Abstract
COLMENA is a microbial culture collection dedicated to the characterization, classification, preservation, and transferal of native microorganisms isolated from various agro-systems and other ecosystems in Mexico. This collection aims to protect microbial diversity, reducing soil degradation, but also exploiting its agro-biotechnological potential. So far, COLMENA has isolated and cryopreserved soil microorganisms from different crops in two major agricultural regions in Mexico, the Yaqui Valley, Sonora, and the Fuerte Valley, Sinaloa. COLMENA has specialized in the identification and characterization of microbial strains with metabolic capacities related to the promotion of plant growth and the biocontrol of phytopathogens. Thus, COLMENA has identified several promising plant growth-promoting microbial (PGPM) strains due to their metabolic and genetic potentials and their beneficial effects in vivo and field trials. These findings demonstrate the biotechnological potential of these strains for their future use in profitable agricultural alternatives focused on enhancing global food security. To share the knowledge and results of the COLMENA team’s scientific research, a virtual platform was created, where the database of the studied and preserved microorganisms is available to professionals, researchers, agricultural workers, and anyone who is interested.
1. Introduction
One of the most pressing challenges that humanity currently faces is global food security, which is threatened by the effects of climate change, the incidence of pests and diseases, the high cost of fertilizers, soil degradation, and loss of fertility [1]. According to the Intergovernmental Panel on Climate Change, projections estimate that global temperatures may rise 1.5 °C by 2040, 2 °C by 2065, and 4 °C by 2100 [2,3]. Several studies have found that climate change causes alterations in plant growth, transpiration, respiration, and photosynthesis rates, which in turn result in a significant decrease in crop yields from the 2030s onwards [4,5]. In the same manner, climate change alters the host-pathogen-environment interaction and increases the incidence of pests and diseases [6,7], which are responsible for the 20–40% decrease in agricultural production [8].
On the other hand, it is projected that the world’s population will increase to almost 10 billion people by 2050, requiring an increase of more than 50% in crop production to satisfy the food demand [9]. Since the green revolution, intensive agricultural practices have been used to increase the productivity of crops, which are characterized by the application of high doses of agrochemicals, large-scale irrigation systems, and new high-yielding disease-resistant varieties [10,11]. Thus, intensive agriculture consumes a large volume of inputs and has contributed to increasing environmental problems such as deforestation, water scarcity, eutrophication, and soil degradation [10].
Soil is of vital importance due to its ability to provide multiple ecosystem services, such as: (a) food, fiber, and energy production; (b) water and nutrient cycling; (c) regulation of climate and greenhouse gases; (d) biological habitat; (e) genetic reserve; and (f) contributing to food security, social and ecological sustainability [12,13,14,15]. Soil harbors the largest population and diversity of microorganisms, which are involved in 80–90% of the processes that occur within the soil [16] and are an important component involved in the maintenance of its fertility [17]. However, it is estimated that 52% of global agricultural land is moderately or severely degraded while rates of soil degradation are increasing [18]. Soil degradation can disturb microbial communities causing loss of genetic and functional diversity of soils and therefore, their fertility [19,20]. Studies have been conducted where the link between microbial diversity and soil fertility was demonstrated [21,22,23]. Lisuma et al. (2020) showed that bacterial diversity was positively correlated with macronutrients (S, P, N, and K) and soil pH, which is a determining factor in the availability of nutrients for plants [24]. Similarly, Lei et al. (2020) reported that the bacterial and fungal community structures in the rhizosphere are positively correlated with the available phosphorus, total nitrogen, sucrose, and soil organic matter [25].
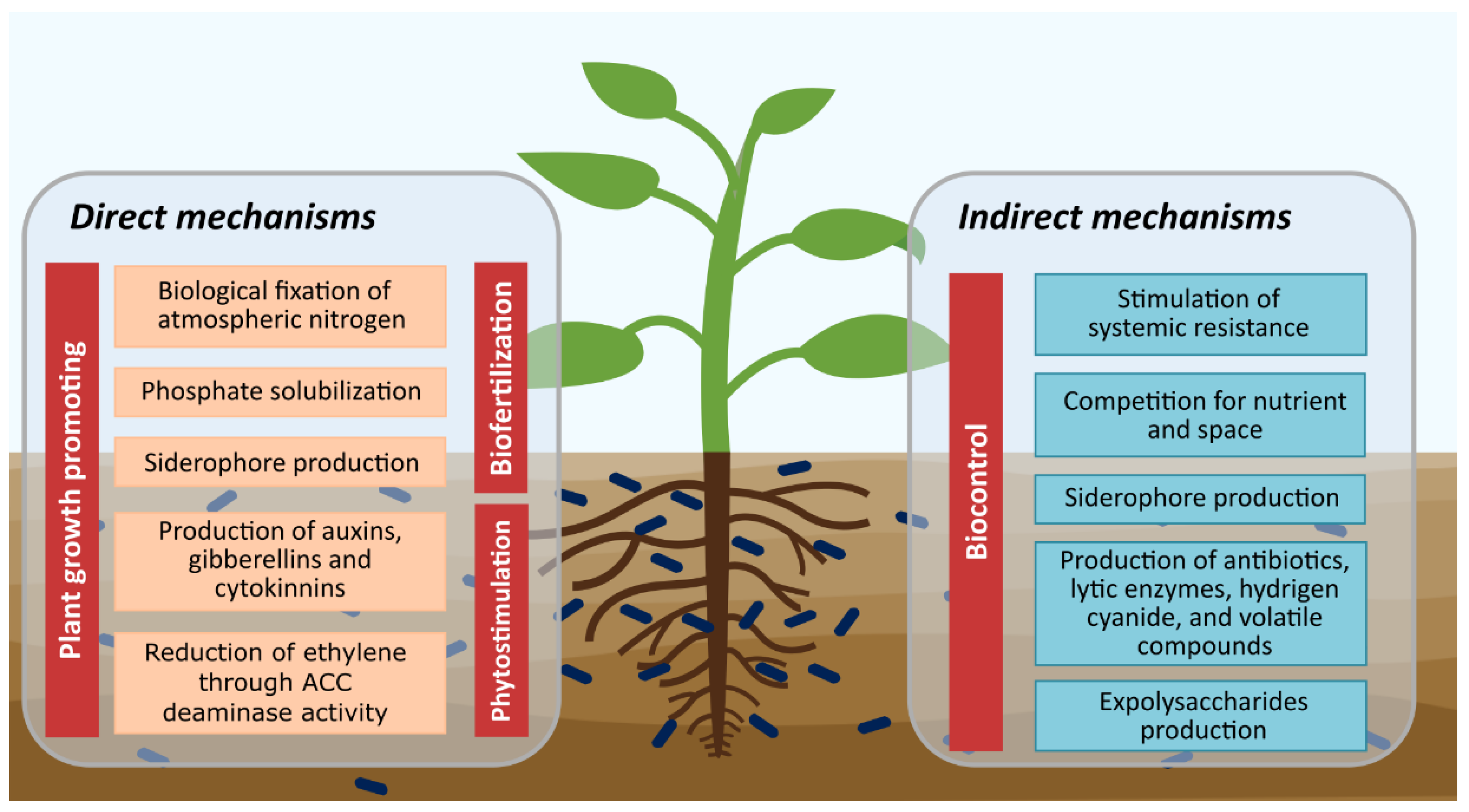
Soil microorganisms contribute to the sustainability of agroecosystems, especially plant growth-promoting microorganisms (PGPM), since they increase crop growth and health, by improving the acquisition of nutrients by plants, mitigating biotic and abiotic stress, and protecting against pests and diseases by various mechanisms [17,26]. Currently, PGPM are used as microbial inoculants for biofertilization through direct mechanisms, such as biological nitrogen fixation, solubilization of organic and inorganic phosphates, and siderophore production. In addition, there are direct mechanisms that mediate the phytostimulation of plants, including phytohormone production such as indole acetic acid, gibberellins, and cytokinins, as well as the production of ACC deaminase that can decrease the stress generated by ethylene in plants. The indirect mechanisms are involved in the biocontrol of phytopathogens including stimulation of systemic resistance, the competition for nutrients and space, competition for iron through the production of siderophores, production of antibiotics, lytic enzymes, hydrogen cyanide, and exopolysaccharides production (Figure 1) [1,27,28,29].
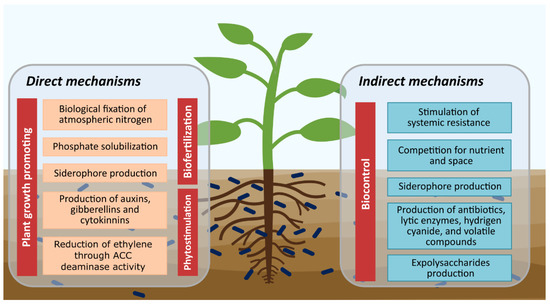
Figure 1.
Plant growth-promoting and biocontrol mechanisms by microorganisms.
Therefore, due to the modification of the native microbial communities of agroecosystems and their metabolic and genetic potentials, the isolation, characterization, and protection of these microorganisms in certified microbial culture collections is essential. The ex situ preservation of microbial diversity associated with crops will further explore the microbial ecology of the current agroecosystems and safeguard their agro-biotechnological potential. However, to promote the study and the extensive use of beneficial microorganisms (such as PGPM), the digitization and dissemination of the biological information of each preserved strain is necessary, as well as easy access for the scientific community, farmers, public-private sector, and any interested person [1,13].
This review aims to describe the Culture Collection of Native Soil and Endophytic Microorganisms (COLMENA) located in Mexico, which is a microbial culture collection that focuses on the characterization, classification, preservation, and transferal of native microorganisms isolated from various agro-systems and other ecosystems, as well as to provide information about its positive impact on food security, and to describe the virtual COLMENA platform for the first time.
2. Culture Collection of Native Soil and Endophytic Microorganisms (COLMENA)
The mission of the Culture Collection of Native Soil and Endophytic Microorganisms (COLMENA) (www.itson.mx/colmena, accessed on 21 July 2021) is to reduce microbial diversity loss related to intensive agricultural practices adopted in Mexican agricultural systems and other ecosystems. COLMENA is dedicated to the preservation of microorganisms as a soil conservation strategy, through the isolation, safeguard, characterization, and typification of cultivable soil microbial resources. The collection also quantifies the potential environmental and economic benefits of the re-incorporation of these strains into ecosystems [12]. COLMENA’s vision is to lead the revolution of the microbial inoculants used in Mexican agriculture, transferring native microorganisms to the field under specific biotic and abiotic conditions [13].
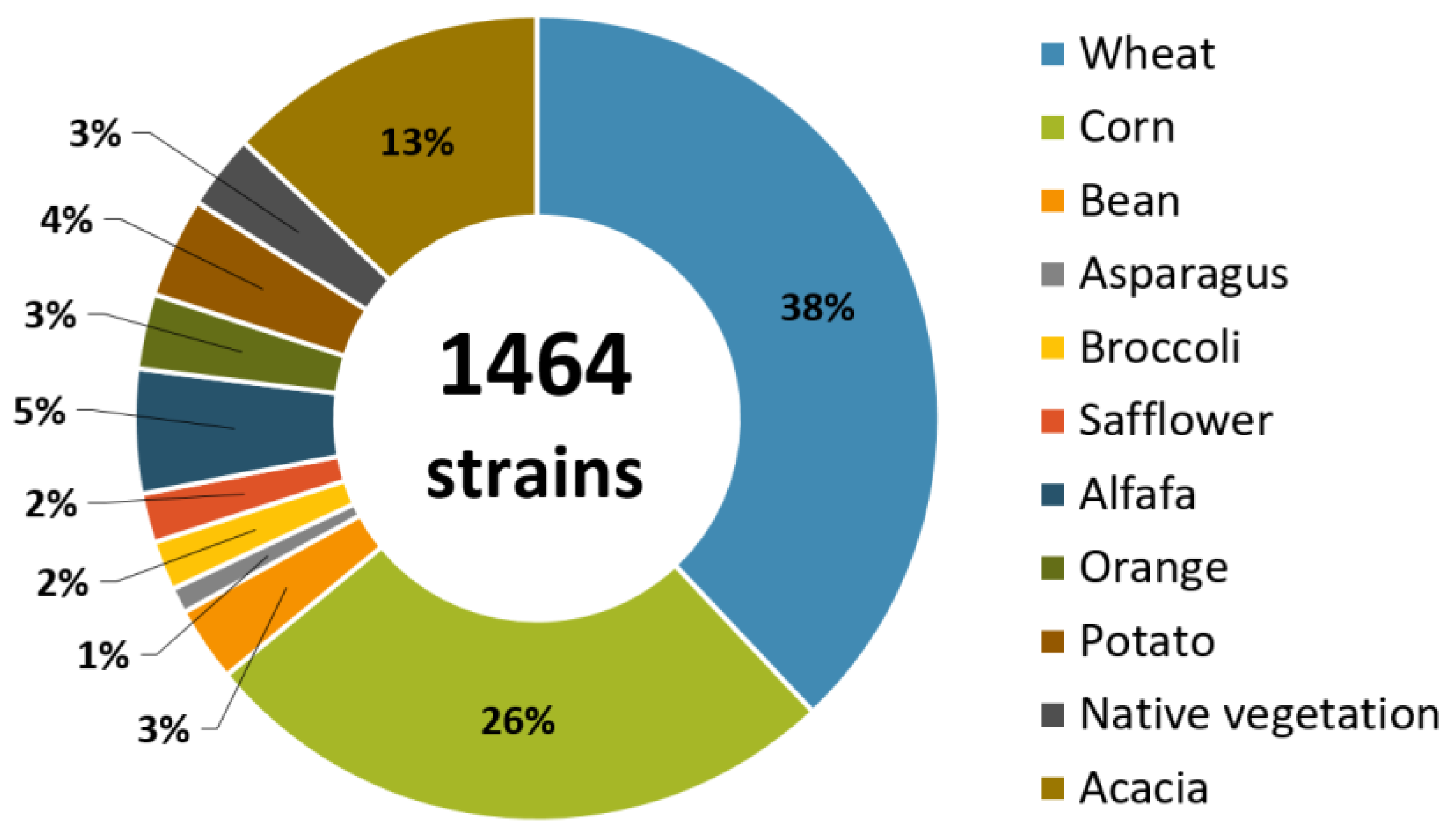
Currently, COLMENA has cryopreserved a collection of 1464 isolated microorganisms (where 70% of these are bacterial and the remaining 30% are fungal strains) from soil associated with various crops of economic importance for Mexico, such as wheat (Triticum spp.) (556), maize (Zea mays) (381), alfalfa (Medicago sativa) (73), potato (Solanum tuberosum) (59), bean (Phaseolus vulgaris) (44), and others (Figure 2).
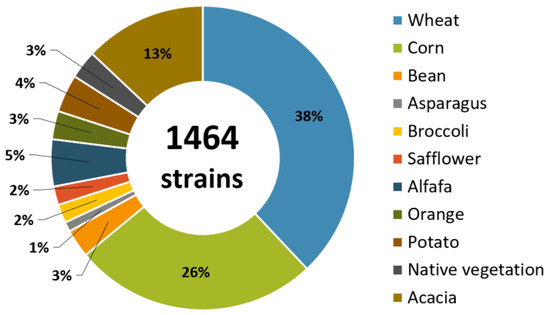
Figure 2.
Percentage of microbial strains preserved in COLMENA isolated from soils associated with various economically important crops located in the Yaqui Valley, Sonora and the Fuerte Valley, Sinaloa, Mexico.
The isolation of the microorganisms was carried out in two main agricultural regions in Mexico: the Yaqui Valley, located in the state of Sonora (26°53′ and 28°37′ N, 108°53′ and 110°37′ E), and the Fuerte Valley, located in Sinaloa (25°53′ and 26°38′ N, 108°16′ and 109°04′ W). These two regions are of great importance due to their contribution to the production of wheat and maize. The Yaqui Valley contributes approximately 50% of the national wheat production [30,31] and the Fuerte Valley with 27% of the state production of maize [32,33,34].
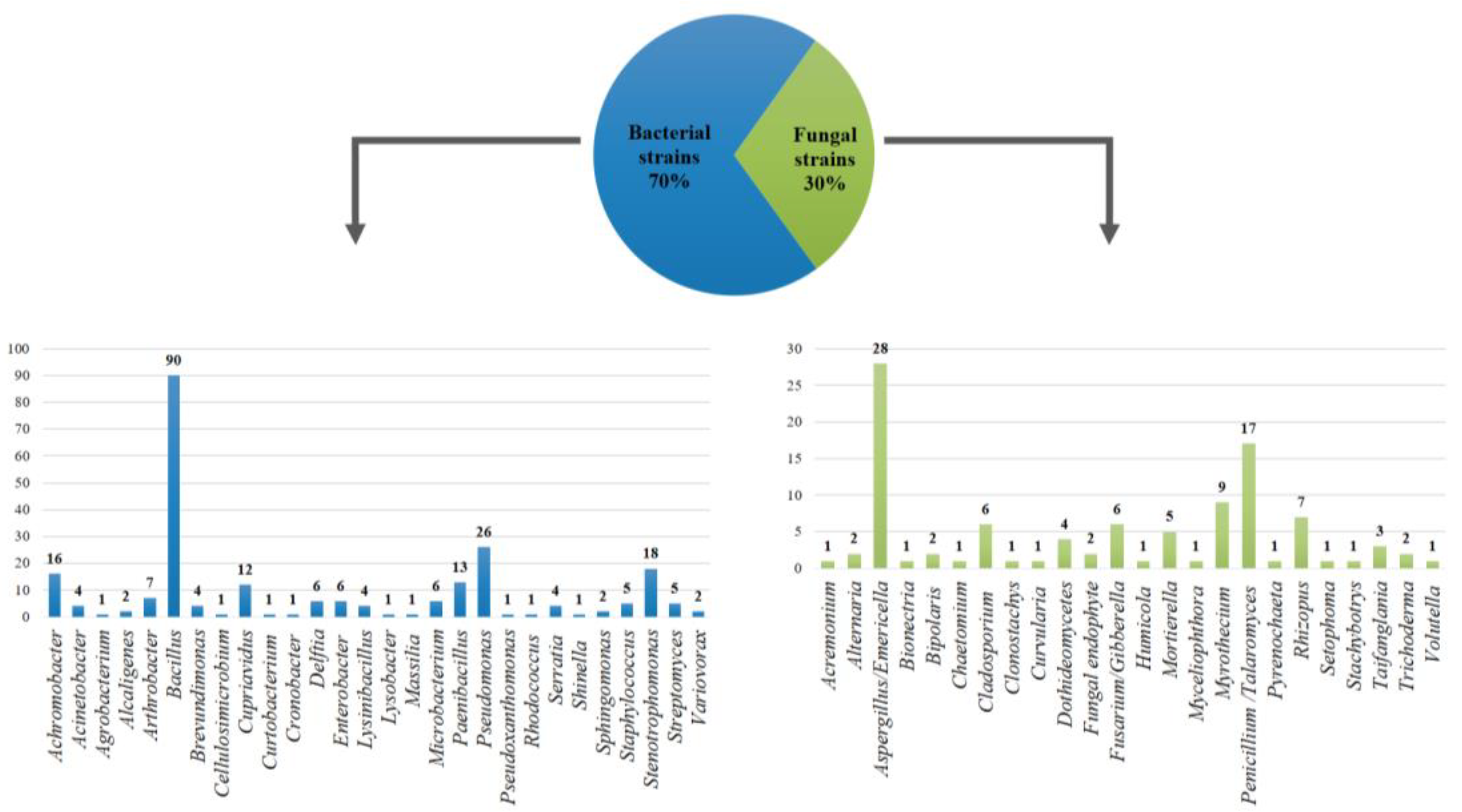
To date, 24% of the 1464 microbial strains preserved in COLMENA have been molecularly characterized by amplifying the 16S rRNA gene for bacteria and 5.8S rRNA gene for fungi. 28 bacterial genera were identified, where Bacillus (27%), Pseudomonas (8%), and Stenotrophomonas (6%) were the most abundant; in addition 24 fungal genera were found, with Aspergillus (8%), Penicillium (3%) and Myrothecium (3%) being the most representative (Figure 3).
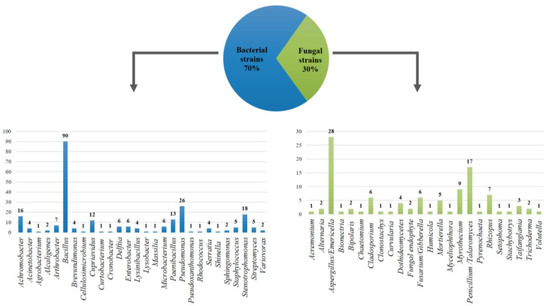
Figure 3.
Bacterial and fungal diversity preserved in COLMENA obtained from the analysis of 16S rRNA and 5.8S rRNA genes, respectively.
These main genera identified are known for their high impact on agricultural production. Bacillus species are the most extensively studied bacteria for the control of phytopathogens and inducing plant systemic resistance through the consumption of leached exudates, production of siderophores, production of antibiotics, the activity of lytic enzymes (glucanases, chitinases, proteases), and biosynthesis of cyclic lipopeptides [29,35]. Likewise, Pseudomonas species are functionally diverse and ecologically remarkable microorganisms, they can be used as plant growth-promoting agents and as bio-remediators due to their ability to fix nitrogen, solubilize phosphates, chelate iron, and produce phytohormones. Pseudomonas can act as biocontrol agents due to their catabolic adaptability, root-colonizing ability, and their capability to produce antifungal metabolites [36,37]. On the other hand, Stenotrophomonas species have presented promising plant growth promotion traits as well as biocontrol of phytopathogens. Some strains of this genus can produce volatile organic compounds, antibiotics, and enzymes that degrade the cell wall of fungi [38].
Several Aspergillus species have been identified as biocontrol agents and plant-growth promoters; some strains can produce extracellular phytases, which can benefit plants in the pretreatment of soils. Aspergillus species can also mineralize phosphate from inaccessible organic sources or increase the availability of inorganic sources for plants, as well as induce growth promotion [39,40]. Some species of the genus Penicillium, such as P. radicum and P. bilaiae can enhance plant growth by increasing phosphorus nutrition [41], however, some species of this genus have been identified as phytopathogens [42]. Furthermore, Myrothecium can help in the biocontrol of insects and diseases through the production of secondary metabolites such as enzymes, antibiotics, sesquiterpenoids, and cyclopeptides, among others. Nevertheless, species Myrothecium such as M. roridum and M. verrucaria can infect different crops [43].
3. COLMENA and the Search for Agro-Biotechnological Alternatives
To achieve sustainability and agricultural profitability in Mexico it is necessary to address the problem of soil degradation in our production systems. The excessive use of agrochemicals (fertilizers and pesticides) has a considerable impact on the soil properties and, with it, on the ecological balance, modifying the metabolic activities of the different microbial populations of the agroecosystem [44,45]. The restoration of the soil microbiota is a fundamental strategy to improve soil quality and sustainably increase agricultural productivity, and thus the use of microbial inoculants formulated from PGPM is now one of the alternatives that achieve the replacement (total or partial) of synthetic agrochemicals [15]. For this, it is important to understand the beneficial biological interactions of these microorganisms in the soil and the diverse components that promote the ecological processes to develop sustainable agroecosystems [46,47].
COLMENA has specialized in the identification and characterization of microbial strains with metabolic capacities related to plant growth promotion and biocontrol of plant diseases, as well as phytopathogenic microorganisms. Thus, to date, 396 strains of the collection have been analyzed, where 12% can solubilize phosphorus, and 20% are capable of synthesizing various types of siderophores. Furthermore, 50 strains in the collection have the ability to produce indoles, a group of phytohormones that includes indole acetic acid, the main natural auxin in plants. Also, 60 microbial strains with the ability to produce cellulolytic enzymes have been identified, these enzymes may have a role in various mechanisms of biocontrol [33,34,48,49,50,51].
Likewise, COLMENA identifies potentially pathogenic strains for humans, this is carried out through taxonomic studies and β-hemolytic activity. To date, 258 microbial strains have been evaluated, where 11% present β-hemolysis activity, restricting their use as microbial inoculants for their application in crops [33,34,51].
Besides the evaluation of the metabolic potential of the cryopreserved strains, COLMENA performs tolerance tests to hydric, thermal, and saline stress [50,52], and fungicides, as is chlorothalonil, a fungicide used according to the Official Mexican Standard on wheat seeds to control partial carbon in wheat by the fungus Tilletia indica Mitra [44]. Conducting studies of susceptibility to biotic and abiotic stress in conjunction with metabolic tests is essential in the development of agro-biotechnological strategies, such as biofertilizers, to ensure success in their implementation in agricultural systems.
COLMENA also preserves microbial species reported as plant pathogens, such as Fusarium verticillioides (causal agent of maize ear rot), Sclerotinia sclerotiorum (causal agent of white mold on beans), and Bipolaris sorokiniana (causal agent of spot blotch in wheat) [13,53,54,55]. The study of these phytopathogenic strains allows us to know the infection mechanisms they carry out, leading to the development of more efficient and sustainable strategies for their control through the use of biological agents.
The detailed molecular and metabolic identification of isolated strains is decisive to address fundamental issues of systematic taxonomy and ecology, allowing us to: (i) identify bioindicator microorganisms of the ecosystem [56]; (ii) identify PGPM strains and biological control agents [12]; (iii) study novel microbial species [57]; (iv) identify pathogenic or harmful strains for humans, plants and animals [58]; and (v) establish quality criteria in products and services based on microorganisms [59]. In addition to metabolic, taxonomic, pathogenicity, and agrochemical compatibility analyzes, other traits should be considered during the screening process related to the scaling up of the biofertilizer production. The strains selected for their bioformulation must be able to grow in artificial media (especially culture media with minimal concentrations or without strict nutritional requirements to reduce costs), survive in carriers, overcome the technological production processes, have genetic stability and the ability to produce beneficial effects on crops [59,60].
Currently, in COLMENA, different promising PGPM strains have been identified for their ability to promote plant growth and control phytopathogenic diseases, based on metabolic [44], molecular [61,62,63], and in vitro pathogenic assays [51]. COLMENA also studies the effects of promising PGPM strains in plants [48,49,50], in order to verify the abilities of these microorganisms in vivo and develop a cost-effective microbial inoculant that improves soil health and crop growth.
COLMENA has studied the metabolic potential of native Bacillus strains with the ability to promote growth, i.e., produce indoles, biosynthesize siderophores, solubilize phosphates, tolerate abiotic stress (saline, thermal, hydric, and chlorothalonil). Some of the strains most studied are B. paralicheniformis TRQ65, B. megaterium TRQ8, B. subtilis TSO9, and B. cabrialesii TE3T. These strains have been reported to be able to solubilize insoluble phosphorus (TRQ8, 38.0 ± 0.9%; TE3T, 43.2 ± 1.7%; TSO9, 54 ± 1%) and produce indoles (TRQ65, 39.29 ± 0.30; TRQ8, 12.03 ± 1.93; TE3T 8.21 ± 1.35 µg mL−1), while TRQ8 is the only one that has been shown to have the ability to produce siderophores (8.1 ± 0.8%) [48,49]. Besides, all these strains have been reported with the ability to tolerate saline, thermal and hydric stress [50] and to tolerate the fungicide chlorothalonil [44].
The potential of COLMENA strains as biocontrol agents has also been evaluated by confrontational tests. Villa-Rodríguez et al. (2019) evaluated 195 bacterial strains against different strains of the phytopathogenic fungus B. sorokiniana [51], which are also cryopreserved in COLMENA. Fourteen strains exhibited antagonistic activity against B. sorokiniana in different degrees, being Bacillus subtilis (strains TSO2, TSO22, TSQ24), a new bacterial species Bacillus cabrialesii (TE3T), and Pseudomonas protegens (TRQ9), the ones with the highest antagonistic activity. However, the strain TE3T was the only non-potentially pathogenic strain for animals and humans, for which a cell-free culture was evaluated against the phytopathogenic fungus. In this way, it was revealed that the strain TE3T produces extracellular antifungal metabolites to suppress the growth of B. sorokiniana (~98% of inhibition), concluding that this strain and its antifungal metabolites (lipopeptides) are an effective and promising treatment to control the causative agent of spot blotch in wheat [64].
Due to the growth-promoting characteristics, stress tolerance, and genetic potential of the strains B. megaterium TRQ8, B. paralicheniformis TRQ65, B. cabrialesii TE3T, and B. subtilis TSO9, COLMENA has performed in vivo assays on wheat plants to evaluate their abilities to promote plant growth. Robles-Montoya et al. (2020) reported a bacterial consortium with the four Bacillus strains (TRQ8, TRQ65, TE3T, and TSO9) and it was inoculated (4 × 107 Colony-forming Units (CFU)) in wheat plants. The consortium inoculation increased the length of the aerial part (28%), root length (25%), stem development (50%), dry weight (72%), and the biovolume index (57%) [48].
Rojas-Padilla et al. (2020) evaluated the effect of individual inoculation and different consortium combinations of the TRQ8, TE3T, and TRQ65 strains to explore their interactions that improve the morphometric variables in wheat plants considering the edaphoclimatic conditions from the Yaqui Valley. In this study, it was determined that the three strains (TRQ8, TE3T, and TRQ65) and the different consortia presented in vivo growth promotion characteristics; however, the co-inoculation of B. megaterium TRQ8 and B. paralicheniformis TRQ65 showed the highest increase in aerial (6%) and root (10%) length, aerial (60%) and root (82%) dry weight, and biovolume index (18%) [49].
To support the potential observed in vitro and in vivo and to know the feasibility of designing a biofertilizer for use in current and future agriculture, inoculation assays have been carried out under field conditions. Ibarra-Villarreal (2020) evaluated the inoculation of five Bacillus strains (B. subtilis TSO9, B. subtilis TSO62, B. subtilis TSO64, B. megaterium TRQ8, and B. paralicheniformis TRQ65) in wheat, along with 3 different doses of nitrogen; 0%, 50%, and 100% of the recommended nitrogen fertilization (250 kg N ha−1). The application of the Bacillus consortium (−1 × 107 CFU plant−1) showed multiple positive effects on wheat, such as larger spike size (2.16%), increase in the number of grains per spike (5.23%), higher yield (+1 t ha−1), and reduction in the amount of nitrogen fertilizer applied (50% of the recommended dose) [65].
Arellano-Wattenbarger (2019) applied a consortium with three of the studied native Bacillus strains (TRQ8, TE3T, TRQ65) in wheat along with different doses of N. In this study, it was determined that inoculation with the consortium (−1 × 106 CFU plant−1) increased the number of spikes/m2 (25%), crop yield (15%), and improved grain quality by reducing the presence of white belly and increased the percentage of protein. In addition, the consortium inoculation reduced the use of nitrogen fertilizers at rates of 0 or 120 kg N ha−1, maintaining the yield of the wheat crop in comparison with the dose of nitrogen fertilization used conventionally (240 kg N ha−1) by optimizing the use of residual and applied nitrogen to the agroecosystem [66].
Likewise, Ayala-Zepeda (2020) showed that the inoculation with the consortium of the 3 Bacillus strains under lower fertilization doses (0 and 120 kg N ha−1) increased the yield (up to 1 ton ha−1) and the quality of the wheat crop, unlike that obtained with 100% of the conventional fertilization without inoculation. In addition, the efficiency in the use of nitrogen from the crop increased by 14.4% when reducing the recommended dose of nitrogen fertilization to 50% (120 kg N ha−1) and by 10.8% under the inoculation of the consortium with 50% fertilization [67].
On the other hand, Valenzuela-Aragon et al. (2019) identified promising PGPM strains through plant-assisted selection (PAS), of which eleven strains (Stenotrophomonas sp. TS1, TS6 and TS7, Enterobacter cloacae TS3, Bacillus sp. TS8, Microbacterium foliorum TS9, Bacillus cereus TS10, Cellulosimicrobium sp. TE6, and Paenibacillus lautus TE8 and TE10) were reported to have the ability to promote wheat growth mainly due to the development of the stem and the increase in the number of leaves through their inoculation in plants. They also showed that these bacteria regulate the expression of the genes involved in the growth of wheat; thus, strains TS1 and TS3 are responsible for the synthesis of chlorophyll through the up-regulation of the GluTR gene; the nitrate transporter (NRT1.4) was slightly down-regulated in wheat leaves by strains TS1, TS8, TS10, TE6, and TE10; while the synthesis of water-soluble carbohydrates (up-regulation of the 6-SFT1 gene) was regulated by all strains except TS6, TS7, and TE6 [50].
Chaparro-Encinas (2020) performed an analysis of the transcriptomic effect of the co-inoculation of B. paralicheniformis TRQ65 and B. megaterium TRQ8 in wheat under conditions of optimal and increased temperature (+2 °C). The gene expression patterns suggest that the studied Bacillus consortium partially inhibited the oxidative stress machinery, and promoted cell division and growth events associated with the progression of developmental stages. Furthermore, the systemic response was simultaneously reprogrammed, suppressing the defense mechanism and inducing central stimuli response (protein kinases). This was carried out as a strategy to facilitate bacterial colonization, but also to promote cell wall strengthening to face the increase in temperature [68].
Until now, COLMENA has sequenced the complete genome of the strains Bacillus megaterium TRQ8 [62], Bacillus paralicheniformis TRQ65 [63], and the strain type Bacillus cabrialesii TE3T [61], to provide a strong taxonomical affiliation based on the Overall Genome Relatedness Index (OGRI), as well as information about their potential metabolic traits. The 3 genomes revealed the presence of genes involved in: (1) tolerance to abiotic factors in agroecosystems (response to osmotic and oxidative stress); (2) biocontrol of phytopathogens (biosynthesis of lipopeptides and antibiotics); and (3) plant growth promotion (auxin biosynthesis, phosphate metabolism, siderophore production).
At present, COLMENA is developing different research projects, focused on the development of fermentation and carrier strategies, as well as carrying out field trials with the studied PGPM consortium to design alternatives that combine the use of PGPM and lower doses of inorganic fertilizer that increase crop yields and the efficiency of the nitrogen and water use by plants. Other current projects are focused on identifying the metabolic and molecular mechanisms of PGPM involved in growth promotion and biocontrol, in addition to the study of comparative evolutionary genomics, metagenomic, metabolomics, and transcriptomic. Furthermore, COLMENA is currently developing strategies to mitigate soil erosion and conserve soil microbial resources using isotopic techniques.
4. Virtual COLMENA Platform
Biological databases play a central role in bioinformatics, providing access to a wide variety of biologically relevant data [69]. Databases archive, store, maintain and share information on sequences, protein structures, metabolites, and microbial diversity, among other essential data required for the development of the science of microbiology [70]. In recent years, technological development has led to an exponential increase in the amount of microbial sequencing and data identification [71], and all of this information needs to be stored and organized in the best possible way, through computer systems that allow us to generate free access platforms, and easy to use.
To make the knowledge and results of the scientific research carried out by the COLMENA team accessible, a virtual platform was created to store the database of the microorganisms studied and preserved. This platform represents a significant mechanism for global dissemination of the potential of these strains, promoting their use for the solution of agro-biotechnological problems. The platform is available to professionals, researchers, agricultural workers, and anyone interested in microorganisms with the agronomic potential to promote crop growth and control plant diseases.
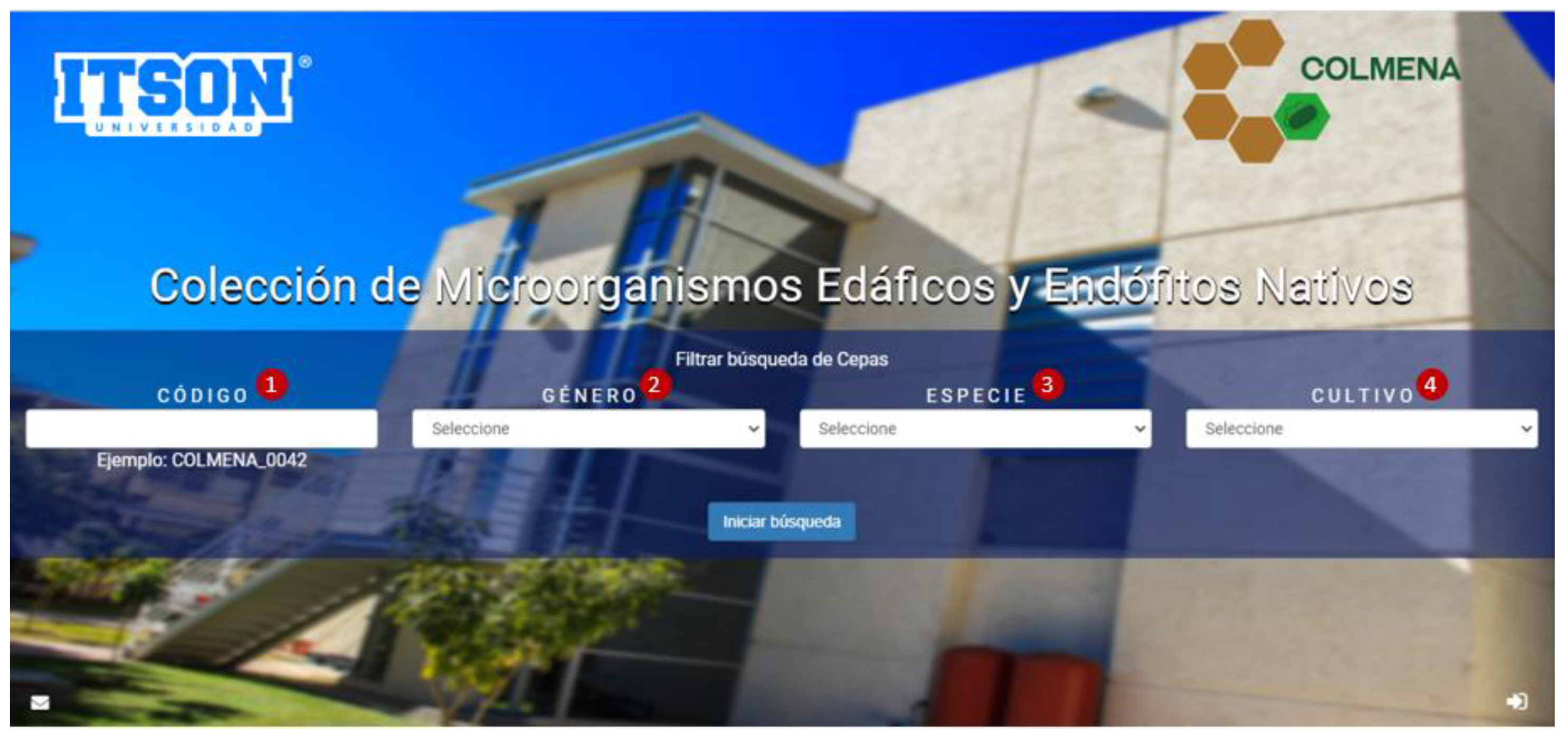
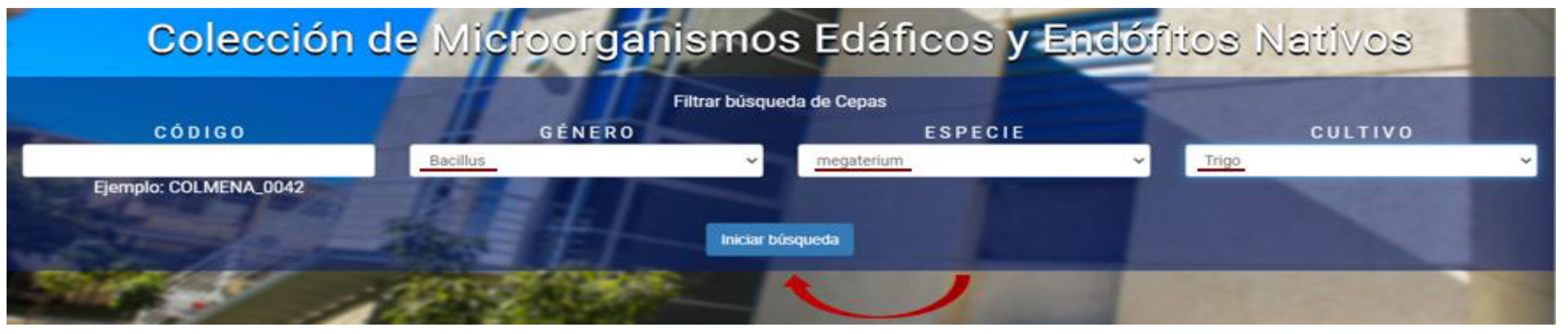
The platform of COLMENA’s virtual database, so far only presented in Spanish, was developed in the C# programming language, with SQL Server 2008 as its database manager. Visual Studio 2010 environment and NET Framework 4 or higher were used. The platform is compatible with Explorer, Firefox, and Google Chrome browsers. This virtual database is published in the following internet link: http://apps2.itson.edu.mx/colmena (released November 2019, accessed on 21 July 2021), in its home page it shows a screen that contains a search categorization system of the strains of interest, as shown in Figure 4. The COLMENA platform is composed of 4 filters for the user to select and access the microorganism of interest. In the first filter (1), the strain code can be entered directly with the nomenclature shown in the example (“COLMENA_0042”); in the second filter (2), users can select between 41 different microbial genera; the third filter (3) consists of 75 species options; and finally, in the fourth filter (4) it is possible to choose between 9 crops associated with the isolated strains. All this information is updated frequently to include more data about strains, location, crops, and metabolic functions.
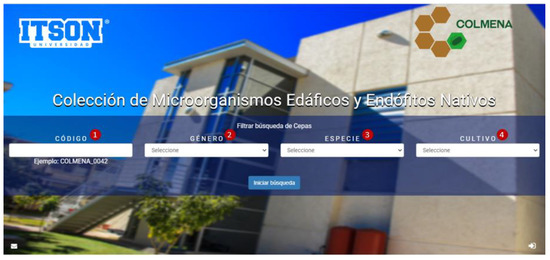
Figure 4.
COLMENA’s platform categorization system (http://apps2.itson.edu.mx/colmena, accessed on 21 July 2021).
The platform is open access and free of charge. The user interface was designed for easy and intuitive use. There are only four types of searches available, but cross-search allows for a wide range of combinations. First, it is necessary to fill in at least one of the four platform filters, depending on the information that is of interest, as shown in Figure 5.
Figure 5.
Filter selection in the COLMENA’s platform.
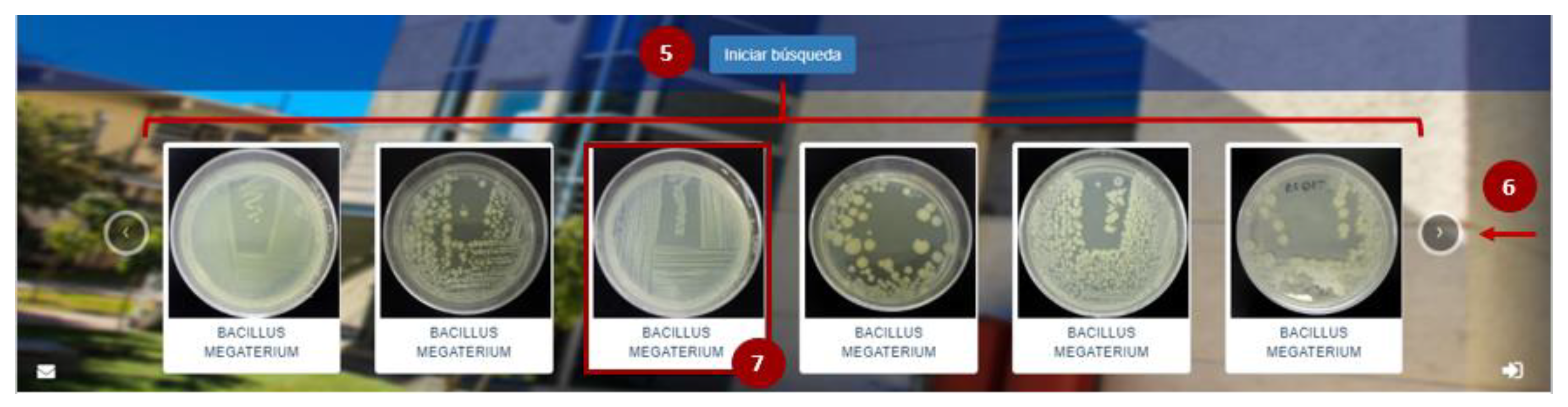
Once the information has been captured in the search system, as indicated in Figure 6, the user must click on the filter search button (5), which will display a series of microorganisms that coincide with the indicated parameters. The platform allows the user to continue reviewing the options by selecting the “next” arrow (6), and finally, to select the strain of interest by clicking on it (7).
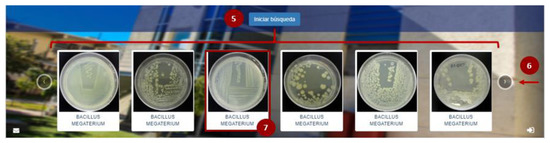
Figure 6.
Strain search results in the COLMENA’s platform.
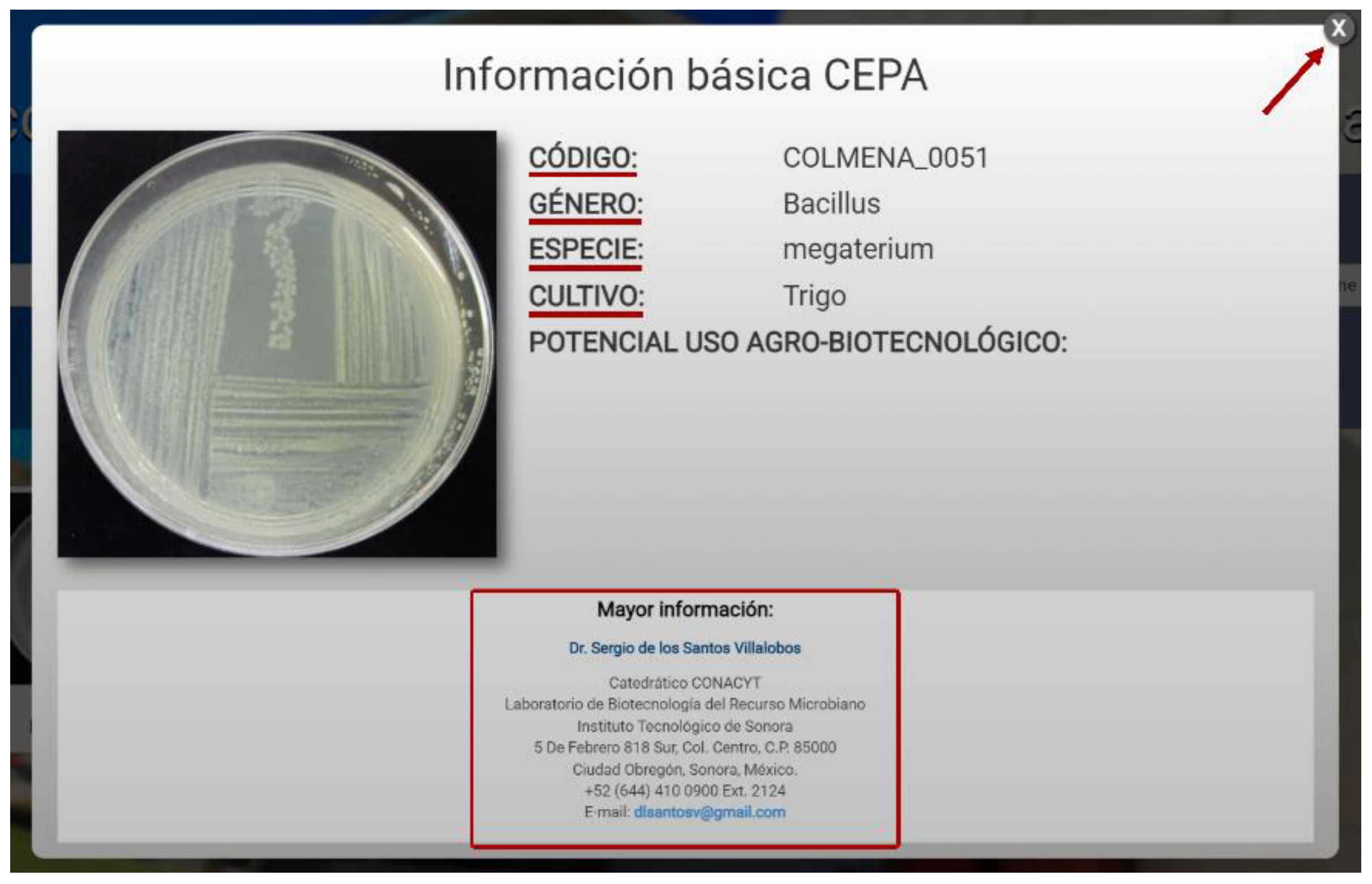
After selecting the strain, a box with information will open as shown in Figure 7. The data displayed for the strain of interest are a macroscopic image, the strain code, genus, species, crop from which it was isolated, and its potential agro-biotechnological use. These strains can be requested from the COLMENA curator using the contact information shown below. To continue reviewing the other strains, the user can close the box in the upper right and select another strain from the list.
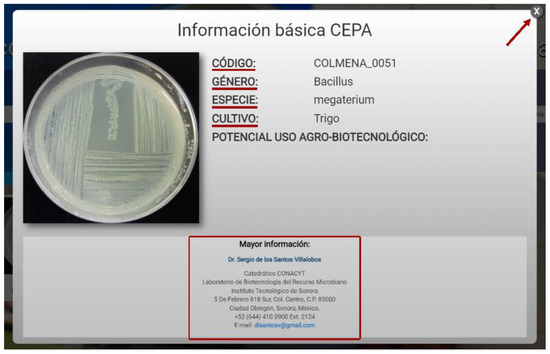
Figure 7.
Basic information about the strain.
Through this platform, COLMENA intends to make more efficient use of its database dissemination to promote the study and extensive use of beneficial microorganisms such as PGPM, and to make it accessible not only to the scientific community, but also to the general public.
5. Conclusions
Due to the increasing need for food as a result of the exponential growth of the world population, there is a growing demand for agricultural productivity, which can be met by employing efficient and sustainable agricultural practices. Soils provide us with a potential agro-biotechnological resource for agriculture, therefore, the use of native microbial diversity associated with crops is a promising alternative. However, the continuous loss of soil microbial diversity due to soil degradation generates the need to preserve and transfer microorganisms to carry out research, teaching, and their biotechnological exploitation. For this reason, microbial culture collections play an important role in the preservation and bioprospection of the microbial resource, providing authentic, stable, and useful biological material for the development of agro-sustainable strategies.
COLMENA represents an alternative for the identification of microorganisms that exhibit characteristics associated with the promotion of plant growth and biocontrol of phytopathogens. Thus, its online catalogs and virtual database are fundamental tools to increase the dissemination of information, results, and discoveries. These tools bring knowledge to other sectors and at the same time promote the study of microorganisms and their potential uses for solving agro-biotechnology problems.
It should be noted that COLMENA is a dynamic project, which is expected in the future to expand to more agricultural areas across the country, as well as increase the metabolic and molecular analysis of the microorganisms preserved in the collection to extend the content of its database and make it accessible to everyone.
Author Contributions
Conceptualization, S.d.l.S.-V. and F.I.P.-C.; writing—original draft preparation and visualization, S.d.l.S.-V., A.M.D.-R., M.F.Á.-M., A.D.M.-V. and F.I.P.-C.; writing—review and editing were developed by S.d.l.S.-V., A.M.D.-R. and F.I.P.-C.; supervision, project administration, funding acquisition, S.d.l.S.-V. and F.I.P.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Consejo Nacional de Ciencia y Tecnología (CONACyT), through the Project 253663 and the Project 257246; the Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP) through the Project 2315932912; the Project PROFAPI_2021_0006; and the NPTC PRODEP Project 511-6/2020-8594.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank all the members and former members of the LBRM-COLMENA research node for their dedication and commitment to the creation of COLMENA. Alondra María Díaz Rodríguez and María Fernanda Avila Mascareño thank CONACyT for the master’s scholarship No. 908966 and No. 1074525, respectively.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Díaz-Rodríguez, A.M.; Salcedo Gastelum, L.A.; Félix Pablos, C.M.; Parra-Cota, F.I.; Santoyo, G.; Puente, M.L.; Bhattacharya, D.; Mukherjee, J.; de los Santos-Villalobos, S. The Current and Future Role of Microbial Culture Collections in Food Security Worldwide. Front. Sustain. Food Syst. 2021, 4. [Google Scholar] [CrossRef]
- IPCC. Summary for Policymakers. In Global Warming of 1.5 °C an IPCC Special Report on the Impacts of Global Warming of 1.5 °C above Pre-Industrial Levels and Related Global Greenhouse Gas Emission Pathways, in the Context of Strengthening the Global Response to the Threat of Climate Change, Sustainable Development, and Efforts to Eradicate Poverty; Masson-Delmotte, V., Zhai, P., Pörtner, H.-O., Roberts, D., Skea, J., Shukla, P.R., Pirani, A., Moufouma-Okia, W., Péan, C., Pidcock, R., et al., Eds.; Intergovernmental Panel on Climate Change: Geneva, Switzerland, 2019; pp. 1–26. [Google Scholar]
- Collins, M.; Knutti, R.; Arblaster, J.; Dufresne, J.-L.; Fichefet, T.; Friedlingstein, P.; Gao, X.; Gutowski, W.J.; Johns, T.; Krinner, G.; et al. Long-Term Climate Change: Projections, Commitments and Irreversibility. In Climate Change 2013 the Physical Science Basis: Working Group I Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Stocker, T.F., Qin, D., Plattner, G.-K., Tignor, M., Allen, S.K., Boschung, J., Nauels, A., Xia, Y., Bex, V., Midgley, P.M., Eds.; Cambridge University Press: Cambridge, MA, USA, 2013; pp. 1029–1136. [Google Scholar] [CrossRef]
- Mall, R.K.; Gupta, A.; Sonkar, G. Effect of Climate Change on Agricultural Crops. In Current Developments in Biotechnology and Bioengineering: Crop Modification, Nutrition, and Food Production; Dubey, S.K., Pandey, A., Sangwan, R.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 23–46. [Google Scholar] [CrossRef]
- Challinor, A.J.; Watson, J.; Lobell, D.B.; Howden, S.M.; Smith, D.R.; Chhetri, N. A Meta-Analysis of Crop Yield under Climate Change and Adaptation. Nat. Clim. Chang. 2014, 4, 287–291. [Google Scholar] [CrossRef]
- Gupta, S.; Sharma, D.; Gupta, M. Climate Change Impact on Plant Diseases: Opinion, Trends and Mitigation Strategies. In Microbes for Climate Resilient Agriculture; Kashyap, P.L., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; pp. 41–56. [Google Scholar] [CrossRef]
- Velásquez, A.C.; Castroverde, C.D.M.; He, S.Y. Plant and Pathogen Warfare under Changing Climate Conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef] [Green Version]
- Cerda, R.; Avelino, J.; Gary, C.; Tixier, P.; Lechevallier, E.; Allinne, C. Primary and Secondary Yield Losses Caused by Pests and Diseases: Assessment and Modeling in Coffee. PLoS ONE 2017, 12, e0169133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FAO. The Future of Food and Agriculture—Trends and Challenges; Food and Agriculture Organization of the United Nations: Rome, Italy, 2017. [Google Scholar]
- Dudley, N.; Alexander, S. Agriculture and Biodiversity: A Review. Biodiversity 2017, 18, 45–49. [Google Scholar] [CrossRef]
- Matson, P.; Jewett, P. Ecosystems and Land-Use Change in the Yaqui Valley: Does Agricultural Intensification Spare Land for Nature? In Seeds of Sustainability: Lessons from the Birthplace of the Green Revolution; Matson, P., Ed.; Island Press: Washington, DC, USA, 2013; pp. 47–62. [Google Scholar] [CrossRef]
- Adhikari, K.; Hartemink, A.E. Linking Soils to Ecosystem Services—A Global Review. Geoderma 2016, 262, 101–111. [Google Scholar] [CrossRef]
- De los Santos-Villalobos, S.; Parra-Cota, F.I.; Herrera-Sepúlveda, A.; Valenzuela-Aragón, B.; Estrada-Mora, J.C. Colmena: Colección de Microorganismos Edáficos y Endófitos Nativos, Para Contribuir a La Seguridad Alimentaria Nacional. Rev. Mex. Cienc. Agrícolas 2018, 9, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Pereira, P.; Bogunovic, I.; Muñoz-Rojas, M.; Brevik, E.C. Soil Ecosystem Services, Sustainability, Valuation and Management. Curr. Opin. Environ. Sci. Health 2018, 5, 7–13. [Google Scholar] [CrossRef]
- Valenzuela-Ruiz, V.; Ayala-Zepeda, M.; Arellano-Wattenbarger, G.L.; Parra-Cota, F.I.; García-Pereyra, G.; Aviña-Martínez, G.N.; de los Santos-Villalobos, S. Las Colecciones Microbianas y Su Potencial Contribución a La Seguridad Alimentaria Actual y Futura. Rev. Latinoam. Rec. Nat. 2018, 14, 18–25. [Google Scholar]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial Diversity and Soil Functions. Eur. J. Soil Sci. 2003, 54, 655–670. [Google Scholar] [CrossRef]
- Sarkar, A.; Saha, M.; Meena, V.S. Plant Beneficial Rhizospheric Microbes (PBRMs): Prospects for Increasing Productivity and Sustaining the Resilience of Soil Fertility. In Agriculturally Important Microbes for Sustainable Agriculture; Meena, V.S., Mishra, P.K., Bisht, J.K., Pattanayak, A., Eds.; Springer Nature: Singapore, 2017; pp. 3–29. [Google Scholar] [CrossRef]
- ELD Initiative. Report for Policy and Decision Makers: Reaping Economic and Environmental Benefits from Sustainable Land Management; ELD Initiative: Bonn, Germany, 2015. [Google Scholar]
- Gupta, G.S. Land Degradation and Challenges of Food Security. Rev. Eur. Stud. 2019, 11, 63–72. [Google Scholar] [CrossRef]
- Tsiafouli, M.A.; Thébault, E.; Sgardelis, S.P.; de Ruiter, P.C.; van der Putten, W.H.; Birkhofer, K.; Hemerik, L.; de Vries, F.T.; Bardgett, R.D.; Brady, M.V.; et al. Intensive Agriculture Reduces Soil Biodiversity across Europe. Glob. Chang. Biol. 2015, 21, 973–985. [Google Scholar] [CrossRef]
- Zhu, Z.; Bai, Y.; Lv, M.; Tian, G.; Zhang, X.; Li, L.; Jiang, Y.; Ge, S. Soil Fertility, Microbial Biomass, and Microbial Functional Diversity Responses to Four Years Fertilization in an Apple Orchard in North China. Hortic. Plant. J. 2020, 6, 223–230. [Google Scholar] [CrossRef]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The Role of Soil Microorganisms in Plant Mineral Nutrition—Current Knowledge and Future Directions. Front. Plant. Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Wu, Z.; Wang, W.; Xu, X.; Liu, X. Rs-198 Liquid Biofertilizers Affect Microbial Community Diversity and Enzyme Activities and Promote Vitis vinifera L. Growth. Biomed. Res. Int. 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- Lisuma, J.B.; Zuberi, Z.; Ndakidemi, P.A.; Mbega, E.R. Linking Rhizosphere Bacterial Diversity and Soil Fertility in Tobacco Plants under Different Soil Types and Cropping Pattern in Tanzania: A Pilot Study. Heliyon 2020, 6. [Google Scholar] [CrossRef]
- Lei, H.; Liu, A.; Hou, Q.; Zhao, Q.; Guo, J.; Wang, Z. Diversity Patterns of Soil Microbial Communities in the Sophora Flavescens Rhizosphere in Response to Continuous Monocropping. BMC Microbiol. 2020, 20. [Google Scholar] [CrossRef] [PubMed]
- Mishra, J.; Prakash, J.; Arora, N.K. Role of Beneficial Soil Microbes in Sustainable Agriculture and Environmental Management. Clim. Chang. Environ. Sustain. 2016, 4, 137. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–Microbiome Interactions: From Community Assembly to Plant Health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Gangwar, M.; Saini, P.; Nikhanj, P.; Kaur, S. Plant Growth-Promoting Microbes (PGPM) as Potential Microbial Bio-Agents for Eco-Friendly Agriculture. In Advances in Soil Microbiology: Recent Trends and Future Prospects, Microorganisms for Sustainability; Adhya, T., Mishra, B., Annapurna, K., Verma, D., Kumar, U., Eds.; Springer: Singapore, 2017; Volume 4, pp. 37–55. [Google Scholar] [CrossRef]
- Valenzuela-Ruiz, V.; Gálvez-Gamboa, G.T.; Villa-Rodríguez, E.D.; Parra-Cota, F.I.; Santoyo, G.; de los Santos-Villalobos, S. Lipopéptidos Producidos por Agentes de Control Biológico del Género Bacillus: Revisión de Herramientas Analíticas Utilizadas para su Estudio. Rev. Mex. Cienc. Agric. 2020, 11, 419–432. [Google Scholar] [CrossRef] [Green Version]
- CIMMYT. Wheat Atlas by CIMMYT. Available online: http://wheatatlas.org/visualizations (accessed on 30 July 2020).
- SAGARPA. Boletín Mensual de La Producción Trigo Grano; Secretaría de Agricultura Ganadería y Desarrollo Rural (SAGARPA): Xalapa-Enríquez, Mexico, 2015. [Google Scholar]
- SIAP. Avance de Siembras y Cosechas. Available online: http://infosiap.siap.gob.mx:8080/agricola_siap_gobmx/ResumenProducto.do (accessed on 17 May 2021).
- Parra-Cota, F.I.; de los Santos-Villalobos, S.; Lugo-Valdez, M.A.; Cruz-Ibarra, R.A.; Fuentes-Dávila, G.; Peinado-Fuentes, L.A. Potencial Agrobiotecnológico de Bacterias Aisladas de Suelos Agrícolas Asociados al Cultivo de Maíz en el Valle del Fuerte, Sinaloa. Rev. Latinoam. Rec. Nat. 2017, 13, 51–57. [Google Scholar]
- Parra-Cota, F.I.; Coronel-Acosta, C.B.; Amézquita-Avilés, C.F.; De los Santos-Villalobos, S.; Escalante-Martínez, D.I. Diversidad Metabólica de Microorganismos Edáficos Asociados al Cultivo de Maíz en el Valle del Yaqui, Sonora. Rev. Mex. Cienc. Agric. 2018, 9, 431–442. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.S.; Lee, Y.; Cheon, W.; Park, J.; Kwon, H.T.; Balaraju, K.; Kim, J.; Yoon, Y.J.; Jeon, Y. Characterization of Bacillus velezensis AK-0 as a Biocontrol Agent against Apple Bitter Rot Caused by Colletotrichum Gloeosporioides. Sci. Rep. 2021, 11. [Google Scholar] [CrossRef]
- Ansari, F.A.; Ahmad, I. Biofilm Development, Plant Growth Promoting Traits and Rhizosphere Colonization by Pseudomonas entomophila FAP1: A Promising PGPR. Adv. Microbiol. 2018, 08, 235–251. [Google Scholar] [CrossRef] [Green Version]
- Panpatte, D.G.; Jhala, Y.K.; Shelat, H.N.; Vyas, R.V. Pseudomonas fluorescens: A Promising Biocontrol Agent and PGPR for Sustainable Agriculture. In Microbial Inoculants in Sustainable Agricultural Productivity: Research Perspectives; Pratap, D., Bahadur, S., Prabha, S., Eds.; Springer: New Delhi, India, 2016; Volume 1, pp. 257–270. [Google Scholar] [CrossRef]
- Mishra, S.; Singh, B.R.; Naqvi, A.H.; Singh, H.B. Potential of Biosynthesized Silver Nanoparticles Using Stenotrophomonas Sp. BHU-S7 (MTCC 5978) for Management of Soil-Borne and Foliar Phytopathogens. Sci. Rep. 2017, 7, 45154. [Google Scholar] [CrossRef] [Green Version]
- Hung, R.; Lee Rutgers, S. Applications of Aspergillus in Plant Growth Promotion. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.K., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 223–227. [Google Scholar] [CrossRef]
- Pandya, N.D.; Desai, P.V.; Jadhav, H.P.; Sayyed, R.Z. Plant Growth Promoting Potential of Aspergillus sp. NPF7, Isolated from Wheat Rhizosphere in South Gujarat, India. Environ. Sustain. 2018, 1, 245–252. [Google Scholar] [CrossRef]
- Wakelin, S.A.; Gupta, V.V.S.R.; Harvey, P.R.; Ryder, M.H. The Effect of Penicillium Fungi on Plant Growth and Phosphorus Mobilization in Neutral to Alkaline Soils from Southern Australia. Can. J. Microbiol. 2007, 53, 106–115. [Google Scholar] [CrossRef]
- Coutinho, T.C.; Ferreira, M.C.; Rosa, L.H.; de Oliveira, A.M.; de Oliveira Júnior, E.N. Penicillium citrinum and Penicillium mallochii: New Phytopathogens of Orange Fruit and Their Control Using Chitosan. Carbohydr. Polym. 2020, 234, 115918. [Google Scholar] [CrossRef]
- Elkhateeb, W.A.; Daba, G.M. Myrothecium as Promising Model for Biotechnological Applications, Potentials and Challenges. Biomed. J. Sci. Tech. Res. 2019, 16. [Google Scholar] [CrossRef]
- Díaz-Rodríguez, A.M.; Parra-Cota, F.I.; Santoyo, G.; de los Santos-Villalobos, S. Chlorothalonil Tolerance of Indole Producing Bacteria Associated to Wheat (Triticum turgidum L.) Rhizosphere in the Yaqui Valley, Mexico. Ecotoxicology 2019, 28, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Escribano-Viana, R.; López-Alfaro, I.; López, R.; Santamaría, P.; Gutiérrez, A.R.; González-Arenzana, L. Impact of Chemical and Biological Fungicides Applied to Grapevine on Grape Biofilm, Must, and Wine Microbial Diversity. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Barea, J.M. Future Challenges and Perspectives for Applying Microbial Biotechnology in Sustainable Agriculture Based on a Better Understanding of Plant-Microbiome Interactions. J. Soil Sci. Plant. Nutr. 2015, 15, 261–282. [Google Scholar] [CrossRef]
- Hernández-Flores, L.; Munive-Hernández, A.; Sandoval-Castro, E.; Martines-Carrera, D.; Villegas-Hernández, M.C. Efecto de las Prácticas Agrícolas sobre las Poblaciones Bacterianas del Suelo en Sistemas de Cultivo en Chihuahua, México. Rev. Mex. Cienc. Agric. 2013, 4, 353–365. [Google Scholar] [CrossRef] [Green Version]
- Robles-Montoya, R.I.; Chaparro-Encinas, L.A.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Improving Biometric Traits of Wheat Seedlings with the Inoculation of a Consortium Native of Bacillus. Rev. Mex. Cienc. Agric. 2020, 11, 229–235. [Google Scholar]
- Rojas-Padilla, J.; Chaparro-Encinas, L.A.; Robles-Montoya, R.I.; de los Santos-Villalobos, S. Promoción de Crecimiento En Trigo (Triticum turgidum L. subsp. durum) por la Co-Inoculación de Cepas Nativas de Bacillus Aisladas del Valle del Yaqui, México. Nov. Sci. 2020, 12, 1–27. [Google Scholar] [CrossRef]
- Valenzuela-Aragon, B.; Parra-Cota, F.I.; Santoyo, G.; Arellano-Wattenbarger, G.L.; de los Santos-Villalobos, S. Plant-Assisted Selection: A Promising Alternative for in Vivo Identification of Wheat (Triticum turgidum L. subsp. durum) Growth Promoting Bacteria. Plant. Soil 2019, 435, 367–384. [Google Scholar] [CrossRef]
- Villa-Rodríguez, E.; Parra-Cota, F.; Castro-Longoria, E.; López-Cervantes, J.; de los Santos-Villalobos, S. Bacillus subtilis TE3: A Promising Biological Control Agent against Bipolaris Sorokiniana, the Causal Agent of Spot Blotch in Wheat (Triticum turgidum L. Subsp. durum). Biol. Control. 2019, 132, 135–143. [Google Scholar] [CrossRef]
- Ibarra-Villarreal, A.L.; Gándara-Ledezma, A.; Godoy-Flores, A.; Díaz-Rodríguez, A.M.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Salt-Tolerant Bacillus Species as a Promising Strategy to Mitigate the Salinity Stress in Wheat (Triticum turgidum Subsp. durum). J. Arid Environ. 2021, 186, 104399. [Google Scholar] [CrossRef]
- Parra-Cota, F.I.; García-Pereyra, J.; Aviña-Martínez, G.N.; de los Santos-Villalobos, S. Primer Reporte de Marchitamiento Por Fusarium en Citrus sinensis Var. Valencia en el Valle del Yaqui, México. Rev. Mex. Fitopatol. Mex. J. Phytopathol. 2018, 37, 193–201. [Google Scholar] [CrossRef]
- Villa-Rodriguez, E.; Lugo-Enríquez, C.; de los Santos-Villalobos, S.; Parra-Cota, F.I.; Figueroa-López, P. First Report of Cochliobolus sativus Causing Spot Blotch on Durum Wheat (Triticum durum) in The Yaqui Valley, Mexico. Plant. Dis. 2016, 100, 2329. [Google Scholar] [CrossRef]
- Sabaté, D.C.; Brandan, C.P.; Petroselli, G.; Erra-Balsells, R.; Audisio, M.C. Biocontrol of Sclerotinia sclerotiorum (Lib.) de Bary on Common Bean by Native Lipopeptide-Producer Bacillus Strains. Microbiol. Res. 2018, 211, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Schloter, M.; Nannipieri, P.; Sørensen, S.J.; van Elsas, J.D. Microbial Indicators for Soil Quality. Biol. Fertil. Soils 2018, 54, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Robles-Montoya, R.I.; Valenzuela-Ruiz, V.; Parra-Cota, F.I.; Santoyo, G.; de los Santos-Villalobos, S. Description of a Polyphasic Taxonomic Approach for Plant Growth-Promoting Rhizobacteria (PGPR). In Microbial Services in Restoration Ecology; Singh, J.S., Vimal, S.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 259–269. [Google Scholar]
- Berg, G.; Zachow, C. Bacteria in Agrobiology: Crop Ecosystems. In Bacteria in Agrobiology: Crop Ecosystems; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 97–109. [Google Scholar] [CrossRef]
- Parnell, J.; Berka, R.; Young, H.; Sturino, J.; Kang, Y.; Barnhart, D.; Dileo, M. From the Lab to the Farm: An Industrial Perspective of Plant Beneficial Microorganisms. Front. Plant. Sci. 2016, 7, 1110. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, L.; Lesueur, D. Challenges of Formulation and Quality of Biofertilizers for Successful Inoculation. Appl. Microbiol. Biotechnol. 2013, 97, 8859–8873. [Google Scholar] [CrossRef] [PubMed]
- De los Santos-Villalobos, S.; Robles-Montoya, R.I.; Parra-Cota, F.I.; Larsen, J.; Lozano, P.; Tiedje, J.M. Bacillus cabrialesii sp. nov., an Endophytic Plant Growth Promoting Bacterium Isolated from Wheat (Triticum turgidum subsp. durum) in the Yaqui Valley, Mexico. Int. J. Syst. Evol. Microbiol. 2019, 69, 3939–3945. [Google Scholar] [CrossRef]
- Robles-Montoya, R.I.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Draft Genome Sequence of Bacillus megaterium TRQ8, a Plant Growth-Promoting Bacterium Isolated from Wheat (Triticum turgidum subsp. durum) Rhizosphere in the Yaqui Valley, Mexico. 3 Biotech 2019, 9, 1–5. [Google Scholar] [CrossRef]
- Valenzuela-Ruiz, V.; Robles-Montoya, R.I.; Parra-Cota, F.I.; Santoyo, G.; Orozco-Mosqueda, M.; Rodríguez-Ramírez, R.; de los Santos-Villalobos, S. Draft Genome Sequence of Bacillus paralicheniformis TRQ65, a Biological Control Agent and Plant Growth-Promoting Bacterium Isolated from Wheat (Triticum turgidum subsp. durum) Rhizosphere in the Yaqui Valley, Mexico. 3 Biotech 2019, 9, 1–7. [Google Scholar] [CrossRef]
- Villa-Rodriguez, E.; Moreno-Ulloa, A.; Castro-Longoria, E.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Integrated Omics Approaches for Deciphering Antifungal Metabolites Produced by a Novel Bacillus Species, B. Cabrialesii TE3T, against the Spot Blotch Disease of Wheat (Triticum turgidum L. subsp. durum). Microbiol. Res. 2021, 251, 126826. [Google Scholar] [CrossRef]
- Ibarra-Villarreal, A.L. El Recurso Microbiano Nativo Como Alternativa Agro-Biotecnológica Para La Producción Sostenible del Trigo (Triticum turgidum L. subsp. durum) Variedad CIRNO C2008 en el Valle del Yaqui, Sonora. Ph.D. Thesis, Instituto Tecnológico de Sonora, Ciudad Obregon, Mexico, 2020. [Google Scholar]
- Arellano-Wattenbarger, G.L. Impacto de la Aplicación de un Consorcio Nativo de Bacillus sobre Características Agronómicas del Cultivo de Trigo (Triticum turgidum L. subsp. durum), Bajo Diferentes Dosis de Urea, en el Valle del Yaqui, México. Master’s Thesis, Instituto Tecnológico de Sonora, Ciudad Obregon, Mexico, 2019. [Google Scholar]
- Ayala-Zepeda, M. Impacto de la Inoculación de un Consorcio Nativo de Bacillus sobre la Eficiencia en el Uso del 15N-Fertilizante por el Trigo (Triticum turgidum L. subsp. durum) en el Valle del Yaqui, México. Master’s Thesis, Instituto Tecnológico de Sonora, Ciudad Obregón, Mexico, 2020. [Google Scholar]
- Chaparro-Encinas, L.A. Transcriptomic Study of the Plant-Plant Growth-Promoting Bacteria Interaction under Increased Temperature to Identify Genes of Agricultural Importance. Ph.D. Thesis, Instituto Tecnológico de Sonora, Ciudad Obregón, Mexico, 2020. [Google Scholar]
- Baxevanis, A.D.; Bateman, A. The Importance of Biological Databases in Biological Discovery. Curr. Protoc. Bioinforma. 2015, 50, 1.1.1–1.1.8. [Google Scholar] [CrossRef] [PubMed]
- Zhulin, I.B. Databases for Microbiologists. J. Bacteriol. 2015, 197, 2458–2467. [Google Scholar] [CrossRef] [Green Version]
- Bertsch, F.; Henríquez, C. 2015: El Año Internacional De Los Suelos. Agron. Costarric. 2015, 39, 149–155. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).