Comparative Plastome Analyses of Ephedra przewalskii and E. monosperma (Ephedraceae)
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction and Sequencing
2.2. Genome Assembly and Annotation
2.3. Comparative Plastomics in Ephedra
2.4. Codon Usage
2.5. Phylogenetic Profiling
3. Results
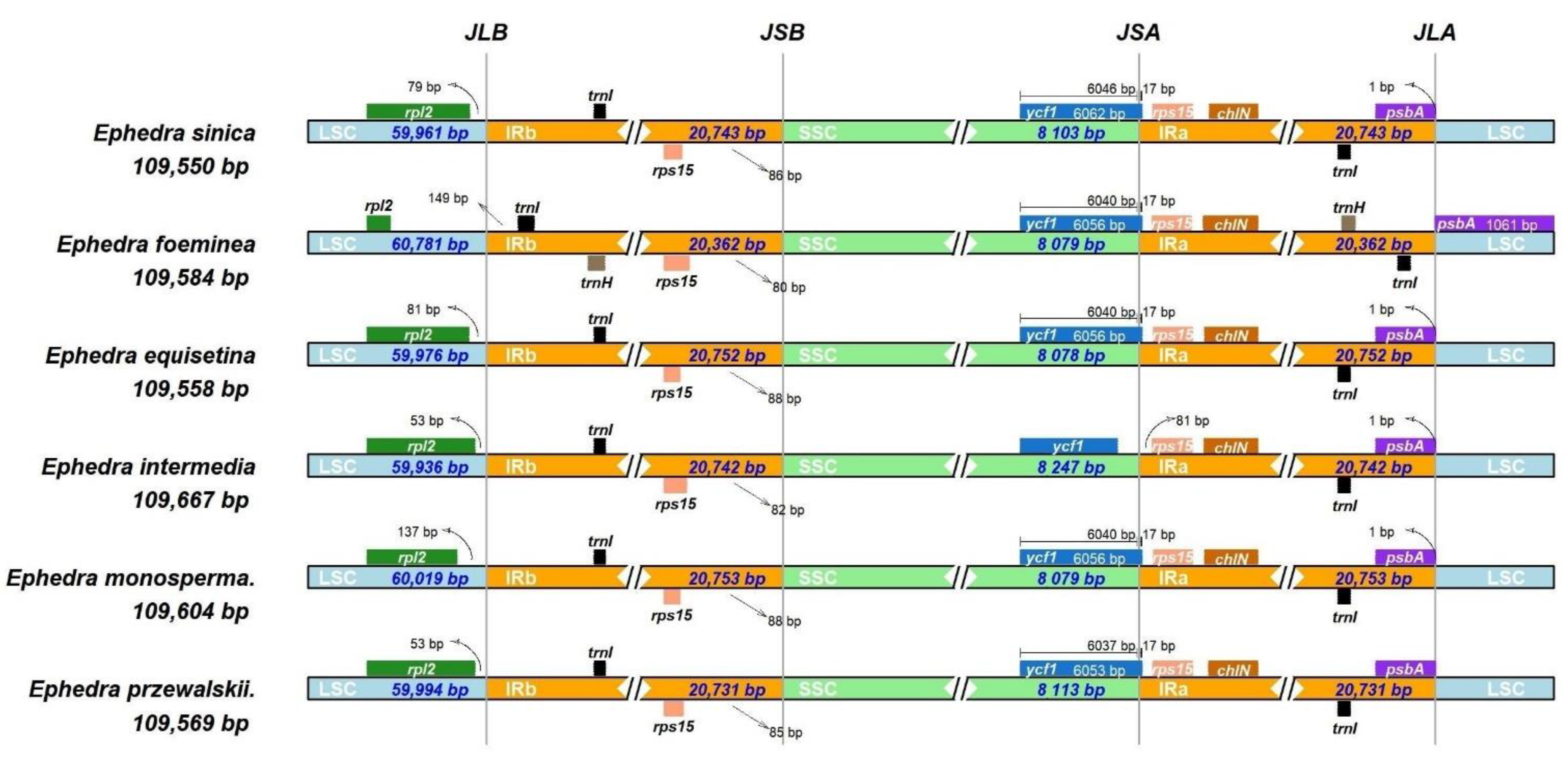
3.1. Ephedra Chloroplast Genome Features
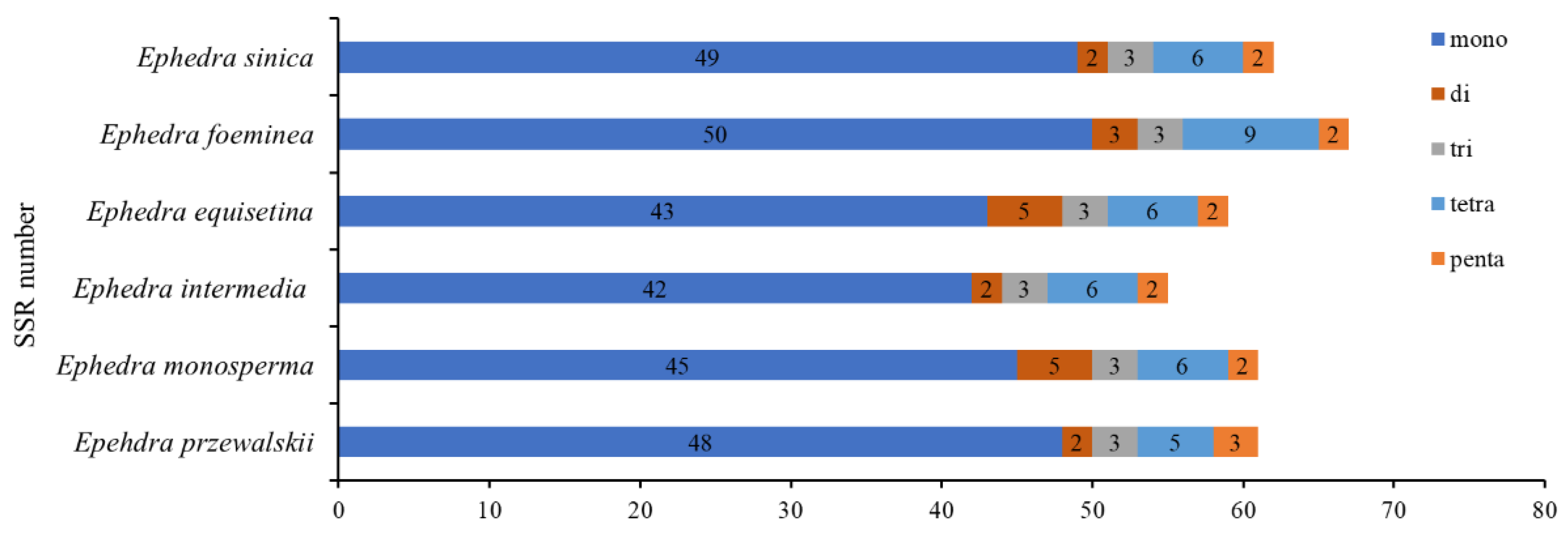
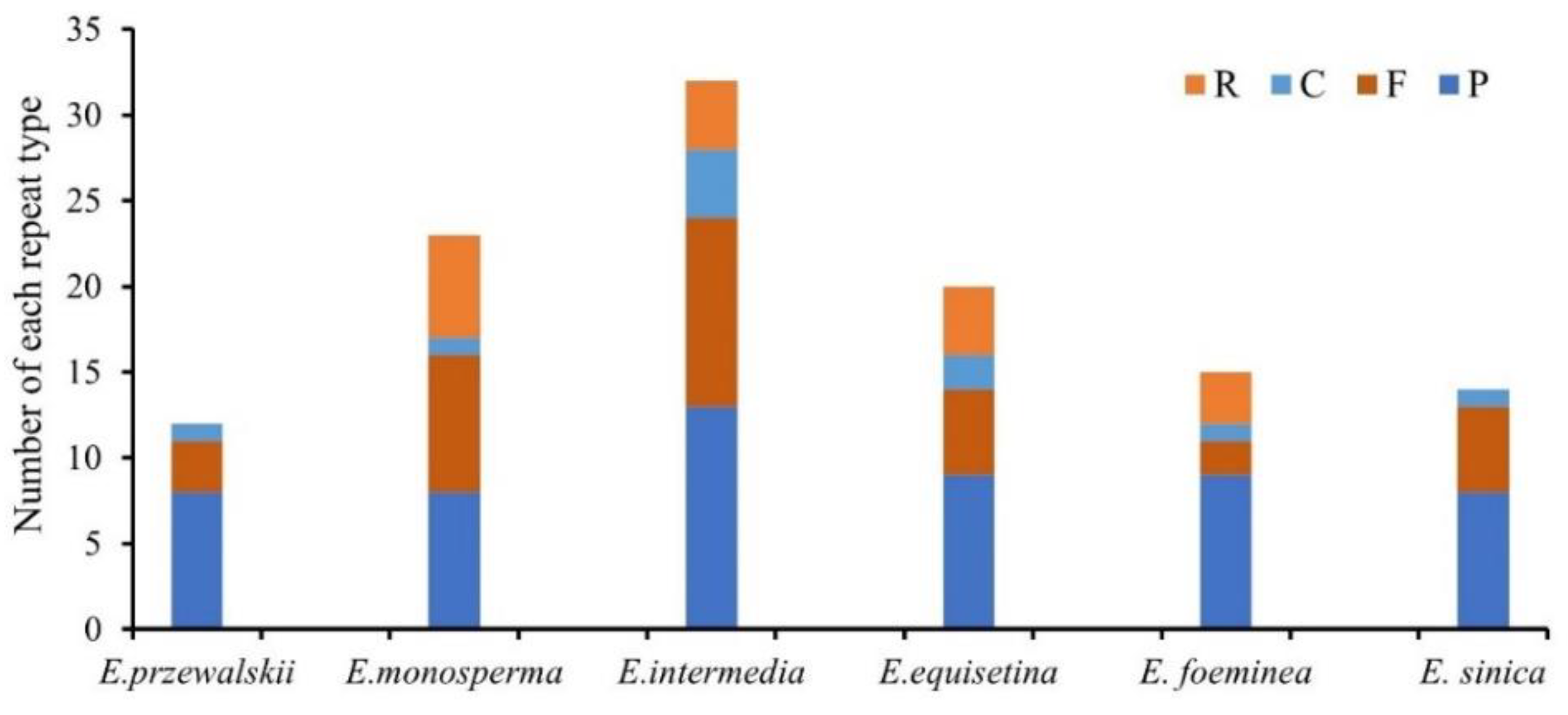
3.2. Repeat Sequences and SSR Analysis
3.3. Codon Usage
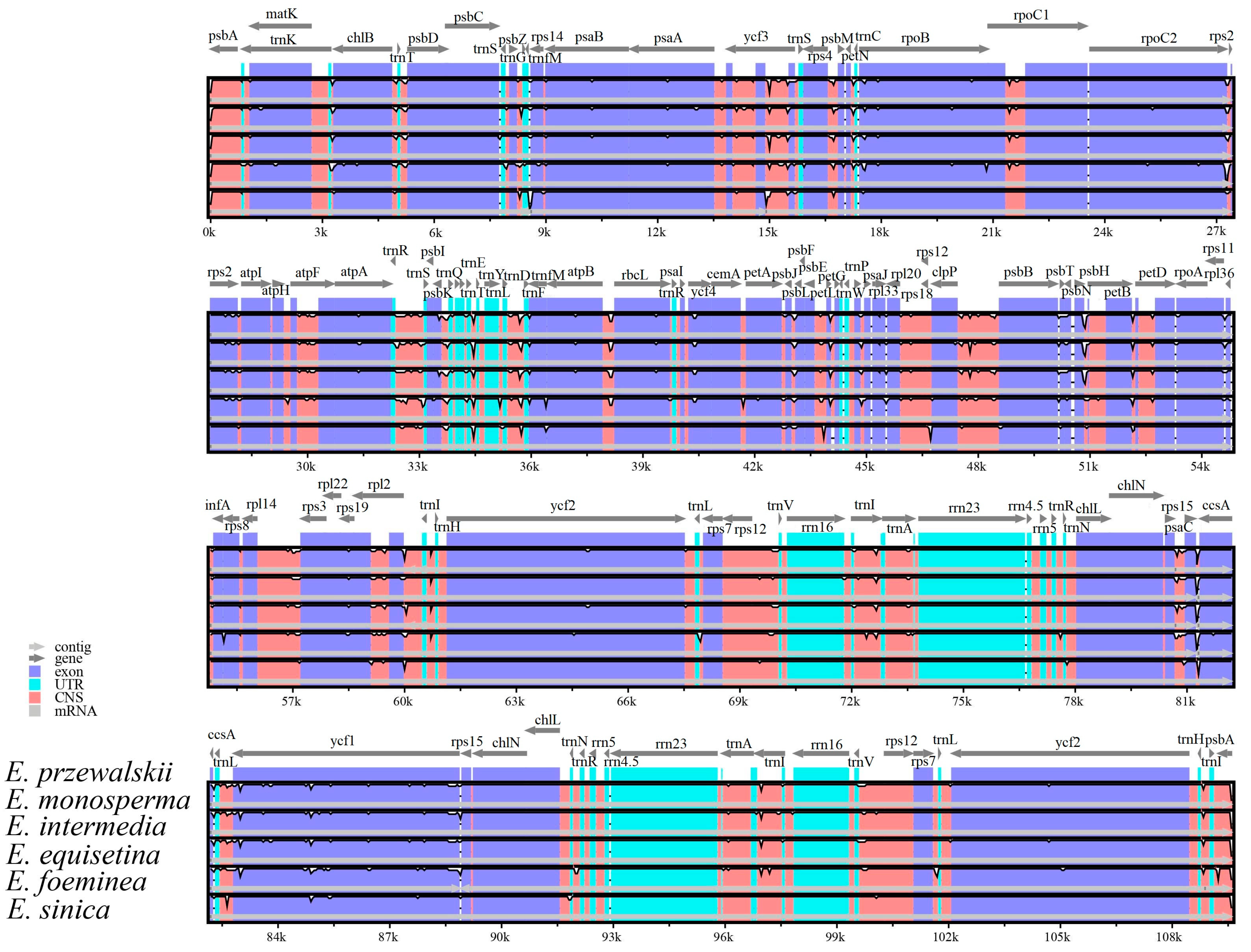
3.4. Divergence in Six Ephedra Chloroplast Genome
3.5. Evolutionary Rates in Protein-Coding Genes of Ephedra species
3.6. Predicted RNA Editing Sites for E. przewalskii and E. monosperma
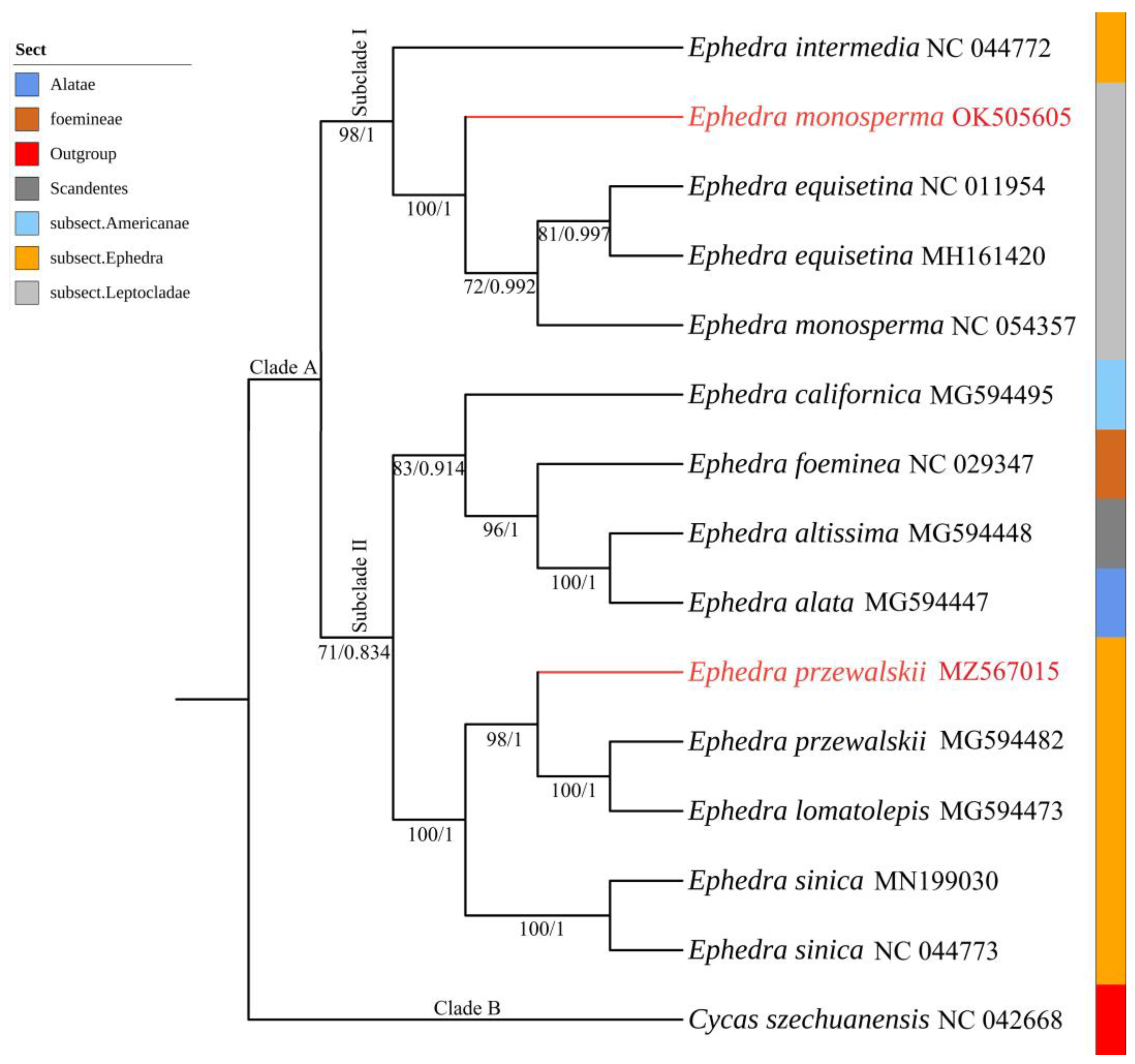
3.7. Phylogenetic Inference
4. Discussion
4.1. Genome Feature in Ephedra
4.2. Comparison of Genomes for Ephedra
4.3. Codon Usage Bias Analysis
4.4. Evolution Analysis
4.5. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, W.J.; Fu, L.K. Gymnospermae. Flora of China; Science Press: Beijing, China, 1978; Volume 7, pp. 471–489. [Google Scholar]
- Qin, A.L.; Wang, M.M.; Cun, Y.Z.; Yang, F.S.; Wang, S.S.; Ran, J.H.; Wang, X.Q. Phylogeographic evidence for a link of species divergence of Ephedra in the Qinghai-Tibetan Plateau and adjacent regions to the Miocene Asian aridification. PLoS ONE 2013, 8, e56243. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.A. Versuch einer Monographie der Gattung Ephedra. Mem. Akad. Imper. Sci. St. Petersburg. Ser.6 (Sci. Nat.) 1846, 5, 225–297. (In German) [Google Scholar]
- Stapf, O. Die Arten der Gattung Ephedra. Denkschr. Math.-Nat. Kl. Akad Wiss. Wien, lvi. 1889, 56, 35. [Google Scholar]
- Shen, G.M. Distribution and evolution of the genus Ephedra in China. Arid. Zone Res. 1993, 10, 39–48. [Google Scholar]
- Yang, Y. Systematics and Evolution of Ephedra L. (Ephedraceae) from China. Chin. Acad. Sci. 2002, 1–231. Available online: https://cir.nii.ac.jp/crid/1572824500456924160 (accessed on 3 January 2021).
- Vishal, S.; Harihara, V.; Mehendale, M. Ephedra. In Encyclopedia of Toxicology, 2nd ed.; Philip, W., Ed.; Elsevier: Amsterdam, The Netherlands, 2005; pp. 223–228. [Google Scholar]
- Barker, W.D.; Antia, U. A study of the use of Ephedra in the manufacture of methamphetamine. Forensic Sci. Int. 2007, 166, 102–109. [Google Scholar] [CrossRef]
- Haller, C.A.; Benowitz, N.L.; Jacob, P. Hemodynamic effects of ephedra-free weight-loss supplements in humans. Am. J. Med. 2005, 118, 998–1003. [Google Scholar] [CrossRef]
- Mei, J.; Zhou, Y.; Yang, X.; Zhang, F.; Liu, X.; Yu, B. Active components in Ephedra sinica Stapf disrupt the interaction between ACE2 and SARS-CoV-2 RBD: Potent COVID-19 therapeutic agents. J. Ethnopharmacol. 2021, 278, 114303. [Google Scholar] [CrossRef]
- Jiang, H.Y.; Li, W.; Li, S.; Shi, L.N.; Gao, Y.H.; Wu, J.L. Relationship analysis of five species in the genus Ephedra L. by RAPD. J. Gansu Agric. Univ. 2006, 41, 49–52. [Google Scholar]
- Deng, N.; Shi, S.Q.; Chang, E.M.; Liu, J.F.; Lan, Q.; Jiang, Z.P. Transcriptomic Analysis of Germinated Seeds of Ephedra przewalskii. J. Northeast. For. Univ. 2015, 43, 28–32. [Google Scholar]
- Xin, G.-Z.; Hu, B.; Shi, Z.-Q.; Zheng, J.-Y.; Wang, L.; Chang, W.-Q.; Li, P.; Yao, Z.-P.; Liu, L.-F. A direct ionization mass spectrometry-based approach for differentiation of medicinal Ephedra species. J. Pharm. Biomed. Anal. 2016, 117, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Gao, H.; Song, M.; Lin, Y.; Fan, J.; Liu, X. Identification of plant materials containing ephedrine alkaloids based on DNA barcoding and TaqMan real-time PCR assay. Acta Physiol. Plant. 2021, 4311, 143. [Google Scholar] [CrossRef]
- Rydin, C.; Pedersen, K.R.; Friis, E.M. On the evolutionary history of Ephedra: Cretaceous fossils and extant molecules. Proc. Natl. Acad. Sci. USA 2004, 101, 16571–16576. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Kakiuchi, N.; Takahashi, A.; Komatsu, K.; Cai, S.; Mikage, M. Phylogenetic analysis of the DNA sequence of the non-coding region of nuclear ribosomal DNA and chloroplast of Ephedra plants in China. Planta Med. 2004, 70, 1080–1084. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Giannasi, D.E.; Price, R.A. Phylogenetic relationships in Ephedra (Ephedraceae) inferred from chloroplast and nuclear DNA sequences. Mol. Phylogenet Evol. 2005, 35, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cui, Y.; Nie, L.; Hu, H.; Xu, Z.; Sun, W.; Gao, T.; Song, J.; Yao, H. Identification and Phylogenetic Analysis of the Complete Chloroplast Genomes of Three Ephedra Herbs Containing Ephedrine. Biomed. Res. Int. 2019, 2019, 5921725. [Google Scholar] [CrossRef] [PubMed]
- Rydin, C.; Blokzijl, R.; Thureborn, O.; Wikström, N. Node ages, relationships, and phylogenomic incongruence in an ancient gymnosperm lineage—Phylogeny of Ephedra revisited. Taxon 2021, 70, 701–719. [Google Scholar] [CrossRef]
- Moore, M.J.; Soltis, P.S.; Bell, C.D.; Burleigh, J.G.; Soltis, D.E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 2010, 107, 4623–4628. [Google Scholar] [CrossRef] [PubMed]
- Samigullin, T.H.; Logacheva, M.D.; Terenteva, E.I.; Degtjareva, G.V.; Vallejo-Roman, C.M. The plastid genome of Seseli montanum: Complete sequence and comparison with plastomes of other members of the Apiaceae family. Biochemistry 2016, 81, 981–985. [Google Scholar] [CrossRef]
- Androsiuk, P.; Jastrzębski, J.P.; Paukszto, Ł.; Makowczenko, K.; Okorski, A.; Pszczółkowska, A.; Chwedorzewska, K.J.; Górecki, R.; Giełwanowska, I. Evolutionary dynamics of the chloroplast genome sequences of six Colobanthus species. Sci. Rep. 2020, 10, 11522. [Google Scholar] [CrossRef] [PubMed]
- Ravi, V.; Khurana, J.P.; Tyagi, A.K.; Khurana, P. An update on chloroplast genomes. Syst. Evol. 2008, 271, 101–122. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, Y.; Landis, J.B.; Lv, Z.; Shen, J.; Zhang, H.; Lin, N.; Li, L.; Sun, J.; Deng, T.; et al. Plastome phylogenomic study of Gentianeae (Gentianaceae): Widespread gene tree discordance and its association with evolutionary rate heterogeneity of plastid genes. BMC Plant Biol. 2020, 20, 340. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.S.; Lai, Y.T.; Lin, C.P.; Wang, Y.N.; Chaw, S.M. Evolution of reduced and compact chloroplast genomes (cpDNAs) in gnetophytes: Selection toward a lower-cost strategy. Mol. Phylogenet Evol. 2009, 52, 115–124. [Google Scholar] [CrossRef] [PubMed]
- McCoy, S.R.; Kuehl, J.V.; Boore, J.L.; Raubeson, L.A. The complete plastid genome sequence of Welwitschia mirabilis: An unusually compact plastome with accelerated divergence rates. BMC Evol. Biol. 2008, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, D.; Albokhari, E.; Yaradua, S.; Abba, A. Complete chloroplast genome sequences of Dipterygium glaucum and Cleome chrysantha and other Cleomaceae Species, comparative analysis and phylogenetic relationships. Saudi J. Biol. Sci. 2021, 28, 2476–2490. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Huang, R.; Li, F.; Tian, E.; Li, C.; Chao, Z. Phylogenetic position of Bupleurum sikangense inferred from the complete chloroplast genome sequence. Gene 2021, 798, 145801. [Google Scholar] [CrossRef] [PubMed]
- Thomson, A.M.; Vargas, O.M.; Dick, C.W. Complete plastome sequence from Bertholletia excelsa and 23 related species yield informative markers for Lecythidaceae. Appl. Plant Sci. 2018, 6, e01151. [Google Scholar] [CrossRef]
- Sun, C.; Chen, F.; Teng, N.; Xu, Y.; Dai, Z. Comparative analysis of the complete chloroplast genome of seven Nymphaea species. Aquat. Bot. 2021, 170, 103353. [Google Scholar] [CrossRef]
- Khan, G.; Zhang, F.; Gao, Q.; Fu, P.C.; Xing, R.; Wang, J.; Liu, H.; Chen, S. Molecular phylogeography and intraspecific divergence of Spiraea alpina (Rosaceae) distributed in the Qinghai-Tibetan Plateau and adjacent regions inferred from nrDNA. Biochem. Syst. Ecol. 2014, 57, 278–286. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bullet. 1987, 19, 11–15. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Simon, A. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Nicolas, D.; Patrick, M.; Guillaume, S. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 4, e18. [Google Scholar]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Jaakko, H.; Peter, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Sebastian, B.; Thomas, T.; Thomas, M.; Uwe, S.; Martin, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.P.; Zhang, Y.B.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Armbruster, U.; Pesaresi, P.; Pribil, M.; Hertle, A.; Leister, D. Update on chloroplast research: New tools, new topics, and new trends. Mol. Plant. 2011, 4, 1–16. [Google Scholar] [CrossRef]
- Khan, A.; Khan, I.A.; Asif, H.; Azim, M.K. Current trends in chloroplast genome research. Afr. J. Biotechnol. 2010, 9, 3494–3500. [Google Scholar]
- Huang, Q.; Liu, Z.X.; Wang, C.; Jing, M.Y.; Liu, J.Q.; Zhou, W.; Kai, G.Y. The Complete Chloroplast Genome Sequences of Anisodus Acutangulus and a Comparison with Other Solanaceae Species. Clin. Complement. Med. Pharmacol. 2021, 1, 100002. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, J.; Xia, M.; Chi, X.; Khan, G.; Chen, S.; Zhang, F. Plastome Sequencing Reveals Phylogenetic Relationships among Comastoma and Related Taxa (Gentianaceae) from the Qinghai-Tibetan Plateau. Ecol. Evol. 2021, 11, 16034–16046. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, J.; Yang, Y.; Fan, C.; Chen, J. Phylogenomic Analysis and Dynamic Evolution of Chloroplast Genomes in Salicaceae. Front. Plant Sci. 2017, 8, 1050. [Google Scholar] [CrossRef] [PubMed]
- González-Juárez, D.E.; Escobedo-Moratilla, A.; Flores, J.; Hidalgo-Figueroa, S.; Martínez-Tagüeña, N.; Morales-Jiménez, J.; Muñiz-Ramírez, A.; Pastor-Palacios, G.; Pérez-Miranda, S.; Ramírez-Hernández, A.; et al. A Review of the Ephedra genus: Distribution, Ecology, Ethnobotany, Phytochemistry and Pharmacological Properties. Molecules 2020, 25, 3283. [Google Scholar] [CrossRef] [PubMed]
- Shahzadi, I.; Abdullah, M.F.; Ali, Z.; Ahmed, I.; Mirza, B. Chloroplast genome sequences of Artemisia maritima and Artemisia absinthium: Comparative analyses, mutational hotspots in genus Artemisia and phylogeny in family Asteraceae. Genomics 2020, 112, 1454–1463. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Tang, D.; Wei, K.; Qin, F.; Li, L.; Lin, Y.; Zhu, Y.; Khan, A.; Kashif, M.H.; Miao, J. The complete chloroplast genome sequence of the medicinal plant Sophora Tonkinensis. Sci. Rep. 2020, 10, 12473. [Google Scholar] [CrossRef]
- Xiong, A.-S.; Peng, R.-H.; Zhuang, J.; Gao, F.; Zhu, B.; Fu, X.-Y.; Xue, Y.; Jin, X.-F.; Tian, Y.-S.; Zhao, W.; et al. Gene duplication, transfer, and evolution in the chloroplast genome. Biotechnol. Adv. 2009, 27, 340–347. [Google Scholar] [CrossRef]
- Shikanai, T. Chloroplast NDH: A different enzyme with a structure similar to that of respiratory NADH dehydrogenase. Biochim. Biophys. Acta. 2016, 1857, 1015–1022. [Google Scholar] [CrossRef]
- Wakasugi, T.; Tsudzuki, J.; Ito, S.; Nakashima, K.; Tsudzuki, T.; Sugiura, M. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA 1994, 91, 9794–9798. [Google Scholar] [CrossRef]
- Ranade, S.S.; García-Gil, M.R.; Rosselló, J.A. Non-functional plastid ndh gene fragments are present in the nuclear genome of Norway spruce (Picea abies L. Karsch): Insights from in silico analysis of nuclear and organellar genomes. Mol. Genet. Genomics. 2016, 291, 935–941. [Google Scholar] [CrossRef]
- Braukmann, T.W.A.; Kuzmina, M.; Stefanović, S. Loss of all plastid ndh genes in Gnetales and conifers: Extent and evolutionary significance for the seed plant phylogeny. Curr. Genet. 2009, 55, 323–337. [Google Scholar] [CrossRef]
- Ruhlman, T.A.; Chang, W.J.; Chen, J.J.; Huang, Y.T.; Chan, M.T.; Zhang, J.; Liao, D.C.; Blazier, J.C.; Jin, X.; Shih, M.C.; et al. NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 2015, 15, 100. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Chen, J.J.; Chiu, C.; Hsiao, H.C.; Yang, C.; Jin, X.; Leebens-Mack, J.; Depamphilis, C.W.; Huang, Y.; Chang, W.; et al. Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 2017, 90, 994–1006. [Google Scholar] [CrossRef] [PubMed]
- Hilu, K.W.; Liang, H.P. The matK gene: Sequence variation and application in plant systematics. Am. J. Bot. 1997, 84, 830–839. [Google Scholar] [CrossRef]
- Plunkett, G.M.; Soltis, D.E.; Soltis, P.S. Clarification of the relationship between Apiaceae and Araliaceae based on matK and rbcL sequence data. Am. J. Bot. 1997, 84, 565–580. [Google Scholar] [CrossRef] [PubMed]
- Cbol Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Steven, G.N.; Subramanyam, R. Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae. Mol. Ecol. Resour. 2009, 9 (Suppl. S1), 172–180. [Google Scholar] [CrossRef]
- Wang, T.; Liu, J.; Guo, Y.; Yuan, N. Phylogenetic relationship among local legumes in Jiangsu Province based on analyses of matK gene and ITS sequence. J. Nanjing Agric. Univ. 2017, 40, 795–803. [Google Scholar]
- Nie, X.; Lv, S.; Zhang, Y.; Du, X.; Wang, L.; Biradar, S.S.; Tan, X.; Wan, F.; Weining, S. Complete Chloroplast Genome Sequence of a Major Invasive Species, Crofton Weed (Ageratina adenophora). PLoS ONE 2012, 7, e36869. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.H. Mathematical Model for Studying Genetic Variation in Terms of Restriction Endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef]
- Tuler, A.C.; Carrijo, T.T.; Nóia, L.R.; Ferreira, A.; Peixoto, A.L.; Ferreira, M.F.D.S. SSR markers: A tool for species identification in Psidium (Myrtaceae). Mol. Biol. Rep. 2015, 42, 1501–1513. [Google Scholar] [CrossRef]
- Mohamed, A. El-Esawi.SSR analysis of genetic diversity and structure of the germplasm of faba bean (Vicia faba L.). Comptes Rendus Biol. 2017, 340, 474–480. [Google Scholar]
- Bi, Y.; Zhang, M.F.; Xue, J.; Dong, R.; Du, Y.P.; Zhang, X.H. Chloroplast genomic resources for phylogeny and DNA barcoding: A case study on Fritillaria. Sci. Rep. 2018, 8, 1184. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Zhang, Q.; Wang, C.; Li, F.; Tian, F.; Lu, Y.; Hu, Y.; Yang, H.; Cui, G. Analysis of codon usage patterns of the chloroplast genome in Delphinium grandiflorum L. reveals a preference for AT-ending codons as a result of major selection constraints. Peer J. 2021, 9, e10787. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.N.; Cui, P.; Zhu, J.; Zhang, Z.H.; Zhang, Z. Translational selection in human: More pronounced in housekeeping genes. Biol Direct 2014, 9, 17. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.H. The codon adaptation index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef]
- Supriyo, C.; Prosenjit, P.; Mazumder, T.H. Codon Usage Bias Prefers AT Bases in Coding Sequences Among the Essential Genes of Haemophilus influenzae. Not. Sci. Biol. 2014, 6, 417–421. [Google Scholar] [CrossRef]
- Wright, F. The ’effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Jiang, S.Z.; Lian, H.; Xiong, Y.F.; Zhang, S.; Chen, S.P. Analysis of Codon Bias in Chloroplast Genome of Castanopsis carlesii. Mol. Plant Breed. 2021, 26, 1–12. [Google Scholar]
- Benne, R. RNA-editing in trypanosome mitochondria. Biochim. Biophys. Acta. 1989, 1007, 131–139. [Google Scholar] [CrossRef]
- Bock, R.; Kössel, H.; Maliga, P. Introduction of a heterologous editing site into the tobacco plastid genome: The lack of RNA editing leads to a mutant phenotype. EMBO J. 1994, 13, 4623–4628. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Q.; He, Y.; Chen, L.; Hao, C.; Jiang, C.; Li, Y.; Dai, Y.; Kang, Z.; Xu, J.R. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 2016, 26, 499–509. [Google Scholar] [CrossRef]
- Gualberto, J.M.; LaMattina, L.; Bonnard, G.; Weil, J.H.; Grienenberger, J.-M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 1989, 341, 660–662. [Google Scholar] [CrossRef]
- Wakasugi, T.; Hirose, T.; Horihata, M.; Tsudzuki, T.; Kössel, H.; Sugiura, M. Creation of a novel protein-coding region at the RNA level in black pine chloroplasts: The pattern of RNA editing in the gymnosperm chloroplast is different from that in angiosperms. Proc. Natl. Acad. Sci. USA 1996, 93, 8766–8770. [Google Scholar] [CrossRef]
- Wang, W.; Yu, H.; Wang, J.; Lei, W.; Gao, J.; Qiu, X.; Wang, J. The Complete Chloroplast Genome Sequences of the Medicinal Plant Forsythia suspensa (Oleaceae). Int. J. Mol. Sci. 2017, 18, 2288. [Google Scholar] [CrossRef] [PubMed]
- Kumbhar, F.; Nie, X.; Xing, G.; Zhao, X.; Lin, Y.; Wang, S.; Weining, S. Identification and characterisation of RNA editing sites in chloroplast transcripts of einkorn wheat (Triticum monococcum). Ann. Appl. Biol. 2018, 172, 197–207. [Google Scholar] [CrossRef]
- Mower, J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009, 37, W253–W259. [Google Scholar] [CrossRef] [PubMed]
- Giegé, P.; Brennicke, A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 1999, 96, 15324–15329. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Horner, D.S.; Chiara, M.; Schiavon, R.; Valle, G.; Pesole, G. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Res. 2010, 38, 4755–4767. [Google Scholar] [CrossRef] [PubMed]
- Grimes, B.T.; Sisay, A.K.; Carroll, H.D.; Cahoon, A.B. Deep sequencing of the tobacco mitochondrial transcriptome reveals expressed ORFs and numerous editing sites out-side coding regions. BMC Geno. 2014, 15, 31. [Google Scholar]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Yin, K.; Zhang, Y.; Li, Y.; Du, F. Different natural selection pressures on the atpF gene in evergreen sclerophyllous and deciduous oak species: Evidence from comparative analysis of the complete chloroplast genome of Quercus aquifolioides with other oak species. Int. J. Mol. Sci. 2018, 19, 1042. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.H. Computational Molecular Evolution; Oxford University Press: Oxford, UK, 2006; p. 284. [Google Scholar]
- Azarin, K.; Usatov, A.; Makarenko, M.; Khachumov, V.; Gavrilova, V. Comparative analysis of chloroplast genomes of seven perennial Helianthus species. Gene 2021, 774, 145418. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Xie, X.; Li, F.; Tian, E.; Chao, Z. Chloroplast genomes of two Mediterranean Bupleurum species and the phylogenetic relationship inferred from combined analysis with East Asian species. Planta 2021, 253, 81. [Google Scholar] [CrossRef] [PubMed]
- Rydin, C.; Pedersen, K.R.; Crane, P.R.; Friis, E.M. Former diversity of Ephedra (Gnetales): Evidence from Early Cretaceous seeds from Portugal and North America. Ann. Bot. 2006, 98, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Rydin, C.; Korall, P. Evolutionary Relationships in Ephedra (Gnetales), with Implications for Seed Plant Phylogeny. Int. J. Plant Sci. 2009, 170, 1031–1043. [Google Scholar] [CrossRef]



| Species | E. przewalskii | E. monosperma | E. intermedia | E. equisetina | E. foeminea | E. sinica |
|---|---|---|---|---|---|---|
| Accession Number | MZ567015 | OK505605 | NC_044772.1 | MH161420 | NC_029347 | NC_044773 |
| Genome size (bp) | 109,569 | 109,604 | 109,667 | 109,558 | 109,584 | 109,550 |
| LSC length (bp) | 59,994 | 60,019 | 59,936 | 59,976 | 60,027 | 59,961 |
| SSC length (bp) | 8113 | 8079 | 8247 | 8078 | 8079 | 8103 |
| IR length (bp) | 20,731 | 20,753 | 20,742 | 20,752 | 20,739 | 20,743 |
| Overall GC content (%) | 36.6 | 36.6 | 36.6 | 36.6 | 36.7 | 36.7 |
| GC content in LSC (%) | 34.2 | 34.2 | 34.2 | 34.2 | 34.1 | 34.2 |
| GC content in SSC (%) | 27.6 | 27.9 | 27.3 | 27.5 | 27.7 | 27.9 |
| GC content in IR (%) | 42 | 42 | 42.1 | 42 | 42 | 42 |
| Total number of genes | 118 | 118 | 118 | 118 | 118 | 118 |
| Protein-coding genes | 73 | 73 | 73 | 73 | 73 | 73 |
| tRNA genes | 37 | 37 | 37 | 37 | 37 | 37 |
| rRNA genes | 8 | 8 | 8 | 8 | 8 | 8 |
| Duplicated genes | 19 | 19 | 18 | 18 | 18 | 18 |
| Category of Genes | Group of Gene | Gene IDs |
|---|---|---|
| Self-replication | Ribosomal RNA genes | rrn23d,i; rrn16d,i; rrn5d,i; rrn4.5d,i |
| Transfer RNA genes | trnY-GUAl; trnW-CCAl; trnV-GACd,i; trnT-UGUl; trnT-GGUl; trnS-UGA l; trnS-GGAl; trnS-GCUl; trnR-UCUl; trnR-CCGl; trnR-ACGd,i; trnQ-UUG l; trnP-UGGl; trnN-GUUd,i; trnL-UAAl; trnL-CAAd,i; trnL-AUGl; trnK-UUU *,l; trnI-GAU d,*,i; trnI-CAU d,i; trnH-GUG d,i; trnG-UCCv l; trnfM-CAU d; trnF-GAA l; trnE-UUC l; trnD-GUC l; trnC-GCA l; trnA-UGC d,*,i; trnL-UAG s | |
| Small subunit of ribosome | rps19l; rps18l; rps15d,i; rps14l; rps12d,**,l&i; rps11l; rps8 l; rps7d,i; rps4l; rps3l; rps2l | |
| Large subunit of ribosome | rpl36l; rpl33l; rpl22l; rpl20l; rpl14l; rpl2 *,l | |
| DNA-dependent RNA polymerase | rpoC2l; rpoC1 *,l; rpoB l; rpoA l | |
| Genes for Photosynthesis | Subunits of photosystem I | psaAl; psaBl; psaCs; psaIl; psaJl |
| Subunits of photosystem II | psbA *,i,l; psbB l; psbC l; psbD l; psbE l; psbF l; psbH l; psbI l; psbJ l; psbK l; psbL l; psbM l; psbN l; psbT l; psbZ l; psbN **,l | |
| Subunits of Cytochrome b/f complex | petAl; petBl; petD *,l; petG l; petL l; petN l | |
| Subunits of ATP synthase | atpAl; atpBl; atpEl; atpF *,l; atpH l; atpI l | |
| Large subunit of RUBISCO | rbcLl | |
| Maturase | matKl | |
| Other genes | Envelope membrane protein | cemAl |
| Photochlorophyllide reductase subunit B/L/N | chlBl; chlLd,i; chlNd,i | |
| C-type cytochrome synthesis gene | ccsAs | |
| Protease | clpPl | |
| Translational initiation factor | infAl | |
| Genes of unknown function | Conserved open reading frames | ycf1s; ycf2d,i |
| Assembly/stability of photosystem I | ycf3 **,l; ycf4l |
| Gene | E. przewalskii | E. monosperma | E. intermedia | E. equisetina | E. foeminea | E. sinica | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exon I/Intron I | Exon II/Intron II | Exon Ⅲ | Exon I/Intron I | Exon II/Intron II | Exon Ⅲ | Exon I/Intron I | Exon II/Intron II | Exon Ⅲ | Exon I/Intron I | Exon II/Intron II | Exon Ⅲ | Exon I/Intron I | Exon II/Intron II | Exon Ⅲ | Exon I/Intron I | Exon II/Intron II | Exon Ⅲ | |
| rps12 * | 113/- | 31/- | 231 | 113/- | 31/- | 231 | 113/- | 31/- | 231 | 113/- | 31/- | 231 | 113/- | 31/- | 231 | 113/- | 31/- | 231 |
| rpl2 | 439/499 | 360/- | - | 439/499 | 360/- | - | 439/493 | 360/- | - | 439/499 | 360/- | - | 439/497 | 360/- | - | 439/494 | 360/- | - |
| rpl16 | 395/604 | 8/- | - | 395/608 | 8- | - | 395/603 | 8/- | - | 395/603 | 8/- | - | 395/604 | 8/- | - | 395/602 | 8/- | - |
| rpoC1 | 457/575 | 1629/- | - | 457/581 | 1629/- | - | 457/580 | 1629/- | - | 457/581 | 1629/- | - | 457/581 | 1629/- | - | 457/574 | 1629/- | - |
| petB | 5/517 | 641/- | - | 5/517 | 641/- | - | 5/517 | 641/- | - | 5/517 | 641/- | - | 5/518 | 641/- | - | 5/517 | 641/- | - |
| petD | 7/521 | 474/- | - | 7/527 | 474/- | - | 7/525 | 477/- | - | 7/524 | 477/- | - | 7/524 | 477/- | - | 7/518 | 477/- | - |
| atpF | 143/588 | 410/- | - | 143/584 | 410/- | - | 143/585 | 410/- | - | 143/584 | 410/- | - | 144/590 | 409/- | - | 143/589 | 410/- | - |
| ycf3 | 154/661 | 225/636 | 125 | 154/661 | 225/615 | 125 | 152/658 | 227/633 | 125 | 152/659 | 227/615 | 125 | 152/654 | 227/636 | 125 | 152/655 | 227/641 | 125 |
| trnK-UUU | 34/2298 | 37/- | - | 34/2298 | 37/- | - | 34/2298 | 37/- | - | 34/2298 | 37/- | - | 34/2294 | 37/- | - | 34/2298 | 37/- | - |
| trnI-GAU | 35/749 | 35/- | - | 35/761 | 35/- | - | 34/757 | 36/- | - | 34/761 | 36/- | - | 34/755 | 36/- | - | 34/749 | 36/- | - |
| trnA-UGC | 35/761 | 39/- | - | 35/759 | 39/- | - | 35/758 | 39/- | - | 35/759 | 39/- | - | 35/762 | 39/- | - | 35/760 | 39/- | - |
| trnL-UAA | 34/291 | 49/- | - | 34/291 | 49/- | - | 34/291 | 49/- | - | 34/291 | 49/- | - | 34/290 | 49/- | - | 34/291 | 49/- | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.; Xia, M.; Yu, J.; Xu, H.; Han, Y.; Zhang, F. Comparative Plastome Analyses of Ephedra przewalskii and E. monosperma (Ephedraceae). Diversity 2022, 14, 792. https://doi.org/10.3390/d14100792
Han S, Xia M, Yu J, Xu H, Han Y, Zhang F. Comparative Plastome Analyses of Ephedra przewalskii and E. monosperma (Ephedraceae). Diversity. 2022; 14(10):792. https://doi.org/10.3390/d14100792
Chicago/Turabian StyleHan, Shuang, Mingze Xia, Jingya Yu, Hao Xu, Yun Han, and Faqi Zhang. 2022. "Comparative Plastome Analyses of Ephedra przewalskii and E. monosperma (Ephedraceae)" Diversity 14, no. 10: 792. https://doi.org/10.3390/d14100792
APA StyleHan, S., Xia, M., Yu, J., Xu, H., Han, Y., & Zhang, F. (2022). Comparative Plastome Analyses of Ephedra przewalskii and E. monosperma (Ephedraceae). Diversity, 14(10), 792. https://doi.org/10.3390/d14100792








