Glyptothorax (Teleostei: Sisoridae) from the Middle East: An Integrated Molecular and Morphological Insight into Its Taxonomic Diversity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Morphological Measurements
2.2. Material Examined in the Morphological Analyses
2.3. DNA Extraction and PCR
2.4. Additional Sequence Data
2.5. Phylogenetic Analyses
2.6. Molecular Species Delimitations
3. Results
3.1. Phyleogenetic Placements and Molecular Species Delimitations
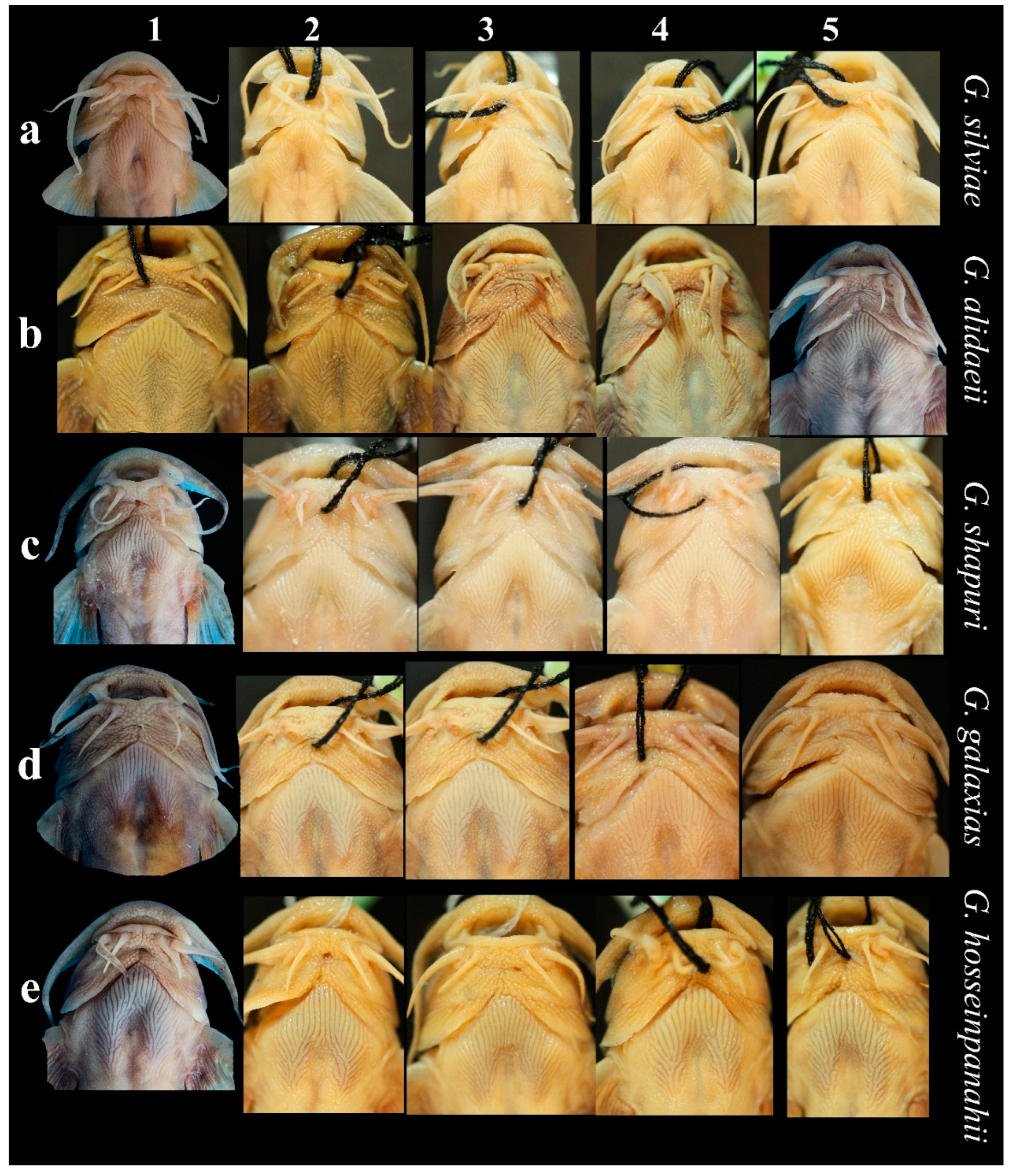
3.2. Morphological Results
3.2.1. The Name-Bearing Iranian Endemic Clades
3.2.2. Glyptothorax cous (Linnaeus, 1766)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fricke, R.; Eschmeyer, W.N.; Van der Laan, R. Eschmeyer’s Catalog of Fishes: Genera, Species, References. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 15 May 2022).
- Ferraris, C.J. Checklist of catfishes, recent and fossil (Osteichthyes: Siluriformes), and catalogue of siluriform primary types. Zootaxa 2007, 1418, 1–628. [Google Scholar] [CrossRef]
- Ng, H.H.; Kottelat, M. The identity of Clarias batrachus (Linnaeus, 1758), with the designation of a neotype (Teleostei: Clariidae). Zool. J. Linn. Soc. 2008, 153, 725–732. [Google Scholar] [CrossRef] [Green Version]
- Freyhof, J.; Kaya, C.; Abdullah, Y.S.; Geiger, M.F. The Glyptothorax catfishes of the Euphrates and Tigris with the description of a new species (Teleostei: Sisoridae). Zootaxa 2021, 4969, 453–491. [Google Scholar] [CrossRef]
- Mousavi-Sabet, H.; Eagderi, S.; Vatandoust, S.; Freyhof, J. Five new species of the sisorid catfish genus Glyptothorax from Iran (Teleostei: Sisoridae). Zootaxa 2021, 5067, 451–484. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.H.; Dodson, J.J. Morphological and genetic descriptions of a new species of catfish, Hemibagrus chrysops, from Sarawak, east Malayasia, with an assessment of phylogenetic relationships (Teleostei: Bagridae). Raffles Bull. Zool. 1999, 47, 45–47. [Google Scholar]
- Bruford, M.W.; Hanotte, O.; Brookfield, J.F.Y.; Burke, T.A. Single-locus and multilocus DNA fingerprinting. In Molecular Genetics Analysis of Populations: A Practical Approach; Hoezel, C., Ed.; Oxford University Press: Oxford, UK, 1992; pp. 225–269. [Google Scholar]
- Jiang, W.; Ng, H.H.; Yang, J.; Chen, X. Monophyly and phylogenetic relationships of the catfish genus Glyptothorax (Teleostei: Sisoridae) inferred from nuclear and mitochondrial gene sequences. Mol. Phylogenet. Evol. 2011, 61, 278–289. [Google Scholar] [CrossRef]
- Zarei, F.; Esmaeili, H.R.; Schliewen, U.K.; Abbasi, K.; Sayyadzadeh, G. Mitochondrial phylogeny, diversity, and ichthyogeography of gobies (Teleostei: Gobiidae) from the oldest and deepest Caspian sub-basin and tracing source and spread pattern of an introduced Rhinogobius species at the tricontinental crossroad. Hydrobiologia 2021, 848, 1267–1293. [Google Scholar] [CrossRef]
- Sullivan, J.P.; Lundberg, J.G.; Hardman, M. A phylogenetic analysis of the major groups of catfishes (Teleostei: Siluriformes) using RAG1 and RAG2 nuclear gene sequences. Mol. Phylogenet. Evol. 2006, 41, 636–662. [Google Scholar] [CrossRef]
- Chen, W.; Ma, X.; Shen, Y.; Mao, Y.; He, S. The fish diversity in the upper reaches of the Salween River, Nujiang River, revealed by DNA barcoding. Sci. Rep. 2015, 5, 17437. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Ho, S.Y.; Zhang, Y.; He, S. Uplift of the Tibetan plateau: Evidence from divergence times of glyptosternoid catfishes. Mol. Phylogenet. Evol. 2006, 39, 568–572. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, X.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef]
- Xia, X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Hart, M.W.; Sunday, J. Things fall apart: Biological species form unconnected parsimony networks. Biol. Lett. 2007, 3, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Kapli, P.; Lutteropp, S.; Zhang, J.; Kobert, K.; Pavlidis, P.; Stamatakis, A.; Flouri, T. Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov Chain Monte Carlo. Bioinformatics 2017, 33, 1630–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Ward, R.D. DNA barcode divergence among species and genera of birds and fishes. Mol. Ecol. Resour. 2009, 9, 1077–1085. [Google Scholar] [CrossRef]
- Yeates, D.K.; Seago, A.; Nelson, L.; Cameron, S.L.; Joseph, L.; Trueman, J.W. Integrative taxonomy, or iterative taxonomy? Syst. Entomol. 2011, 36, 209–217. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Phelps, S.R. Loss of genetic variation in hatchery stock of cutthroat trout. Trans. Am. Fish. Soc. 1988, 109, 537–543. [Google Scholar] [CrossRef]
- Swain, D.P.; Foote, C.J. Stocks and chameleons: The use of phenotypic variation in stock identification. Fish. Res. 1999, 43, 113–128. [Google Scholar] [CrossRef]
- Wimberger, P.H. Plasticity of fish body shape, the effects of diet, development, family and age in two species of Geophagus (Pisces: Cichlidae). Biol. J. Linn. Soc. 1992, 45, 197–218. [Google Scholar] [CrossRef]
- Turan, C.; Oral, M.; Ozturk, B.; Duzgunes, E. Morphometric and meristic variation between stocks of bluefish (Pomatomus saltatrix) in the Black, Marmara, Aegean and northeastern Mediterranean Seas. Fish. Res. 2005, 79, 139–147. [Google Scholar] [CrossRef]
- Hashemzadeh Segherloo, I.; Bernatchez, L.; Golzarianpour, K.; Abdoli, A.; Primmer, C.R.; Bakhtiary, M. Genetic differentiation between two sympatric morphs of the blind Iran cave barb Iranocypris typhlops. J. Fish Biol. 2012, 81, 1747–1753. [Google Scholar] [CrossRef] [PubMed]
- Hashemzadeh Segherloo, I.; Normandeau, E.; Benestan, L.; Rougeux, C.; Coté, G.; Moore, J.S.; Ghaedrahmati, N.; Abdoli, A.; Bernatchez, L. Genetic and morphological support for possible sympatric origin of fish from subterranean habitats. Sci. Rep. 2018, 8, 2909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashemzadeh Segherloo, I.; Najafi Chaloshtory, S.; Naser, M.D.; Yasser, A.G.; Tabatabaei, S.N.; Piette-Lauziere, G.; Mashtizadeh, A.; Elmi, A.; Sedighi, O.; Changizi, A.; et al. Sympatric morphotypes of the restricted-range Tashan Cave Garra: Distinct species or a case of phenotypic plasticity? Environ. Biol. Fishes 2022, 105, 1251–1260. [Google Scholar] [CrossRef]
- Esmaeili, H.R.; Teimori, A.; Sayyadzadeh, G.; Masoudi, M.; Reichenbacher, B. Phylogenetic relationships of the tooth-carp Aphanius (Teleostei: Cyprinodontidae) in the river systems of southern and south-western Iran based on mtDNA sequences. Zool. Middle East 2014, 60, 29–38. [Google Scholar] [CrossRef]
- Ryder, O.A. Species conservation and systematics: The dilemma of subspecies. Trends Ecol. Evol. 1986, 1, 9–10. [Google Scholar] [CrossRef]
- Moritz, C. Defining “evolutionarily significant units” for conservation. Trends Ecol. Evol. 1994, 9, 373–375. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Zarei, F.; Esmaeili, H.R.; Sadeghi, R.; Schliewen, U.K.; Kovačić, M.; Abbasi, K.; Gholamhosseini, A. An integrative insight into the diversity, distribution and biogeography of the freshwater endemic clade of the Ponticola syrman group (Teleostei: Gobiidae). Ecol. Evol. 2022, 12, e9300. [Google Scholar] [CrossRef]








| Species | Source | COI | Cyt b | RAG2 | Latitude | Longitude | Locality Code ‡ | Locality |
|---|---|---|---|---|---|---|---|---|
| G. shapuri | A | MZ959031 | 29.762 | 51.551 | Iran: Fars prov., Shapur River at Eslamabad | |||
| G. shapuri | A | MZ959032 | 29.762 | 51.551 | Iran: Fars prov., Shapur River at Eslamabad | |||
| G. shapuri | C | GY292 [I] | 29.394 | 52.161 | 1 | Iran: Helleh, Kohmareh Sorkhi stream | ||
| G. shapuri | C | G396 [I, II] | P396 [II] | G396R [II] | 29.394 | 52.161 | 1 | Iran: Helleh, Kohmareh Sorkhi stream |
| G. shapuri | C | G970F | 29.467 | 51.395 | 2 | Iran: Konar Takhteh, Helleh, Dalaki River | ||
| G. shapuri | C | G971F | 29.467 | 51.395 | 2 | Iran: Konar Takhteh, Helleh, Dalaki River | ||
| G. shapuri | C | G333 [III] | GY333 [III] | 29.394 | 52.161 | 1 | Iran: Helleh, Kohmareh Sorkhi stream | |
| G. shapuri | C | G401 [IV] | G401 [IV] | G401R [IV] | 29.394 | 52.161 | 1 | Iran: Helleh, Kohmareh Sorkhi stream |
| G. silviae | C | G395 | 31.376 | 49.730 | 3 | Iran: Khuzestan, Ab-e Aala River, Baghmalek | ||
| G. silviae | C | G410 [V] | 31.376 | 49.730 | 3 | Iran: Khuzestan, Ab-e Aala River, Baghmalek | ||
| G. silviae | C | GY330 [V] | 30.949 | 50.904 | 4 | Iran: Kohgiluyeh and Boyer-Ahmad prov., Maroun River at Garab-e-Lodab | ||
| G. silviae | C | G589 [VI] | GY295 [VI] | G589R [VI] | 31.376 | 49.730 | 3 | Iran: Khuzestan, Ab-e Aala River, Baghmalek |
| G. silviae | C | G411 [VII] | G411 [VII] | G411R [VII] | 31.376 | 49.730 | 3 | Iran: Khuzestan, Ab-e Aala River, Baghmalek |
| G. silviae | A | MZ959028 | 30.946 | 50.904 | Iran: Kohgiluyeh-va-Boyer Ahmad prov., Maroun River at Garab-e-Lodab | |||
| G. silviae | A | MZ959029 | 30.946 | 50.904 | Iran: Kohgiluyeh-va-Boyer Ahmad prov., Maroun River at Garab-e-Lodab | |||
| G. silviae | A | MZ959030 | 30.959 | 50.615 | Iran: Kohgiluyeh-va-Boyer Ahmad prov., Maroun River at Qale-Gol | |||
| G. hosseinpanahii | C | G597 [VIII] | GY294 [VIII] | G597R [VIII] | 30.328 | 51.251 | 5 | Iran: Fars prov., Zohreh, Fahlian River |
| G. hosseinpanahii | C | G598 [IX] | GY332 [IX] | G598R [IX] | 30.328 | 51.251 | 5 | Iran: Fars prov., Zohreh, Fahlian River |
| G. hosseinpanahii | A | MZ959033 | 30.461 | 51.353 | Iran: Kohgiluyeh-va-Boyer Ahmad prov., Zohreh River at Tange-Shiv | |||
| G. hosseinpanahii | A | MZ959034 | 30.530 | 50.416 | Iran: Khuzestan prov., Zohreh River at Kheirabad | |||
| G. hosseinpanahii | A | MZ959035 | 30.461 | 51.353 | Iran: Kohgiluyeh-va-Boyer Ahmad prov., Zohreh River at Tange-Shiv | |||
| G. alidaeii | C | ZM-CBSU Ex58F4 | 30.852 | 51.342 | 6 | Iran: Kohgiluyeh and Boyer-Ahmad prov., Khersaan River at Betari | ||
| G. alidaeii | C | G606 | 31.040 | 51.218 | 7 | Iran: Kohgiluyeh and Boyer-Ahmad prov., Khersaan River | ||
| G. alidaeii | C | ZM-CBSU Ex58F3 | 30.852 | 51.342 | 6 | Iran: Kohgiluyeh and Boyer-Ahmad prov., Khersaan River at Betari | ||
| G. alidaeii | C | G414 [X, XI, XII] | G414 [X] | G414R [X] | 30.676 | 51.532 | 8 | Iran: Kohgiluyeh and Boyer-Ahmad prov., Beshar River at Aliabade-e Satrol |
| G. alidaeii | C | GY326 [XI] | 31.670 | 50.769 | 9 | Iran: Chaharmahal and Bakhtiari prov., Armand River | ||
| G. alidaeii | C | GY329 [XII] | 31.184 | 51.450 | 10 | Iran: Isfahan prov., Bibi Seyedan | ||
| G. alidaeii | B | MW770731 | 30.852 | 51.342 | Iran: Beshar at Doruhan | |||
| G. alidaeii | B | MW770718 | 30.852 | 51.342 | Iran: Beshar at Doruhan | |||
| G. alidaeii | A | MZ959036 | 33.688 | 47.064 | Iran: Lorestan prov., Seimare River at Zirkhaki | |||
| G. alidaeii | A | MZ959037 | 33.688 | 47.064 | Iran: Lorestan prov., Seimare River at Zirkhaki | |||
| G. alidaeii | C | G728F [XIII] | G728b [XIII] | G835R [XIII] | 33.458 | 47.930 | 11 | Iran: Lorestan prov., Kashkan River, Ghalebi-e-Sofla village Betwwen Khoramabad and Mamulan Lorestan |
| G. alidaeii | C | G726F | 33.458 | 47.930 | 11 | Iran: Lorestan prov., Kashkan River, Ghalebi-e-Sofla village Betwwen Khoramabad and Mamulan Lorestan | ||
| G. alidaeii | C | G729F | 33.458 | 47.930 | 11 | Iran: Lorestan prov., Kashkan River, Ghalebi-e-Sofla village Betwwen Khoramabad and Mamulan Lorestan | ||
| G. alidaeii | C | G727F | 33.458 | 47.930 | 11 | Iran: Lorestan prov., Kashkan River, Ghalebi-e-Sofla village Betwwen Khoramabad and Mamulan Lorestan | ||
| G. cf. galaxias | C | G323 [XIV] | GY323 [XIV] | G323R [XIV] | 34.371 | 47.912 | 12 | Iran: Hamadan prov., Gamasiab River at Do Ab |
| G. cf. galaxias | C | G558 [XV, XVI] | G558b [XV] | G558R [XV] | 33.694 | 46.883 | 13 | Iran: Ilam prov., Chardaval River |
| G. cf. galaxias | C | G557b [XVI] | G557R [XVI] | 33.694 | 46.883 | 13 | Iran: Ilam prov., Chardaval River | |
| G. cf. galaxias | C | G296 [XVII] | GY296 [XVII] | G296R [XVII] | 34.371 | 47.912 | 12 | Iran: Hamadan prov., Gamasiab River at Do Ab |
| G. galaxias | A | MZ959025 [XVIII] | 32.029 | 50.627 | Iran: Chaharmahal-va-Bakhtiari prov., stream Beheshtabad at Beheshtabad | |||
| G. galaxias | C | GY327 [XVIII] | 31.078 | 51.518 | 14 | Iran: Isfahan prov., Khak Daneh | ||
| G. galaxias | A | MZ959026 | 32.165 | 50.423 | Iran: Chaharmahal-va-Bakhtiari prov., stream Afsarabad at Afsarabad | |||
| G. galaxias | A | MZ959027 [XIX] | 31.280 | 51.266 | Iran: Chaharmahal-va-Bakhtiari prov., stream Mal-e-Khalife at Mal-e-Khalife | |||
| G. galaxias | C | GY325 [XIX] | G331R [XIX] | 31.186 | 51.256 | 15 | Iran: Kohgiluyeh and Boyer-Ahmad prov., Kata | |
| G. galaxias | B | MW770721 | 32.034 | 50.633 | Iran: Behesht Abad River north of Ardal, 75 km south-west of Shahr-e-kord | |||
| G. armeniacus | B | MW770712 | 37.173 | 41.270 | Türkiye: Tunceli prov., stream Pülümür at Pülümür | |||
| G. armeniacus | B | MW724503 | 37.292 | 37.573 | Türkiye: Gaziantep prov., stream Merzimen 3 km south of Yavuzeli | |||
| G. armeniacus | C | G834 | Türkiye: Pueluemuer | |||||
| G. armeniacus | ZFMK | ZFMK FSJF-2634 | 37.837 | 37.685 | Türkiye: upper River Göksu 5 km northeast of Gölbaşı | |||
| G. armeniacus | ZFMK | ZFMK FSJF-2634-2 | 37.837 | 37.685 | Türkiye: upper River Göksu 5 km northeast of Gölbaşı | |||
| G. armeniacus | C | G369 | Türkiye: Pueluemuer | |||||
| G. armeniacus | B | MW770726 | 37.837 | 37.685 | Türkiye: upper River Göksu, 5 km northeast of Gölbaşı | |||
| G. armeniacus | B | MW770714 | 37.837 | 37.685 | Türkiye: upper River Göksu, 5 km northeast of Gölbaşı | |||
| G. armeniacus | B | MW770723 | 37.837 | 37.685 | Türkiye: Tunceli prov., stream Pülümür at Pülümür | |||
| G. daemon 1 | C | G931F | Türkiye: Greater Zab | |||||
| G. daemon 1 | B | MW724515 | 37.672 | 43.863 | Türkiye: Hakkari prov., stream Eziki 6 km northeast Konak | |||
| G. daemon 1 | B | MW724520 | 38.355 | 41.781 | Türkiye: Bitlis prov., stream Çıratan 3 km southwest of Üçadım | |||
| G. daemon 1 | B | MW724519 | 38.355 | 41.781 | Türkiye: Bitlis prov., stream Çıratan 3 km southwest of Üçadım | |||
| G. daemon 1 | B | MW724518 | 38.355 | 41.781 | Türkiye: Bitlis prov., stream Çıratan 3 km southwest of Üçadım | |||
| G. daemon 1 | B | MW724514 | 37.672 | 43.863 | Türkiye: Hakkari prov., stream Eziki 6 km northeast Konak | |||
| G. daemon 1 | B | MW770713 | 38.385 | 41.782 | Türkiye: stream Çıratan about 5 km east of Gümüşkanat | |||
| G. daemon 1 | B | MW770729 | 38.385 | 41.782 | Türkiye: stream Çıratan about 5 km east of Gümüşkanat | |||
| G. daemon 1 | C | G559 | Türkiye Yukeskora Hakkari prov Yukeskora | |||||
| G. daemon 1 | B | MW724516 | 37.672 | 43.863 | Türkiye: Hakkari prov., stream Eziki 6 km northeast Konak | |||
| G. daemon 1 | B | MW770722 | 37.666 | 44.139 | Türkiye: Hakkari prov., stream Dilektaşı 16 km northeast Yüksekova | |||
| G. daemon 2 † | B | MW724512 | 37.666 | 44.139 | Türkiye: Hakkari prov., stream Dilektaşı 16 km northeast Yüksekova | |||
| G. daemon 2 † | B | MW724513 | 37.666 | 44.139 | Türkiye: Hakkari prov., stream Dilektaşı 16 km northeast Yüksekova | |||
| G. daemon 2 † | B | MW724517 | 37.666 | 44.139 | Türkiye: Hakkari prov., stream Dilektaşı 16 km northeast Yüksekova | |||
| G. daemon 2 † | B | MW770730 | 36.611 | 44.838 | Iraq: stream Choman at Qubay Galala | |||
| G. daemon 2 † | B | MW770732 | 36.611 | 44.838 | Iraq: stream Choman at Qubay Galala | |||
| G. daemon 2 † | B | MW770733 | 36.611 | 44.838 | Iraq: stream Choman at Qubay Galala | |||
| G. daemon 2 † | B | MW770724 | 36.611 | 44.838 | Iraq: stream Choman at Qubay Galala | |||
| G. daemon 2 † | B | MW770725 | 36.611 | 44.838 | Iraq: stream Choman at Qubay Galala | |||
| G. cous | B | MW724510 | 35.915 | 45.368 | Iraq: Qal’ah Chwalan River at Bardbard | |||
| G. cous | B | MW724509 | 35.915 | 45.368 | Iraq: Qal’ah Chwalan River at Bardbard | |||
| G. cous | B | MW724504 | 35.915 | 45.368 | Iraq: Qal’ah Chwalan River at Bardbard | |||
| G. cous | B | MW724511 | 35.915 | 45.368 | Iraq: Qal’ah Chwalan River at Bardbard | |||
| G. cous | B | MW724508 | 35.915 | 45.368 | Iraq: Qal’ah Chwalan River at Bardbard | |||
| G. cous | B | MW724507 | 35.915 | 45.368 | Iraq: Qal’ah Chwalan River at Bardbard | |||
| G. cous | B | MW770716 | 35.751 | 45.479 | Iraq: Aw-e Shiler River at Khewata | |||
| G. cous | B | MW770715 | 35.751 | 45.479 | Iraq: Aw-e Shiler River at Khewata | |||
| G. cous | B | MW770719 | 35.751 | 45.479 | Iraq: Aw-e Shiler River at Khewata | |||
| G. cous | B | MW770727 | 36.748 | 44.300 | Iraq: Great Zab about 2 km upriver of confluence with Rawanduz River | |||
| G. cous | B | MW724505 | 35.915 | 45.368 | Iraq: Qal’ah Chwalan River at Bardbard | |||
| G. cous | B | MW724506 | 35.915 | 45.368 | Iraq: Qal’ah Chwalan River at Bardbard | |||
| G. cous | C | G324 [XX] | GY324 [XX] | G324R [XX] | 33.680 | 47.055 | 16 | Iran: Ilam prov., Seimare River |
| G. cous | C | G368 [XXI] | G368 [XXI] | G368R [XXI] | 33.183 | 47.417 | 17 | Iran: Ilam prov., Seimare River |
| G. kurdistanicus | C | G510 [XXII] | G510b [XXII] | G510R [XXII] | 35.118 | 46.257 | 18 | Iran: Kermanshah prov., Sirwan River |
| G. kurdistanicus | B | MW724521 | 35.915 | 45.368 | Iraq: Qal’ah Chwalan River at Bardbard | |||
| G. kurdistanicus | B | MW770728 | 35.751 | 45.480 | Iraq: Aw-e Shiler at Khewata | |||
| G. kurdistanicus | B | MW770720 | 35.751 | 45.480 | Iraq: Aw-e Shiler at Khewata | |||
| G. kurdistanicus | B | MW770717 | 35.751 | 45.480 | Iraq: Aw-e Shiler at Khewata | |||
| G. kurdistanicus | B | MW724523 | 38.355 | 41.781 | Türkiye: Bitlis prov., stream Çıratan 3 km southwest of Üçadım | |||
| G. kurdistanicus | B | MW724522 | 38.355 | 41.781 | Türkiye: Bitlis prov., stream Çıratan 3 km southwest of Üçadım | |||
| G. kurdistanicus | B | MW724524 | 38.355 | 41.781 | Türkiye: Bitlis prov., stream Çıratan 3 km southwest of Üçadım | |||
| G. pallens | A | MZ959042 | 34.481 | 45.756 | Iran: Kermanshah prov., stream Alvand near Qasr-e-Shirin | |||
| G. pallens | A | MZ959038 | 34.645 | 46.286 | Iran: Kermanshah prov., stream Zemkan 3 km north of Zamkan-e Olya | |||
| G. pallens | A | MZ959041 | 34.228 | 46.004 | Iran: Kermanshah prov., stream Golain at Sare-Baghe-Golain | |||
| G. pallens | A | MZ959039 [XXIII] | 35.160 | 46.339 | Iran: Kermanshah prov., Sirvan River at Hajij | |||
| G. pallens | C | G511b [XXIII] | G511R [XXIII] | 35.118 | 46.257 | 19 | Iran: Kermanshah prov., Sirwan River | |
| G. pallens | A | MZ959040 | 34.645 | 46.286 | Iran: Kermanshah prov., stream Zemkan 3 km north of Zamkan-e Olya |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | G. galaxias † | 0.50 | 0.39 | 0.50 | 0.36 | 0.68 | 0.60 | 0.56 | 0.46 | 0.64 | 0.62 | |
| 2 | G. silviae | 2.06 | 0.37 | 0.54 | 0.43 | 0.72 | 0.63 | 0.59 | 0.57 | 0.66 | 0.68 | |
| 3 | G. shapuri | 1.40 | 1.22 | 0.50 | 0.36 | 0.63 | 0.63 | 0.53 | 0.53 | 0.61 | 0.62 | |
| 4 | G. hosseinpanahii | 1.97 | 1.97 | 1.76 | 0.47 | 0.71 | 0.66 | 0.66 | 0.61 | 0.73 | 0.69 | |
| 5 | G. alidaeii | 1.43 | 1.66 | 1.30 | 1.73 | 0.60 | 0.56 | 0.57 | 0.50 | 0.61 | 0.55 | |
| 6 | G. pallens | 3.45 | 3.81 | 3.27 | 3.71 | 2.98 | 0.68 | 0.68 | 0.63 | 0.83 | 0.65 | |
| 7 | G. cous | 2.38 | 2.77 | 2.89 | 3.00 | 2.37 | 3.55 | 0.61 | 0.43 | 0.74 | 0.63 | |
| 8 | G. daemon 1 | 2.32 | 2.85 | 2.32 | 3.08 | 2.76 | 3.96 | 2.61 | 0.50 | 0.71 | 0.70 | |
| 19 | G. daemon 2 ‡ | 1.44 | 2.28 | 2.08 | 2.51 | 1.88 | 3.05 | 1.10 | 1.80 | 0.70 | 0.64 | |
| 10 | G. armeniacus | 2.93 | 2.93 | 2.69 | 3.49 | 2.67 | 4.37 | 3.65 | 3.71 | 3.16 | 0.69 | |
| 11 | G. kurdistanicus | 2.71 | 3.25 | 2.72 | 3.16 | 2.44 | 3.56 | 2.84 | 3.32 | 2.75 | 3.48 |
| Species | G. alidaeii | G. galaxias | G. hosseinpanahii | |||
|---|---|---|---|---|---|---|
| Range | Mean ± SD | Range | Mean ± SD | Range | Mean ± SD | |
| SL | 59.7–148.8 | 100.2 | 27.6–76.3 | 62.1 | 46.8–71.5 | 62.5 |
| % SL | ||||||
| HL | 24.1–27.0 | 25.4 ± 0.8 | 25.6–30.3 | 27.7 ± 1.6 | 25.3–27.2 | 26.3 ± 0.6 |
| Pdl | 32.8–37.9 | 35.3 ± 1.7 | 34.1–40.9 | 37.1 ± 2.4 | 34.6–36.8 | 35.6 ± 0.6 |
| Pol | 68.4–82.2 | 77.4 ± 3.4 | 70.1–78.7 | 74.7 ± 3.5 | 73.8–79.6 | 76.3 ± 1.6 |
| Dl | 10.8–14.9 | 13.1 ± 0.9 | 10.1–13.9 | 12.3 ± 1.2 | 11.5–14.2 | 12.6 ± 0.9 |
| Dd | 18.0–25.6 | 21.2 ± 1.8 | 19.4–23.9 | 21.1 ± 1.6 | 19.9–23.0 | 21.3 ± 0.9 |
| Al | 10.9–14.2 | 12.5 ± 1.0 | 11.1–14.8 | 12.7 ± 1.6 | 11.2–13.8 | 12.2 ± 0.9 |
| Ad | 18.6–23.5 | 21.2 ± 1.6 | 20.2–24.2 | 22.2 ± 1.5 | 19.8–24.0 | 21.1 ± 1.2 |
| Pal | 66.0–74.5 | 69.2 ± 2.0 | 70.3–73.3 | 71.8 ± 1.3 | 66.9–71.3 | 69.5 ± 1.3 |
| Pl | 23.1–28.3 | 25.3 ± 1.4 | 22.8–24.9 | 23.9 ± 0.9 | 23.9–26.8 | 25.1 ± 0.9 |
| Vl | 15.6–18.9 | 17.2 ± 0.9 | 16.0–17.1 | 16.5 ± 0.5 | 16.4–18.1 | 16.9 ± 0.5 |
| Pvl | 47.4–54.6 | 51.6 ± 1.7 | 53.4–61.0 | 55.8 ± 2.7 | 50.1–53.4 | 51.5 ± 0.9 |
| Mxd | 17.1–25.5 | 19.8 ± 2.4 | 18.6–23.7 | 21.2 ± 2.2 | 18.7–22.3 | 20.1 ± 1.2 |
| Mid | 10.9–15.0 | 13.1 ± 1.1 | 13.0–16.4 | 14.7 ± 1.2 | 11.7–13.0 | 12.6 ± 0.4 |
| PV | 28.4–40.1 | 32.2 ± 2.7 | 32.3–38.5 | 34.5 ± 2.4 | 29.8–31.7 | 30.7 ± 0.5 |
| VA | 15.9–21.0 | 18.3 ± 1.4 | 13.6–19.3 | 16.3 ± 2.1 | 14.9–18.9 | 17.6 ± 1.2 |
| DAd | 6.2–17.2 | 13.4 ± 2.5 | 7.9–14.7 | 11.8 ± 2.6 | 9.4–14.1 | 11.8 ± 1.7 |
| Cl | 22.7–32.0 | 27.2 ± 2.7 | 21.2–30.8 | 25.0 ± 3.3 | 26.1–30.9 | 27.9 ± 1.2 |
| Cp | 15.4–23.5 | 20.5 ± 2.4 | 18.7–20.5 | 19.5 ± 0.7 | 19.8–22.2 | 20.8 ± 0.7 |
| CPI | 1.2–1.8 | 1.6 ± 1.4 | 1.2–1.6 | 1.3 ± 0.1 | 1.6–1.7 | 1.6 ± 0.0 |
| TAI | 1.1–1.6 | 1.3 ± 0.1 | 1.0–1.4 | 1.2 ± 0.1 | 1.2–1.5 | 1.3 ± 0.1 |
| Adl/DAd | 1.3–3.6 | 1.8 ± 0.5 | 1.6–2.7 | 1.9 ± 0.5 | 1.5–2.4 | 1.8 ± 0.3 |
| % HL | ||||||
| Hd | 50.4–67.9 | 61.1 ± 4.4 | 52.1–71.7 | 61.7 ± 7.8 | 54.0–72.1 | 62.5 ± 6.0 |
| Hw | 75.9–90.5 | 84.2 ± 3.7 | 87.6–93.2 | 90.6 ± 2.3 | 58.2–83.4 | 79.1 ± 7.4 |
| Pr | 46.0–56.3 | 52.8 ± 3.1 | 41.9–49.0 | 46.7 ± 2.7 | 50.2–59.4 | 53.0 ± 2.6 |
| Po | 35.0–44.7 | 40.3 ± 2.5 | 43.3–47.3 | 45.1 ± 1.6 | 36.1–50.5 | 40.1 ± 4.1 |
| I | 20.7–32.2 | 25.6 ± 2.3 | 26.6–31.1 | 28.5 ± 1.6 | 25.2–31.1 | 27.5 ± 1.9 |
| E | 8.0–12.1 | 10.0 ± 1.2 | 7.3–10.9 | 9.2 ± 1.2 | 6.3–11.2 | 8.3 ± 1.5 |
| Mw | 25.7–36.1 | 29.8 ± 2.7 | 30.5–35.2 | 33.1 ± 1.8 | 26.4–35.2 | 30.2 ± 2.9 |
| Nbl | 25.5–38.2 | 30.4 ± 3.4 | 37.5–49.4 | 44.3 ± 4.3 | 28.3–37.1 | 32.0 ± 2.4 |
| Mbl | 78.3–121.5 | 95.4 ± 10.0 | 79.2–94.8 | 86.2 ± 5.9 | 88.0–100.7 | 95.5 ± 3.9 |
| Omb | 42.3–67.1 | 54.4 ± 7.6 | 56.1–60.1 | 57.9 ± 1.3 | 48.8–57.6 | 52.0 ± 2.8 |
| Imb | 22.6–39.3 | 29.9 ± 4.6 | 31.6–36.6 | 34.3 ± 1.8 | 26.2–29.5 | 27.8 ± 1.2 |
| G. shapuri | G. silviae | G. cous | ||||
| Range | Mean ± SD | Range | Mean ± SD | Range | Mean ± SD | |
| SL | 50.6–81.4 | 65.5 | 33.5–64.9 | 47.4 | 68.4–97.9 | 83.1 |
| % SL | ||||||
| HL | 24.7–28.1 | 26.3 ± 1.0 | 26.6–28.9 | 27.8 ± 0.9 | 31.0–31.4 | 31.2 ± 0.3 |
| Pdl | 33.6–36.8 | 35.1 ± 1.0 | 35.2–37.9 | 36.4 ± 1.0 | 38.6–40.8 | 39.7 ± 1.6 |
| Pol | 75.0–82.4 | 77.3 ± 1.8 | 79.4–82.8 | 81.1 ± 1.0 | 71.2–71.2 | 71.2 ± 0.3 |
| Dl | 12.0–15.7 | 13.6 ± 1.0 | 11.5–13.2 | 12.3 ± 0.6 | 13.1–13.4 | 13.3 ± 0.2 |
| Dd | 18.2–22.1 | 20.0 ± 1.2 | 21.1–23.6 | 22.2 ± 0.7 | 21.3–24.2 | 22.7 ± 2.0 |
| Al | 11.4–13.9 | 12.4 ± 0.6 | 10.6–14.4 | 11.8 ± 1.2 | 13.0–13.3 | 13.2 ± 0.2 |
| Ad | 18.7–21.1 | 19.8 ± 0.7 | 21.3–22.9 | 22.0 ± 0.5 | 21.2–21.4 | 21.3 ± 0.1 |
| Pal | 66.4–70.8 | 68.1 ± 1.3 | 68.2–70.5 | 69.1 ± 0.8 | 69.9–72.1 | 71.0 ± 1.6 |
| Pl | 21.5–25.6 | 23.5 ± 1.1 | 24.2–26.9 | 25.8 ± 0.7 | 20.6–24.2 | 22.4 ± 2.5 |
| Vl | 14.0–17.5 | 15.8 ± 0.9 | 16.4–18.3 | 17.2 ± 0.7 | 15.0–17.1 | 16.0 ± 1.5 |
| Pvl | 48.6–54.0 | 51.1 ± 1.4 | 50.2–53.9 | 52.3 ± 1.3 | 55.5–56.4 | 56.0 ± 0.6 |
| Mxd | 18.2–21.5 | 19.9 ± 1.2 | 15.9–19.5 | 17.1 ± 1.2 | 18.7–19.3 | 19.0 ± 0.4 |
| Mid | 11.5–15.2 | 13.4 ± 0.9 | 10.2–13.1 | 11.4 ± 0.8 | 8.7–9.1 | 8.9 ± 0.3 |
| PV | 26.0–33.3 | 30.4 ± 1.7 | 27.2–31.1 | 30.1 ± 1.4 | 32.0–33.3 | 32.7 ± 0.9 |
| VA | 15.8–18.6 | 17.4 ± 0.9 | 14.8–17.5 | 16.0 ± 1.0 | 16.9–18.6 | 17.7 ± 1.2 |
| DAd | 10.4–17.0 | 14.7 ± 1.7 | 10.9–14.5 | 13.1 ± 1.2 | 10.7–10.7 | 10.7 ± 0.5 |
| Cl | 24.5–30.0 | 26.7 ± 1.5 | 27.2–29.9 | 28.5 ± 0.9 | 25.4–25.5 | 25.4 ± 0.1 |
| Cp | 20.4–23.3 | 22.0 ± 0.9 | 19.6–21.7 | 20.6 ± 0.6 | 16.9–17.7 | 17.3 ± 0.6 |
| CPI | 1.5–1.8 | 1.6 ± 0.1 | 1.6–2.0 | 1.8 ± 0.1 | 1.9–2.0 | 1.9 ± 0.1 |
| TAI | 1.0–1.2 | 1.1 ± 0.1 | 1.0–1.2 | 1.5 ± 0.1 | 1.0–1.1 | 1.1 ± 0.1 |
| Adl/DAd | 1.1–2.2 | 1.4 ± 0.3 | 1.5–2.1 | 1.7 ± 0.2 | 2.0–2.0 | 2.0 ± 0.1 |
| % HL | ||||||
| Hd | 53.8–71.9 | 61.7 ± 4.4 | 47.1–53.4 | 50.9 ± 2.2 | 48.2–55.9 | 52.0 ± 5.4 |
| Hw | 73.1–84.6 | 80.5 ± 2.5 | 73.5–81.4 | 77.2 ± 2.4 | 77.0–82.2 | 79.6 ± 3.7 |
| Pr | 46.4–53.5 | 50.8 ± 2.2 | 48.6–54.0 | 51.1 ± 1.7 | 47.1–50.1 | 48.6 ± 2.1 |
| Po | 40.0–44.1 | 41.7 ± 0.9 | 36.6–40.5 | 38.7 ± 1.4 | 43.1–45.0 | 44.1 ± 1.3 |
| I | 21.9–28.3 | 26.4 ± 1.7 | 24.1–27.7 | 25.6 ± 1.1 | 23.9–24.9 | 24.4 ± 0.7 |
| E | 6.5–11.4 | 9.7 ± 1.1 | 7.4–9.7 | 8.8 ± 0.7 | 6.1–8.6 | 7.3 ± 1.8 |
| Mw | 26.4–33.3 | 29.7 ± 2.3 | 25.2–33.9 | 28.4 ± 2.5 | 37.4–40.1 | 38.8 ± 1.9 |
| Nbl | 27.7–38.6 | 33.0 ± 2.8 | 34.0–44.4 | 37.0 ± 3.3 | 34.3–35.2 | 34.7 ± 0.6 |
| Mbl | 87.9–109.4 | 96.1 ± 6.2 | 103.3–117.4 | 108.6 ± 5.2 | 78.5–85.6 | 82.0 ± 5.0 |
| Omb | 50.3–58.7 | 54.9 ± 2.6 | 54.4–65.1 | 59.0 ± 2.9 | 50.0–51.9 | 51.0 ± 1.3 |
| Imb | 23.6–30.8 | 26.5 ± 2.2 | 26.7–37.5 | 30.2 ± 3.2 | 26.5–29.2 | 27.8 ± 1.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sayyadzadeh, G.; Zarei, F.; Esmaeili, H.R. Glyptothorax (Teleostei: Sisoridae) from the Middle East: An Integrated Molecular and Morphological Insight into Its Taxonomic Diversity. Diversity 2022, 14, 884. https://doi.org/10.3390/d14100884
Sayyadzadeh G, Zarei F, Esmaeili HR. Glyptothorax (Teleostei: Sisoridae) from the Middle East: An Integrated Molecular and Morphological Insight into Its Taxonomic Diversity. Diversity. 2022; 14(10):884. https://doi.org/10.3390/d14100884
Chicago/Turabian StyleSayyadzadeh, Golnaz, Fatah Zarei, and Hamid Reza Esmaeili. 2022. "Glyptothorax (Teleostei: Sisoridae) from the Middle East: An Integrated Molecular and Morphological Insight into Its Taxonomic Diversity" Diversity 14, no. 10: 884. https://doi.org/10.3390/d14100884
APA StyleSayyadzadeh, G., Zarei, F., & Esmaeili, H. R. (2022). Glyptothorax (Teleostei: Sisoridae) from the Middle East: An Integrated Molecular and Morphological Insight into Its Taxonomic Diversity. Diversity, 14(10), 884. https://doi.org/10.3390/d14100884






