3 | 0.2 | 0.2 |
| AcTh | Actinobacteria; Thermoleophilia | 2.1 | 0.4 | 1.8 | 1.1 | 1.1 | 0.3 |
| Ar | Armatimonadetes; Armatimonadia | 0.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| ArC | Armatimonadetes; Chthonomonadetes | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| ArFi | Armatimonadetes; Fimbriimonadia | 0.6 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 |
| Bac | Bacteroidetes; Bacteroidia | 2.6 | 0.0 | 0.2 | 0.0 | 6.1 | 0.4 |
| Chl | Chloroflexi; Chloroflexia | 0.0 | 0.0 | 0.0 | 0.0 | 0.7 | 0.0 |
| CyOt | Cyanobacteria; Oxyphotobacteria | 0.0 | 0.0 | 0.0 | 0.0 | 0.3 | 0.0 |
| FirB | Firmicutes; Bacilli | 0.0 | 0.0 | 0.3 | 0.0 | 10.7 | 0.7 |
| FirCl | Firmicutes; Clostridia | 0.0 | 0.0 | 0.0 | 0.7 | 4.9 | 1.8 |
| Gem | Gemmatimonadetes; Gemmatimonadetes | 0.0 | 0.0 | 0.0 | 0.0 | 0.6 | 0.0 |
| GLon | Gemmatimonadetes; Longimicrobia | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 |
| Ot | Others; Others | 13.3 | 17.5 | 2.1 | 0.6 | 1.9 | 0.4 |
| PatS | Patescibacteria; Saccharimonadia | 0.0 | 0.6 | 1.3 | 0.0 | 0.1 | 0.0 |
| PlPh | Planctomycetes; Phycisphaerae | 3.9 | 0.3 | 0.0 | 0.0 | 0.2 | 0.0 |
| PlPl | Planctomycetes; Planctomycetacia | 5.7 | 0.0 | 0.7 | 0.0 | 0.7 | 0.0 |
| ProA | Proteobacteria; Alphaproteobacteria | 9.4 | 6.3 | 9.7 | 50.2 | 24.6 | 1.7 |
| ProD | Proteobacteria; Deltaproteobacteria | 2.9 | 0.6 | 0.0 | 0.0 | 1.6 | 0.1 |
| ProG | Proteobacteria; Gammaproteobacteria | 12.7 | 1.6 | 1.1 | 1.8 | 17.7 | 87.5 |
| ProOt | Proteobacteria; Others | 1.2 | 0.0 | 0.0 | 0.0 | 0.4 | 0.0 |
| Ver | Verrucomicrobia; Verrucomicrobiae | 10.3 | 1.0 | 0.1 | 0.0 | 5.3 | 0.3 |
| Parameter | Abbreviated in Figure 6 | F.1 (52.0%) | Parameter | Abbreviated in Figure 6 | F.2 (27.3%) | Parameter | Abbreviated in Figure 6 | F.3 (13.2%) |
|---|---|---|---|---|---|---|---|---|
| Coordinate a* according to CIE lab (red color) | a* | −0.96 | Verrucomicrobia; Verrucomicrobiae | * Ver | −0.99 | N peat | N | −0.86 |
| Depth peat (negative values) | Depth | −0.94 | Proteobacteria; Deltaproteobacteria | * ProD | −0.98 | Mo peat | MoP | −0.86 |
| O-alkyl C | COalk | −0.93 | Ti peat | TiP | −0.95 | P ice water | PW | −0.74 |
| SUVA peat ice/water | IW SUVA | −0.88 | Proteobacteria; Others | * ProOt | −0.95 | Sr peat | SrP | 0.74 |
| Coordinate b* according to CIE lab (yellow color) | b* | −0.86 | Armatimonadetes; Fimbriimonadia | * ArFi | −0.89 | Actinobacteria; Others | * AcOt | 0.81 |
| L* CIE lab | L* | −0.85 | K peat | KP | −0.88 | δ15N | 15N | 0.84 |
| Mg peat | MgP | −0.82 | La peat | LaP | −0.86 | C/N | C/N | 0.93 |
| S peat | S | −0.82 | Proteobacteria; Gammaproteobacteria | * ProG | −0.85 | |||
| P peat | PP | −0.77 | Rb peat | RbP | −0.85 | |||
| Cd peat | CdP | −0.75 | V peat | VP | −0.85 | |||
| Plab peat | Plab | −0.71 | Cs peat | CsP | −0.85 | |||
| Maximum temperature in the horizon | MaxAn | −0.70 | Planctomycetes; Phycisphaerae | * PlPh | −0.85 | |||
| Zr peat | ZrP | 0.71 | Mean temperature in the horizon | MeAn | −0.85 | |||
| Mo ice water | MoW | 0.72 | Ce peat | CeP | −0.84 | |||
| Nb peat | NbP | 0.76 | Mn peat | MnP | −0.84 | |||
| Be ice water | BeW | 0.76 | Na peat | NaP | −0.84 | |||
| U peat | UP | 0.76 | Dy peat | DyP | −0.84 | |||
| aromatic C | Carom | 0.79 | δ13C peat | 13C | −0.84 | |||
| B ice water | BW | 0.80 | pH ice water | IW pH | −0.84 | |||
| Euryarchaeota; Methanobacteria | * EuMeth | 0.81 | Y peat | YP | −0.82 | |||
| As ice water | AsW | 0.81 | Planctomycetes; Planctomycetacia | * PlPh | −0.82 | |||
| carboxylic C | Ccarb | 0.82 | Pb peat | PbP | −0.82 | |||
| Dy ice water | DyW | 0.82 | As peat | AsP | −0.81 | |||
| Ce ice water | CeW | 0.83 | Armatimonadetes; Chthonomonadetes | * ArC | −0.80 | |||
| Table 3, continued. | ||||||||
| Parameter | Abbreviated in Figure 6 | F.1 (52.0%) | Parameter | Abbreviated in Figure 6 | F.2 (27.3%) | Parameter | Abbreviated in Figure 6 | F.3 (13.2%) |
| Y ice water | YW | 0.83 | Armatimonadetes; Armatimonadia | * Ar | −0.80 | |||
| Minimum temperature in the horizon | MinAn | 0.84 | Sb peat | SbP | −0.79 | |||
| Sb ice water | SbW | 0.84 | Yb peat | YbP | −0.77 | |||
| Ba peat | BaP | 0.85 | Li peat | LiP | −0.74 | |||
| Cu ice water | CuW | 0.85 | Bacteroidetes; Bacteroidia | * Bac | −0.71 | |||
| La ice water | LaW | 0.85 | Acidobacteria; Acidobacteriia | * Ac | 0.76 | |||
| Cu peat | CuP | 0.86 | Bulk density peat | BD | 0.79 | |||
| U ice water | UW | 0.88 | DIC ice water | IW DIC | 0.83 | |||
| Decomposition of peat | DP | 0.89 | UV254 | UV254 | 0.84 | |||
| Alkyl C | Calkyl | 0.90 | ||||||
| Nb ice water | NbW | 0.90 | ||||||
| Cyanobacteria; Oxyphotobacteria | * CyOt | 0.91 | ||||||
| Gemmatimonadetes; Gemmatimonadetes | * Gem | 0.91 | ||||||
| Thaumarchaeota; Nitrososphaeria | * ThNit | 0.91 | ||||||
| Acidobacteria; Subgroup 6 | * Ac6 | 0.91 | ||||||
| Chloroflexi; Chloroflexia | * Chl | 0.91 | ||||||
| Zr ice peat | ZrW | 0.91 | ||||||
| Yb ice peat | YbW | 0.91 | ||||||
| Firmicutes; Bacilli | * FirB | 0.91 | ||||||
| Cr ice peat | CrW | 0.92 | ||||||
| B peat | BP | 0.92 | ||||||
| Cr peat | CrP | 0.92 | ||||||
| Ti ice peat | TiW | 0.93 | ||||||
| Specific conductivity of peat ice waters | IW EC | 0.93 | ||||||
| Mn ice peat | MnW | 0.93 | ||||||
| Ba ice peat | BaW | 0.94 | ||||||
| Firmicutes; Clostridia | * FirCl | 0.94 | ||||||
| Parameter | Abbreviated in Figure 6 | F.1 (52.0%) | ||||||
| V ice peat | VW | 0.95 | ||||||
| Cs ice peat | CsW | 0.95 | ||||||
| Na ice peat | NaW | 0.96 | ||||||
| Ca ice peat | CaW | 0.96 | ||||||
| Li ice peat | LiW | 0.96 | ||||||
| Fe ice peat | FeW | 0.96 | ||||||
| Mg ice peat | MgW | 0.97 | ||||||
| Cd ice peat | CdW | 0.97 | ||||||
| K ice peat | KW | 0.97 | ||||||
| Rb ice peat | RbW | 0.98 | ||||||
| Zn ice peat | ZnW | 0.98 | ||||||
| Co ice peat | CoW | 0.99 | ||||||
| Ni ice peat | NiW | 0.99 | ||||||
| Sr ice peat | SrW | 0.99 | ||||||
| DOC ice peat | IW DOC | 0.99 | ||||||
| Si ice peat | SiW | 1.00 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aksenov, A.S.; Shirokova, L.S.; Kisil, O.Y.; Kolesova, S.N.; Lim, A.G.; Kuzmina, D.; Pouillé, S.; Alexis, M.A.; Castrec-Rouelle, M.; Loiko, S.V.; et al. Bacterial Number and Genetic Diversity in a Permafrost Peatland (Western Siberia): Testing a Link with Organic Matter Quality and Elementary Composition of a Peat Soil Profile. Diversity 2021, 13, 328. https://doi.org/10.3390/d13070328
Aksenov AS, Shirokova LS, Kisil OY, Kolesova SN, Lim AG, Kuzmina D, Pouillé S, Alexis MA, Castrec-Rouelle M, Loiko SV, et al. Bacterial Number and Genetic Diversity in a Permafrost Peatland (Western Siberia): Testing a Link with Organic Matter Quality and Elementary Composition of a Peat Soil Profile. Diversity. 2021; 13(7):328. https://doi.org/10.3390/d13070328
Chicago/Turabian StyleAksenov, Andrey S., Liudmila S. Shirokova, Oksana Ya. Kisil, Sofiia N. Kolesova, Artem G. Lim, Darya Kuzmina, Sophie Pouillé, Marie A. Alexis, Maryse Castrec-Rouelle, Sergey V. Loiko, and et al. 2021. "Bacterial Number and Genetic Diversity in a Permafrost Peatland (Western Siberia): Testing a Link with Organic Matter Quality and Elementary Composition of a Peat Soil Profile" Diversity 13, no. 7: 328. https://doi.org/10.3390/d13070328
APA StyleAksenov, A. S., Shirokova, L. S., Kisil, O. Y., Kolesova, S. N., Lim, A. G., Kuzmina, D., Pouillé, S., Alexis, M. A., Castrec-Rouelle, M., Loiko, S. V., & Pokrovsky, O. S. (2021). Bacterial Number and Genetic Diversity in a Permafrost Peatland (Western Siberia): Testing a Link with Organic Matter Quality and Elementary Composition of a Peat Soil Profile. Diversity, 13(7), 328. https://doi.org/10.3390/d13070328